Spectral Optimized Multiderivative Hybrid Block Method for Fitzhugh–Nagumo Equations
Abstract
The Fitzhugh–Nagumo equation, a key model for excitable systems in biology and neuroscience, requires efficient numerical methods due to its nonlinear nature. A spectral optimized multiderivative hybrid block method is proposed, constructed using a multistep collocation and interpolation technique with an approximated power series as the basis function. Incorporating two optimal intra-step points, the method demonstrates improved accuracy, with its consistency, convergence, and absolute stability rigorously analyzed. By combining the optimized multiderivative hybrid block method in time with a spectral collocation method in space, the approach demonstrates potency and flexibility in solving partial differential equations. Prior to using the spectral method, the partial differential equation is linearized using a linear partition technique. Numerical experiments confirm the accuracy and efficiency of the method compared to existing methods, demonstrating the potential of the method as a robust framework for solving partial differential equations requiring both high accuracy and stability.
1. Introduction
Several methods have been investigated for the direct solution of equation (1), such as the adomian decomposition method (ADM) [11], variational iteration method (VIM) [12, 13], shifted Legendre polynomial (SLP) [14] and differential transform method (DTM) [15], including the Haar wavelet method presented in [16]. According to [17], analytical solutions for PDEs related to Nagumo reaction-diffusion have been derived using the variational method. Additionally, the homotopy analysis technique has effectively been utilized to provide approximate solutions for the standard F-N equation [18]. Furthermore, the approximate conditional symmetry method as proposed in [19], has been utilized to find approximate solutions for the perturbation of equation (1). Exact solutions have also been obtained using the first integral method [20] and Jacobi elliptic function [21]. While some of these methods require many terms for accurate analytical solutions, others face challenges in determining a suitable initial guess.
Among the diverse array of approaches used to solve PDEs (see [22–24]), the nonstandard finite difference (NSFD) techniques have recently emerged as one of the most effective approaches [25]. The exact finite-difference method (see [26–29]) is a special NSFD scheme. These methods produce reliable results across a wide range of initial and boundary conditions (BCs). However, they can be computationally demanding and prone to numerical instability.
Spectral methods are a class of numerical techniques that approximate solutions to differential equations (DEs). The three traditional spectral methods are the Galerkin, Tau and collocation methods [30]. Spectral techniques have attracted interest due to their computational efficiency and the relative simplicity with which they are used to solve nonlinear DEs [31, 32]. These methods are based on expanding the solution to the equation in terms of a set of basis functions like polynomial kind or Fourier basis [33]. Spectral collocation methods (SCMs), such as the Lagrange pseudo-spectral approach [34], the uniform Haar wavelets and quasilinearization process [35], and the Chebyshev SCMs (see [36–38]), have been presented for solving time-dependent DEs, including nonlinear and stochastic Burgers’ equation. The reaction-diffusion Brusselator system and several nonlinear Burgers’ type equations were effectively solved numerically utilizing the differential quadrature method (DQM) [39, 40]. Motsa et al. [41] introduced the bivariate SCM with the use of Chebyshev and Lagrange cardinal polynomials. This approach involves applying SCM to the time variable to transform the PDE into an ODE, followed by the spatial direction to form systems of linear equations and simplifying the solution process. Initially applied to evolutionary parabolic PDEs, this method has been adapted in [42] to solve various classes of PDEs with two independent variables. The precision of SCMs relies on the choice of grid points and the type of basis functions used to construct the interpolating polynomial [43]. However, their effectiveness decreases when applied to PDEs over extended time intervals. By using fewer grid points, SCMs not only enhance accuracy but also significantly reduce computational costs [44]. The ongoing pursuit of greater precision and computational efficiency continues to drive the development of new techniques. Spectral methods have been combined with other numerical techniques to enhance performance when utilized to solve various types of DEs. One such approach, presented in [45], integrates the finite difference method for time discretization with the spectral method for spatial discretization to solve DEs. Despite the advancements, existing methods often struggle to balance high accuracy, stability, and computational efficiency for nonlinear PDEs like the F-N equation.
Hybrid block method (HBM) has been utilized to solve equation (1) by transforming it into an equivalent system of ordinary DEs through the method of lines [46]. It was noted that as the value of time increases, the error also increases, indicating that shorter time is required to achieve more accurate solution approximations. The Hermite block method introduced in [47] was developed using Hermite polynomials as basis functions to solve second-order nonlinear elliptic PDEs, aiming to enhance numerical accuracy and efficiency. Olaoluwa Omole et al. [48] employed an implicit block method to solve PDEs, achieving greater precision and faster convergence.
