ZDHHC9 as a Potential Biomarker for Prognostic Evaluation and Diagnostic Identification in BRCA, CESC, HNSC, and KIRP Tumors
Abstract
An expanding corpus of evidence suggests that zinc finger DHHC-type palmitoyltransferase 9 (ZDHHC9) may represent a promising biomarker in various malignancies. Noteworthy is the fact that ZDHHC9 has been discovered to be significantly upregulated in specific varieties of human gastrointestinal cancers. Nevertheless, the precise role it plays in tumor prognostic evaluation and diagnostic identification remains ambiguous. In this study, we commenced with a thorough investigation of ZDHHC9 expression by leveraging the extensive and comprehensive TCGA database. This resource offers a wealth of valuable information and data samples, which enabled a more comprehensive and thorough analysis. The expression levels of ZDHHC9 in BRCA, CESC, HNSC, and KIRP were thoroughly investigated, revealing a significant elevation compared to normal tissues. This significant escalation was observed to be intimately linked with an adverse prognosis. A comprehensive analysis of the data revealed that the increased expression of ZDHHC9 in these particular cancer types was associated with poorer outcomes for patients. The extensive study examining various cancer types has yielded significant insights regarding the function of ZDHHC9, as well as its potential implications for prognosis and therapeutic strategies. Functional predictions indicate that immune or metabolic disorders, as well as the activation of carcinogenic signaling pathways due to abnormal expression of ZDHHC9, may significantly contribute to tumor progression and poor prognosis in patients. This further corroborates the influence of ZDHHC9 on the migration and invasion capabilities of CESC cell lines. ZDHHC9 is a key target for therapeutic and diagnostic use in BRCA, CESC, HNSC, and KIRP tumors, contributing significantly to the understanding of molecular tumor treatments.
1. Introduction
Cancer continues to be a critically important contributor to mortality, presenting considerable challenges for public health with extensive implications. It negatively impacts health in various ways and places significant economic strains on individuals, families, and society. The detrimental impact of cancer extends to a marked reduction in the quality of life for those affected, resulting in physical discomfort, emotional distress, and limitations in daily activities as well as social interactions. It is projected that by 2022, there will be approximately 4.82 million new cancer cases and 3.21 million cancer-related deaths in China alone, with lung cancer being the most prevalent type [1, 2]. Although current clinical treatments for cancer primarily involve surgical intervention combined with radiotherapy, chemotherapy, immunotherapy, and targeted drug therapy to enhance patient survival rates, the cytotoxic effects and drug resistance associated with these therapies still result in relatively low survival rates among patients. Consequently, the discovery of innovative pan-cancer biomarkers and therapeutic targets holds an extremely crucial and significant position for the substantial advancement and improvement of human health.
Zinc finger DHHC-type palmitoyltransferase 9 (ZDHHC9) is a complete membrane protein that encodes palmitoyltransferase, facilitating the palmitoylation of proteins. It is a member of the illustrious zinc finger DHHC protein family [3]. Previous studies have extensively documented a substantial association between ZDHHC9 and a range of neurodevelopmental disorders. [4]. Loss-of-function mutations in ZDHHC9 may be intricately associated with X-linked intellectual disabilities and the concomitant manifestation of epilepsy [5, 6]. In bladder cancer, ZDHHC9 has been identified as a significantly upregulated oncogene; it inhibits tumor proliferation, promotes apoptosis in tumor cells, and augments the therapeutic efficacy of gemcitabine and cisplatin [7]. In a scholarly investigation undertaken by Li and colleagues, it was found that overexpression of ZDHHC9 in lung adenocarcinoma inhibits PD-L1 protein degradation by increasing its palmitoylation levels, and this elevation in PD-L1 protein levels can jeopardize antitumor immunity and facilitate the proliferation of LUAD cells [7, 8]. Furthermore, ZDHHC9 has been shown to enhance colon cancer progression by upregulating PD-L1 expression while inhibiting CD8 T cell functionality [9]. Significantly, ZDHHC9 serves as a pivotal facilitator in the advancement of breast cancer proliferation through its role in the palmitoylation of PD-L1, which affects its stability and protects tumor cells evading T-cell immune surveillance [10]. ZDHHC9 has revealed profound associations with an array of pivotal molecules and intricate pathways that play a critical role in the regulation of cancer, accentuating its promising potential as a groundbreaking therapeutic target within the realm of oncology. However, despite these findings, the exact role of ZDHHC9 in solid tumors remains unclear.
This study revealed that ZDHHC9 expression was significantly upregulated in the tissues of patients afflicted with BRCA, CESC, HNSC, and KIRP when contrasted with their corresponding normal tissues. This upregulation correlated with poor prognosis. Protein–protein interaction (PPI) analysis and gene set enrichment analysis (GSEA) were used to investigate the signaling pathway of ZDHHC9 and its possible underlying mechanism. Additionally, therapeutic strategies targeting ZDHHC9 were explored by examining molecules involved in its signaling pathway. In vitro experiments further confirmed that ZDHHC9 may assume a pivotal role in the migration and invasion of tumor cells. These findings deliver fresh and profound perspectives for diagnosis.