Another approach is the multiderivative (MDHBMs), which offer a significant advancement in computational techniques for solving DEs. These methods incorporate higher-order derivatives in the formulation to enhance accuracy and stability without significantly increasing computational complexity [49–51]. By utilizing second or higher-order derivative terms, multiderivative methods reduce truncation errors and improve the convergence rates [51–53]. By merging the strengths of linear multistep methods and block techniques, these methods can efficiently solve both linear and nonlinear problems. Kayode et al. [52] introduced a predictor-corrector technique utilizing higher derivative to solve IVPs. To achieve optimal accuracy in block hybrid methods (BHMs), various optimized BHMs for the numerical integration of IVPs have been presented in [54–57]. A one-step optimized HBM for solving systems of IVPs was presented in [58]. An efficient optimized HBM for integrating second-order IVPs was presented in [59]. Recently, Yakubu and Sibanda [60] used a power series polynomial as a basis function to develop an optimized second-derivative HBM to maximize accuracy when utilized to solve IVPs. While these methods have shown promise for initial value problems (IVPs), their application to nonlinear PDEs have not been extensively studied. This gap highlights the need for innovative numerical techniques to tackle nonlinear PDEs with greater efficiency and accuracy.
To address these challenges, this study introduces a spectral optimized MDHBM (SOMDHBM) for solving the F-N equations (1) and (2). By combining the high accuracy of spectral methods with the stability and computational efficiency of multiderivative hybrid block techniques, the SOMDHBM offers a robust framework for solving nonlinear and diffusive characteristics of the equation. The F-N equation is first linearized using a linear partition mapping technique before applying the SOMDHBM. The effectiveness of the SOMDHBM is validated through numerical experiments, demonstrating the accuracy and efficiency compared to existing techniques such as the polynomial DQM (PDQM) [61], compact finite difference block method (CFDBM) [49], ADM [11], VIM [12, 13], DTM [15], exact finite-difference and NSFD scheme [62], İnan et al. [63] and exponential finite difference method (ExpFDM) [64].
2. Derivation of the Method
The Mathematica 13.0 code presented in Algorithm 1, for equations (8)–(10), is used to determine the unknown coefficients ηn,j.
-
Algorithm 1: Mathematica code for obtaining the coefficients ηn,j.
- 1.
Define Points: pi = {0, c, 1 − c, 1}; Set: J = Length[p];
- 2.
Collocate: , for i = 0, …, J;
- 3.
Set:
-
;
-
, for i = 1, …, J;
-
, for i = J;
- 4.
Identify Unknowns: Unknowns = Table[ηn,i, {i, 0, J + 1}];
- 5.
Apply Initial Condition: Initial = (W/.t⟶tn) = = wn;
- 6.
Combine All Equations: AllEquations = Join[W’, W”, {Initial}];
- 7.
Solve for Unknowns:
-
Solutions = Simplify[Flatten[Solve[AllEquations, Unknowns]]];
3. Analysis of the Method
In this section, we present the truncation error, order, zero-stability and absolute stability of the OMDHBM.
3.1. Local Truncation Error (LTE)
3.2. Zero Stability
By equating ρ(λ) = 0, the roots of the polynomials are λ = 0 and λ = 1. Thus, following [66], the OMDHBM is zero stable. Since the order of the OMDHBM is greater than 1 (), the method is consistent. When the method is zero stable and consistent, this leads to the conclusion that the method converges.
3.3. Proof of Absolute Stability
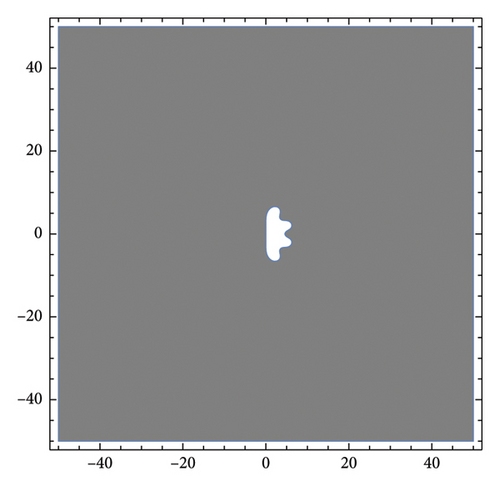
As shown in Figure 1, the stability region for the OMDHBM is the shaded area, which encompasses the entire left side of the plane (A-stable), while the unshaded area is the unstable region. It is evident that the method is not L-stable since . A-stable method is more suitable for a wider range of problems compared to methods with limited regions of absolute stability. It ensures stability regardless of the step size.
4. Implementation
The next section discusses the applications of the SOMDHBM on the F-N equations (1) and (2).
5. Applications in F-N Equations
-
Algorithm 2: Algorithmic implementation of the SOMDHBM.
-
Input: Parameter δ, step size h, number of spatial points Nx, number of iterations R
- 1.
Generate Chebyshev collocation points and compute D, D1, D2 over spatial domain [a, b];
- 2.
Define the time grid Nt with step size h over an interval [0, T];
- 3.
Define initial condition w(x, 0);
- 4.
Define boundary functions w(a, t), w(b, t), and exact solution w(x, t);
- 5.
Define first-derivative parameters: Φ, Ψ;
- 6.
Define second-derivative parameters: Θ, Ω, K;
- 7.
Forn = 1 to Nt:
-
a. Initialize guesses: Wr(:, n + i) ← W(:, n) for i = 0 to 3;
-
b. Forr = 1 to R:
-
i. Fori = 0 to 3:
-
- Evaluate Ψ, Φ, Θ, Ω, K at tn + h·points(i + 1);
-
ii. Fori = 1 to 3:
-
- Forj = 1 to 3:
-
∗ Assemble coefficient matrices Ei,j, Ai,j, Ci,j, Bi,j;
-
∗ Impose boundary conditions on rows 1 and Nx + 1 of Ei,j;
-
- Adjust diagonals in Ei,i for boundary rows;
-
iii. Flatten block matrices into matrix system: E1·W = Wn + A0·Fn + C1·Ψ + B1·K + B1·Θ·Ψ;
-
iv. Apply boundary values in RHS vector;
-
v. Solve the linear system for Wn+i,r;
-
vi. Compute error: ∥Wn+3,r − exact(x, tn + h·points(4))∥∞;
-
c. Update solution: W(:, n + 1) ← Wr(:, n + 3);
-
Output: Final numerical solution W
Example 1. Consider the F-N equation (1) in [62], subject to the BCs
The exact solution of (1) is
The BCs are imposed on the first and last rows of equation (53) in the form of equation (54). For different values of δ, the SOMDHBM for the current problem is evaluated and compared to the calculated absolute error for accuracy and efficiency. The absolute error is defined as
When δ = 0.01, Nx = 50, h = 0.01 and fifteen iterations, Table 1 shows a comparison of the absolute error (AEn) in the computed solutions for the F-N equation (1) obtained using ADM [11], VIM [12, 13], DTM [15], NSFD [62] and SOMDHBM. The results demonstrate that SOMDHBM consistently achieves the lowest AEn error across all test cases, with errors significantly lower than those obtained using other methods. For instance, at x = 0.1 and t = 1.0, the SOMDHBM achieves an AEn error in the order of 10−13, whereas NSFD produces errors around 10−12 and ADM, VIM and DTM yield errors in the range of 10−6 to 10−5. This confirms the high accuracy of SOMDHBM, as it outperforms all compared methods. The NFEval column in table indicates the number of function evaluations required by SOMDHBM. The values show that SOMDHBM maintains its high accuracy while using a relatively small number of function evaluations. Based on the observed numerical results, the SOMDHBM is highly competitive with existing methods and demonstrates efficiency in finding solutions for the F-N equation (1).
When δ = 0.001, Nx = 50, h = 0.01 and fifteen iterations, Table 2 presents the AEn error for different values of x and t in the F-N (1), comparing the performance of the SOMDHBM and NSFD [62]. The data reveal that SOMDHBM consistently produces more accurate results than NSFD, achieving significantly lower AEn error. At all tested spatial points (x = 0.2, 0.5, 0.8), the AEn error of SOMDHBM remain within the order of 10−15, while NSFD exhibits errors in the order of 10−14, which are approximately ten times larger. This trend is observed across different time steps, confirming that SOMDHBM maintains its precision over time. The NFEval for SOMDHBM increases as t progresses, from 1800 at t = 0.3 to 5400 at t = 0.9. This suggests that while computational effort increases over time, SOMDHBM retains its superior accuracy. These results show that the SOMDHBM not only improves accuracy compared to NSFD but also provides a stable and efficient approach for solving the F-N (1).
The numerical results demonstrate that the SOMDHBM accurately captures the propagation of traveling wavefronts, which represent electrical signals, across a spatial domain over time. The extremely low error magnitudes (on the order of 10−15) indicate that SOMDHBM preserves sharp wave profiles, essential for realistic simulations of nerve activity. The ability of the method to handle small δ values (which correspond to near-threshold excitability) shows its robustness in modeling sensitive nerve dynamics.