2. Materials and Methods
2.1. Gene Expression Analysis
We strictly adhered to the methodologies proposed by Xiangjian Zhang et al. in 2024 without any deviation. The comprehensive ZDHHC9 mRNA data of 33 distinct cancer types, along with their corresponding adjacent and normal samples, were meticulously retrieved from the highly reliable Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases [11]. The expression levels of the ZDHHC9 protein throughout different organs and tissues in the human body were obtained using the authoritative Human Protein Atlas (HPA). Statistical analyses were performed with precision using the advanced R software, while visualizations were generated utilizing the sophisticated “ggplot2” package. The Wilcoxon rank-sum test, a widely recognized and reliable statistical method, was deliberately employed for detailed comparisons between groups. Specifically, the Wilcoxon rank-sum test was precisely utilized for meticulous and comprehensive comparisons between the various groups under investigation. Moreover, a p value less than 0.05 was stringently regarded as the definitive and conclusive criterion for statistically significant differences. This strict threshold ensures the accuracy and validity of the identified differences, providing a solid foundation for drawing meaningful and scientifically sound conclusions in the research context.
2.2. Survival Prognosis Analysis
The Kaplan–Meier survival analysis software (Version 3.2-10) was employed to generate survival curves. The association between ZDHHC9 expression and key outcomes, including overall survival (OS), disease-specific survival (DSS), and progression-free survival (PFS), was investigated. To enhance clarity and efficiency in visualization, the survminer package was utilized. Furthermore, the utilization of forest plots to systematically distill and vividly illustrate the findings of univariate Cox regression analyses not only enhances the meticulous evaluation of the data but also elucidates the intricate relationships that are being scrutinized.
2.3. Correlation of ZDHHC9 Expression With Mutational Tumor Heterogeneity
The intricate relationship between ZDHHC9 and tumor mutation burden (TMB), alongside microsatellite instability (MSI), was meticulously examined through the application of Spearman correlation analysis. In this process, the powerful fmsb R software package (Version 0.7.5) facilitated the generation of a clear and intuitive radar map, which provided significant visual support and a robust data foundation for subsequent research and analysis.
2.4. Immunoinfiltration Assessment
We painstakingly acquired the comprehensive immune dataset from the well-known and authoritative TCGA database. Simultaneously, we effectively utilized the highly functional R packages GSVA and GGPLOT to meticulously assess the abundance of tumor-infiltrating immune cells. Furthermore, we conducted a detailed analysis to explore the variations in infiltration levels, aiming to obtain a more in-depth and comprehensive understanding of the related immune cell dynamics.
2.5. GSEA and STRING Method
In the realm of biomedical research, GSEA is extensively employed as a crucial data mining tool to elucidate potential biological pathways within gene expression profiles. By concentrating on and analyzing groups of genes that share common biological functions, chromosomal locations, or regulatory mechanisms, GSEA can effectively assess its efficacy and assist researchers in gaining a deeper understanding of the pathogenesis and prognostic implications associated with various diseases.
This study utilized the latest version of GSEA software (v4.0.3) to thoroughly investigate the potential impact mechanism underlying the differential expression of the ZDHHC9 gene on both onset and prognosis across tumor types such as BRCA, CESC, HNSC, and KIRP (TCGA). Through systematic analysis of large-scale gene expression data, we aimed to comprehend the intricate and nuanced relationship between ZDHHC9 and these tumor types from a comprehensive perspective.
Furthermore, regarding PPI information, the STRING database is dedicated to collecting, scoring, and integrating relevant data from all public sources while constructing PPI networks and predicting potential interactions through computational methods. We employed the STRING database to construct a PPI network that encompasses interaction relationships among differentially expressed genes. Additionally, we combined this with tools such as GEPIA 2.0 for a comprehensive and nuanced exploration of the intricate relationship between ZDHHC9 and NRAS in BRCA, CESC, KIRP, and HNSC.
2.6. Cell Cultures and Transfection
The normal epithelial cell line HEEC alongside the human endometrial carcinoma cell lines HeLa and CaSKi were procured from BNCC. These cells were maintained in DMEM medium (Gibco, USA), enriched with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), complemented by 1% penicillin/streptomycin within a humidified incubator set to an atmospheric composition of 5% carbon dioxide at a constant temperature of 37°C. For the comparative analysis of negative controls, Lipofectamine 2000 (Invitrogen, USA) was employed si-NC, si-ZDHHC9#1, (5H′ -CTGTTACATGCAAGATCT-3′) and si-ZDHHC9#2, (5′-GAAGTCCTCATTTGCTTCTTT-3′) were transfected into HeLa and CaSKi cells.