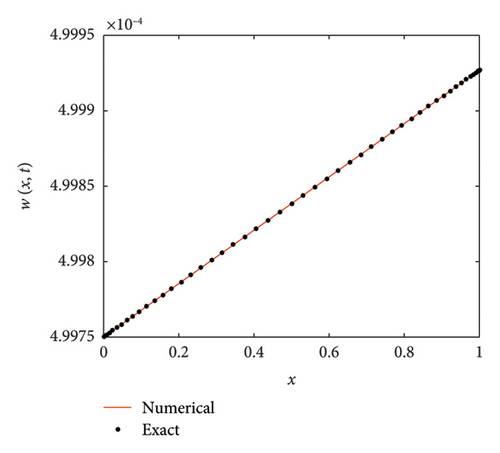
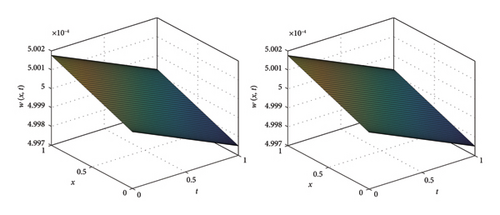
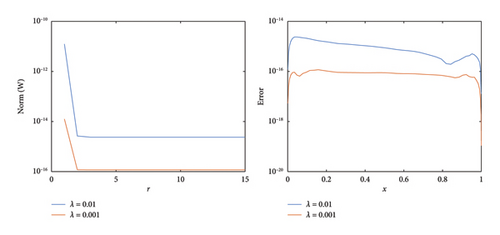
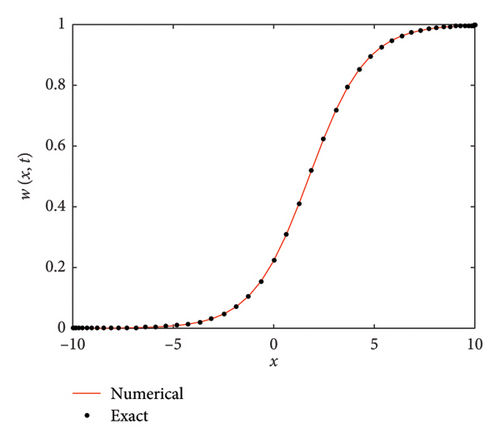
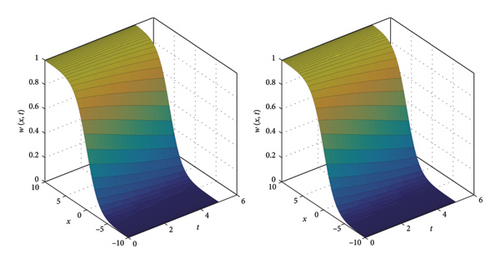
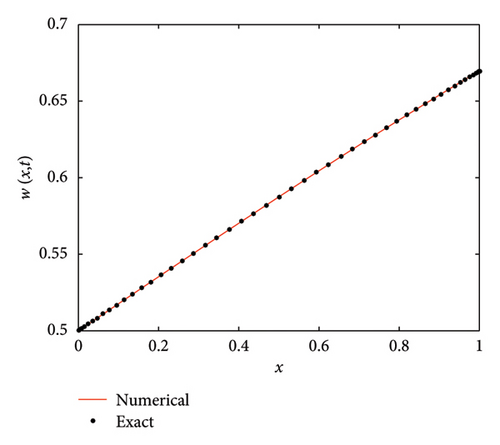
The numerical and exact solutions of (1) for δ = 0.001 with h = 0.01 are compared and presented in Figure 2. The graph shows that the numerical solution aligns closely with the exact solution throughout the entire domain, highlighting the effectiveness of the SOMDHBM. Figure 3 shows the exact solution alongside the numerical solution. The convergence and error profile of the numerical solution for SOMDHBM when δ = 0.01, 0.001, h = 0.01 and Nx = 50 are illustrated in Figure 4. As the δ decreases, the SOMDHBM converges faster.
| x | t | SOMDHBM | NFEval | NSFD [62] | ADM [11] | VIM [12, 13] | DTM [15] |
|---|---|---|---|---|---|---|---|
| 0.1 | 0.05 | 3.4425E − 13 | 300 | 5.0915E − 12 | 2.4853E − 06 | 2.4880E − 06 | 2.4875E − 06 |
| 0.1 | 1.3896E − 13 | 600 | 7.4482E − 12 | 4.9728E − 06 | 4.9750E − 06 | 4.9750E − 06 | |
| 1.0 | 2.0949E − 13 | 6000 | 1.1137E − 12 | 4.9744E − 05 | 4.9750E − 05 | 4.9750E − 05 | |
| 0.5 | 0.05 | 6.9701E − 15 | 300 | 1.1430E − 11 | 2.4763E − 06 | 2.4875E − 06 | 2.4875E − 06 |
| 0.1 | 6.5494E − 15 | 600 | 1.8999E − 11 | 4.9638E − 06 | 4.9750E − 06 | 4.9750E − 06 | |
| 1.0 | 6.2910E − 15 | 6000 | 3.0936E − 11 | 4.9738E − 05 | 4.9749E − 05 | 4.9749E − 05 | |
| 0.9 | 0.05 | 3.1650E − 15 | 300 | 5.0915E − 12 | 2.4674E − 06 | 2.4875E − 06 | 2.4874E − 06 |
| 0.1 | 3.3081E − 15 | 600 | 7.4481E − 12 | 4.9549E − 06 | 4.9750E − 06 | 4.9750E − 06 | |
| 1.0 | 3.4564E − 15 | 6000 | 7.8174E − 12 | 4.9730E − 05 | 4.9789E − 05 | 4.9749E − 05 | |
| t | SOMDHBM | NSFD [62] | |||||
|---|---|---|---|---|---|---|---|
| x = 0.2 | x = 0.5 | x = 0.8 | NFEval | x = 0.2 | x = 0.5 | x = 0.8 | |
| 0.3 | 4.6113E − 15 | 6.0347E − 16 | 4.2761E − 16 | 1800 | 1.8987E − 14 | 1.9838E − 14 | 1.9974E − 14 |
| 0.5 | 4.4390E − 15 | 5.8384E − 16 | 4.2696E − 16 | 3000 | 2.9530E − 14 | 3.0978E − 14 | 3.1209E − 14 |
| 0.9 | 4.2354E − 15 | 5.2259E − 16 | 4.2577E − 16 | 5400 | 1.8987E − 14 | 1.9838E − 14 | 1.9974E − 14 |
Example 2. Consider the F-N (1) in [49], subject to the BCs
The exact solution is
By imposing the BCs from Example 2 on (53) in the form of (54), we analyze the numerical accuracy and efficiency of different methods. With a = −10, b = 10, δ = 0.75, Nx = 50, h = 0.02 over fifteen iterations, Table 3 presents the maximum absolute error (ME∞), error norm (EN2), computation time and number of function evaluations (NFEval) for different values of t in the F-N equation (1). The table compares the performance of SOMDHBM, PDQM [61] and CFDBM [49].
The numerical results show that SOMDHBM consistently achieves the lowest ME∞ error ranging from 10−9 to 10−8, while PDQM exhibits much higher errors, reaching values as large as 10−3 to 10−4. Similarly, the EN2 norm for SOMDHBM remains significantly smaller than that of PDQM, confirming the effectiveness of the SOMDHBM. Compared to CFDBM, SOMDHBM achieves at least one order of magnitude lower error across all time steps. The SOMDHBM demonstrates higher computational efficiency by achieving lower computation times while maintaining superior accuracy.
The CPU time for SOMDHBM remains below 0.2 s even for large values of t, whereas PDQM requires significantly longer processing time, reaching 0.55 s at t = 5.0. This indicates that SOMDHBM not only produces more accurate results but also operates with significantly lower computation time, making it suitable for long-term and large-scale simulations in applied fields like neuroscience and chemical reaction-diffusion systems. The numerical findings confirm that the SOMDHBM outperforms PDQM and CFDBM in both accuracy and efficiency, making it a highly effective method for solving the F-N equation (1). The maximum absolute error is calculated using the following equation:
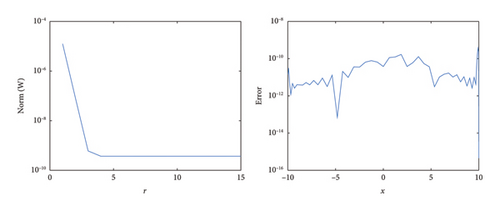
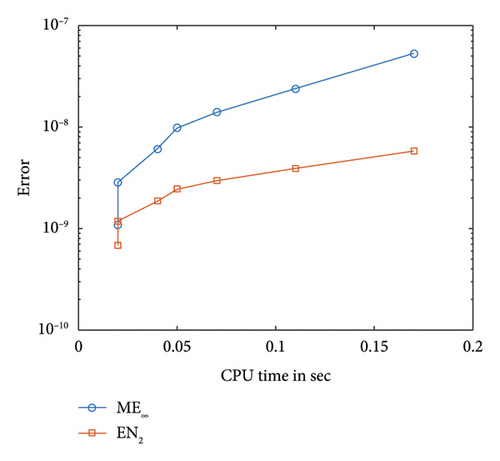
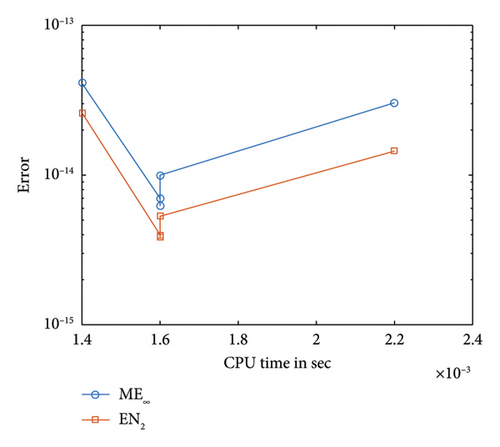
Figure 5 illustrates the comparison between the numerical and exact solutions of the F-N equation (1) for δ = 0.75 with a step-size of h = 0.02. Figure 6 presents a proximity of the exact solution against the numerical solution. The convergence and corresponding error profile of the SOMDHBM are displayed in Figure 7, indicating that the numerical method is accurate and that convergence is achieved after the 3rd iteration. The efficiency curves in Figure 8 illustrate the relationship between CPU time and error measures (ME∞ and EN2) for the SOMDHBM. The plot suggests that as CPU time increases, the error decreases, indicating the accuracy of the method improves with more computational effort.