2.6.1. RNA Extraction and Quantitative RT-PCR
Total RNA was meticulously extracted utilizing TRIzol (Invitrogen, Carlsbad, CA, USA). Subsequently, the total RNA underwent reverse transcription into complementary DNA (cDNA) employing the PrimeScript RT Reagent Kit (TaKaRa, Otsu, Shiga, Japan), in strict accordance with established protocols. Quantitative PCR (qPCR) was conducted following TaKaRa’s comprehensive system guidelines. The relative expression levels of mRNA were precisely quantified using SYBR Premix Ex Taq II (TaKaRa, Otsu, Shiga, Japan) in real-time PCR assays. The RT-PCR primers utilized for this process were sourced from Gene Pharma (Shanghai Gene Pharma, Shanghai, China). To ensure accuracy and consistency in results, mRNA expression levels were normalized to GAPDH, thus enabling the calculation of relative gene expression through the 2-ΔΔCT method.
RT-PCR primers ZDHHC9: Forward: 5′-AAGGTGACACGGAAATGGGAG-3′, Reverse: 5′-CGACACTCGAGAGCAAGAAGAA-3′
2.6.2. Protein Extraction and Western Blot Analysis
Total proteins were extracted by using RIPA buffer (Beyotime Biotechnology, Shanghai, China) and protease inhibitors (Roche Diagnostics, Indianapolis, IN, USA). The protein concentrations were quantified with a diquinolinic acid (BCA) assay kit (Thermo Fisher Scientific, Waltham, MA, USA). 40 μg of protein was loaded onto a 10% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) and then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membranes were blocked in 5% fat-free milk at 25°C for 1 h and incubated with primary antibodies overnight at 4°C. The primary antibodies were from Abcam (Abcam, Cambridge, MA, USA), as shown in Table 1. After that, the membranes were washed and incubated for 1 h with matching secondary antibodies (1:50,000). Finally, immunoreactive proteins were detected by Pierce ECL Western blot substrate (Thermo Fisher Scientific, Waltham, MA, USA) using an ECL detection system. Quantification was also carried out using ImageJ software (NIH, Bethesda, MD, USA).
| Proteins | Host | Catalog | Antibody dilution |
|---|---|---|---|
| ZDHHC9 | Rabbit | ab74504 | 1:1000 |
| GAPDH | Rabbit | 81640-5-RR | 1:50,000 |
2.6.3. Transwell
In summary, 5 × 104 cells were inoculated into a chamber coated with an invasion and migration matrix gel (BD Biosciences, USA). Serum-free medium was added to the upper compartment, while complete DMEM medium was placed in the lower compartment. After 24 h incubation, cells were fixed with 4% paraformaldehyde for assessment of migration or invasion and stained with 0.1% crystal violet.
2.6.4. Wound-Healing Assay
Cell migration was evaluated using a wound-healing assay. Transfected cells (5 × 104) were seeded in a 6-well plate and incubated until reaching approximately 90% confluence. Cells were washed with PBS through a 20 μL straw tip to create linear scratches. After 24 h, the cells were fixed and stained sequentially with 4% paraformaldehyde and then with 0.1% crystal violet (Servicebio) for 15 min each. Wound closure extent was imaged and quantified using an inverted microscope (Leica) in combination with ImageJ software.
3. Ethical Statement
Since the clinical data employed in this study were retrieved from a publicly accessible database, there were absolutely no ethical issues or concerns at either the local or state level. This is because the data were made available for public use and research purposes. Furthermore, given that this retrospective analysis was based on public data obtained from the TCGA database, the requirement for informed consent was not applicable. The nature of the data being public and intended for such analyses eliminated the need for obtaining individual consent from the patients or participants. This approach is in line with the established norms and regulations for using publicly accessible data in scientific research.
3.1. Statistical Analysis
The statistical analysis for this study was conducted automatically through the aforementioned online databases. The results were determined to be statistically significant at the subsequent levels: ∗p < 0.05, indicating a moderately significant difference; ∗∗p < 0.01, suggesting a highly significant difference; ∗∗∗p < 0.001, highlighting an extremely significant difference; and ∗∗∗∗p < 0.0001, demonstrating an exceptionally and profoundly significant difference.