| t | SOMDHBM | CFDBM [49] | PDQM [61] | |||||
|---|---|---|---|---|---|---|---|---|
| ME∞ | EN2 | CPU time | NFEval | ME∞ | ME∞ | EN2 | CPU time | |
| 0.2 | 1.0905E − 09 | 6.8424E − 10 | 0.02 | 600 | 5.4996E − 08 | 4.7416E − 05 | 2.3012E − 06 | 0.10 |
| 0.5 | 2.8575E − 09 | 1.1878E − 09 | 0.02 | 1500 | 1.0653E − 07 | 1.2312E − 04 | 5.5695E − 06 | 0.10 |
| 1.0 | 6.1097E − 09 | 1.8586E − 09 | 0.04 | 3000 | 1.5822E − 07 | 2.6261E − 04 | 1.1864E − 05 | 0.18 |
| 1.5 | 9.7815E − 09 | 2.4373E − 09 | 0.05 | 4500 | 1.9190E − 07 | 4.2096E − 04 | 1.9400E − 05 | 0.24 |
| 2.0 | 1.3928E − 08 | 2.9581E − 09 | 0.07 | 6000 | 2.1710E − 07 | 5.9999E − 04 | 2.8162E − 05 | 0.31 |
| 3.0 | 2.3928E − 08 | 3.9140E − 09 | 0.11 | 9000 | 2.5517E − 07 | 1.0324E − 03 | 4.9735E − 04 | 0.35 |
| 5.0 | 5.3186E − 08 | 5.7975E − 09 | 0.17 | 15,000 | 3.1252E − 07 | 2.3050E − 03 | 1.1395E − 04 | 0.55 |
Example 3. Consider the F-N equation (1), as presented in [49], subject to the BCs
The exact solution is
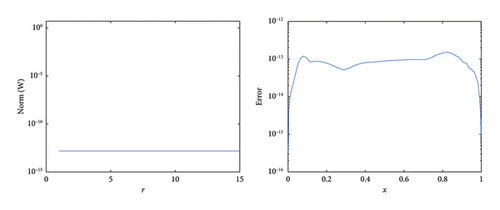
Table 4 displays the absolute error (AEn) and computation time for different values of x at t = 0.04 over one iteration for N = 10 in equation (1). It compares the performance of the SOMDHBM with CFDBM [49], İnan et al. [63] and ExpFDM [64]. The results indicate that SOMDHBM achieves the highest accuracy, with AEn error ranging from 10−15 to 10−16, while CFDBM produces errors on the order of 10−9 to 10−8. Although CFDBM is more accurate than İnan et al. and ExpFDM, SOMDHBM still outperforms CFDBM by at least six orders of magnitude across all spatial points. The SOMDHBM, like CFDBM, converges in just one iteration, whereas İnan et al. and ExpFDM require eight iterations, reinforcing the computational efficiency of the SOMDHBM.
Table 5 further supports these findings by presenting the NFEval, ME∞ error and EN2 norm for SOMDHBM. The results show that ME∞ and EN2 remain stable across all x, with values between 10−14 and 10−15, confirming that SOMDHBM maintains its accuracy across the domain. Overall, the numerical findings confirm that SOMDHBM surpasses CFDBM, İnan et al., and ExpFDM in both accuracy and efficiency, making it an optimal approach for solving the F-N equation (1). Its ability to capture sharp gradients and preserve wave profiles with minimal iterations also makes it ideal for real-time simulations and high-fidelity modeling, enhancing its practical relevance in both theoretical and applied contexts.
Figure 9 compares the numerical solution to the exact solution of the F-N equation (1) when δ = 0.5, h = 0.02 over fifteen iterations. Figure 10 shows the closeness of the exact solution and the numerical solution. The convergence and corresponding error profile of the SOMDHBM are depicted in Figure 11. The efficiency curves in Figure 12 show the relationship between CPU time, ME∞ and EN2 for the SOMDHBM. The graph shows that the method efficiently reduces computational errors with minimal processing time.
| x | SOMDHBM | CPU time | CFDBM [49] | İnan et al. [63] | ExpFDM [64] |
|---|---|---|---|---|---|
| 0.2 | 4.3299E − 15 | 0.0014 | 1.72E − 08 | 2.00E − 07 | 3.00E − 06 |
| 0.4 | 7.7716E − 16 | 0.0016 | 4.63E − 09 | 5.00E − 07 | 1.00E − 05 |
| 0.6 | 7.7716E − 16 | 0.0016 | 1.63E − 09 | 7.00E − 07 | 2.00E − 05 |
| 0.8 | 1.2212E − 15 | 0.0016 | 7.26E − 09 | 6.00E − 07 | 4.00E − 05 |
| 1 | 3.1086E − 15 | 0.0022 | 0 | 0 | 0 |
| Iterations | 1 | 1 | 8 | 8 |
| SOMDHBM | |||
|---|---|---|---|
| x | ME∞ | EN2 | NFEval |
| 0.2 | 4.1078E − 14 | 2.6128E − 14 | 40 |
| 0.4 | 6.9944E − 15 | 3.9893E − 15 | 40 |
| 0.6 | 6.2172E − 15 | 3.8373E − 15 | 40 |
| 0.8 | 9.9920E − 15 | 5.3106E − 15 | 40 |
| 1 | 3.0420E − 14 | 1.4457E − 14 | 40 |
| Iteration | 1 | ||

Example 4. Consider the F-N equation (2), with the exact solutions given by
With parameters A1 = 1, A2 = 1, ε = 0.3, Λ = 1.03, Δ = 2, Nx = 50, and h = 0.1, Table 6 presents the accuracy and efficiency of the SOMDHBM in solving a nonlinear neuronal PDE system. The method achieves very high accuracy, with ME∞ and EN2 remaining extremely small (around 10−13 to 10−12) across time steps. The errors increase slowly and smoothly, indicating good long-term stability. Computationally, the method is efficient, with CPU time rising moderately and the number of function evaluations increasing linearly. Overall, SOMDHBM is shown to be accurate, stable, and reliable for solving complex neurodynamic systems.
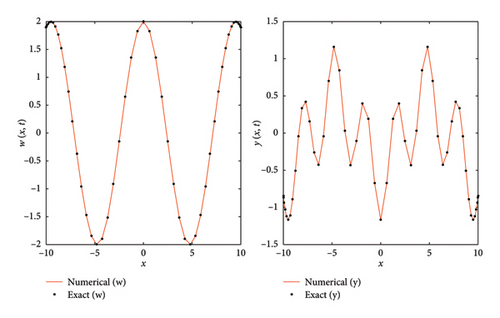
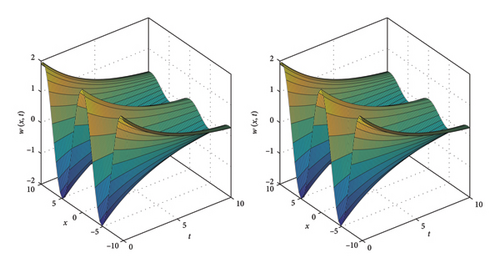
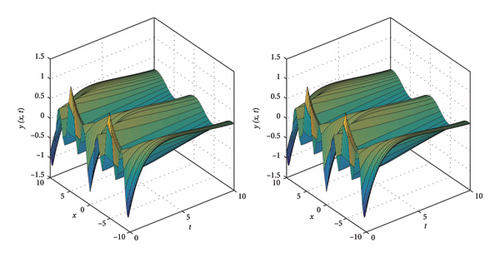
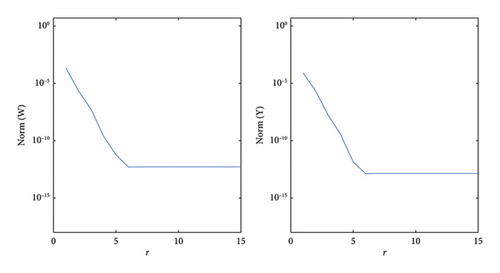
Figures 13, 14, and 15 illustrate the comparison between the numerical and exact solutions of the variables w and y for Example 4, using parameters Nx = 50, h = 0.1 and t = 10. Figure 13 displays both the numerical and exact solutions of w and y over the spatial domain x ∈ [−10, 10]., demonstrating a close agreement and validating the accuracy of SOMDHBM. Figures 14 and 15 separately present the numerical and exact solutions for w and y, respectively. The near-perfect alignment between the numerical and exact profiles confirms the reliable performance of the method for each component. Figure 16 illustrates the convergence behavior of the numerical solutions w and y over fifteen iterations. After approximately the sixth iteration, the norms of both solutions decrease rapidly and nearly exponentially, confirming strong stability and rapid convergence of the SOMDHBM.
| t | w | y | CPU time | NFEval | ||
|---|---|---|---|---|---|---|
| ME∞ | EN2 | ME∞ | EN2 | |||
| 1.0 | 6.8345E − 13 | 9.2908E − 13 | 9.2584E − 13 | 1.4409E − 12 | 0.31 | 750 |
| 2.0 | 1.1466E − 12 | 1.1409E − 12 | 2.4992E − 12 | 2.6659E − 12 | 0.40 | 1500 |
| 3.0 | 1.2450E − 12 | 1.1523E − 12 | 3.9191E − 12 | 3.3491E − 12 | 0.29 | 2250 |
| 4.0 | 1.5676E − 12 | 1.2269E − 12 | 4.9390E − 12 | 3.6615E − 12 | 0.45 | 3000 |
| 5.0 | 2.2278E − 12 | 1.4265E − 12 | 5.5462E − 12 | 3.7777E − 12 | 0.46 | 3750 |
| 6.0 | 3.1541E − 12 | 1.7283E − 12 | 5.9411E − 12 | 3.8094E − 12 | 0.56 | 4500 |
| 7.0 | 4.3344E − 12 | 2.1448E − 12 | 6.2205E − 12 | 3.8135E − 12 | 0.66 | 5250 |
| 8.0 | 5.7888E − 12 | 2.8069E − 12 | 6.4206E − 12 | 3.8143E − 12 | 0.88 | 6000 |
| 9.0 | 8.0435E − 12 | 4.0106E − 12 | 6.6980e − 12 | 3.8204E − 12 | 0.89 | 6750 |
| 10.0 | 1.1832E − 11 | 6.1989E − 12 | 7.6159E − 12 | 3.8380E − 12 | 0.89 | 7500 |
6. Conclusions
In this study, the efficiency and accuracy of the SOMDHBM were demonstrated for the F-N equation over large space and time intervals. The method utilized the optimized MDHBM in time and the SCM in space to solve F-N equation, a widely studied model in neuroscience and mathematical biology. The SOMDHBM results were compared with those obtained from the ADM, İnan et al. [63], CFDBM, ExpFDM, DTM, NSFD, PDQM and VIM scheme. Details of the numerical results were presented and it was evident from the numerical experiments that the SOMDHBM solutions performed better than other schemes in terms of accuracy, computation efficiency and agrees with the exact solutions. Future work may extend this approach to higher-dimensional or stochastic versions of the F-N model, or apply it to other nonlinear PDEs arising in applied sciences.
Nomenclature
-
- ADM
-
- Adomian decomposition method
-
- AE
-
- Absolute error
-
- BCs
-
- Boundary conditions
-
- BHM
-
- Block hybrid method
-
- CFDBM
-
- Compact finite difference block method
-
- CPU time
-
- Computation time
-
- DQM
-
- Difference quadrature method
-
- DTM
-
- Differential transform method
-
- EN2
-
- Error norm
-
- ExpFDM
-
- Exponential finite difference method
-
- F-N
-
- Fitzhugh–Nagumo
-
- HBM
-
- Hybrid block method
-
- IC
-
- Initial condition
-
- MDHBM
-
- Multiderivative hybrid block method
-
- ME∞
-
- Maximum absolute error
-
- NFEval
-
- Number of function evaluations
-
- NSFD
-
- Nonstandard finite difference
-
- OMDHBM
-
- Optimized multiderivative hybrid block method
-
- PDQM
-
- Polynomial differential quadrature method
-
- SCM
-
- Spectral collocation method
-
- SOMDHBM
-
- Spectral optimized multiderivative hybrid block method
-
- VIM
-
- Variational iteration method
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
This research was funded by the University of KwaZulu-Natal, South Africa.
Acknowledgments
The authors are grateful to the University of Kwazulu-Natal, South Africa, for assistance.
Open Research
Data Availability Statement
The data that support the findings of this study are available within the article.