4. Results
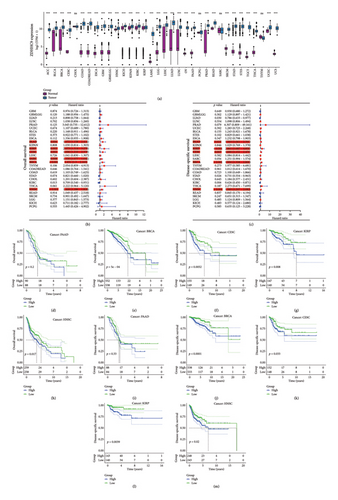
4.1. Analysis of the Expression and Prognosis Associated With ZDHHC9
Due to the insufficiency of normal tissue samples within the TCGA database, we integrated normal tissue data sourced from the GTEx database with tumor tissue data obtained from TCGA. This comprehensive approach enabled us to scrutinize ZDHHC9 expression across 34 distinct cancer types. The research unveiled that the expression of ZDHHC9 was markedly heightened across 30 distinct types of cancer (all p values < 0.05) (Figure 1(a)). To evaluate the significance of ZDHHC9 in cancer prognosis, we employed a Cox proportional hazards model to examine the relationship between ZDHHC9 expression levels and survival outcomes, specifically OS (Figure 1(b)), DSS (Figure 1(c)), and DFI and PFI (Figure S1). The results from the Cox proportional hazards models indicated significant associations between ZDHHC9 expression levels and both OS and DSS across various cancer types, including PAAD, KIRP, CESC, SARC, BRCA, and HNSC. When the clinical data of individual tumors were analyzed, Kaplan–Meier survival curves (Figures 1(d), 1(e), 1(f), 1(g), 1(h), 1(i), 1(j), 1(k), 1(l), and 1(m)) demonstrated that the OS and DSS of patients with high ZDHHC9 expression were significantly higher than those of patients with low ZDHHC9 expression in BRCA, CESC, KIRP, and HNSC.
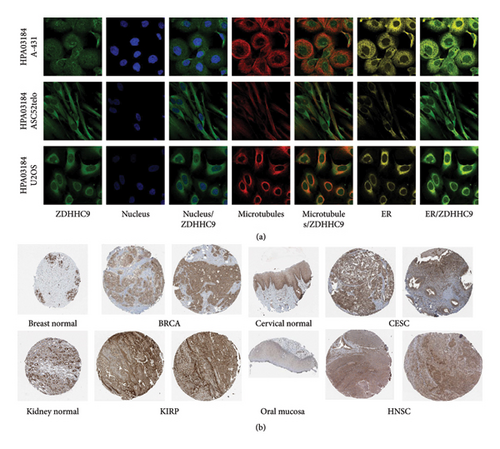
4.2. Cell Localization and Immunohistochemistry (IHC) Analyses of ZDHHC9
We investigated the protein expression pattern of ZDHHC9 using the HPA database. Our analysis reveals that ZDHHC9 is primarily situated within the intricate networks of the endoplasmic reticulum and Golgi apparatus, while also exhibiting a significant presence in the cytosolic environment (Figure 2(a)). Furthermore, we evaluated the IHC results for ZDHHC9 available in the HPA database (Figure 2(b)), which revealed an upregulation of ZDHHC9 in BRCA, CESC, KIRP, and HNSC.
4.3. The Relationship Between ZDHHC9 Expression and TMB, MSI, Tumor Microenvironment, Immune Cell Infiltration, and Immune Checkpoints
TMB and MSI are emerging biomarkers of mutant tumor heterogeneity that have the considerable potential to predict the efficacy of immune checkpoint inhibitor immunotherapy. In this comprehensive study, we meticulously evaluated the intricate association between ZDHHC9, TMB, and MSI (Figures 3(a) and 3(b)). Our in-depth analysis uncovered a highly significant and positive correlation between ZDHHC9 and TMB in the context of BRCA. Moreover, ZDHHC9 exhibited a strikingly notable positive correlation with MSI not only in BRCA but also in HNSC tumors. These remarkable findings strongly suggest that ZDHHC9 is profoundly and significantly associated with TMB and MSI in BRCA, indicating its potential substantial influence on the progression of tumors through its close relationship with these crucial biomarkers.
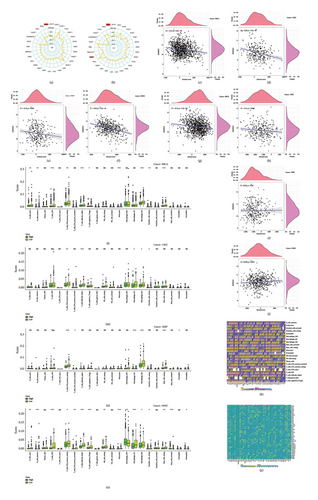
In our correlation analysis of the tumor microenvironment, we found that the expression levels of ZDHHC9 were negatively correlated with immune and stromal cell densities. Specifically, lower levels of ZDHHC9 were associated with higher immune cell infiltration in BRCA (r = −0.17; r = −0.14), CESC (r = −0.29; r = −0.11), KIRP (r = −0.13; r = 0.012), and HNSC (r = −0.35; r = 0.079) (Figures 3(c), 3(d), 3(e), 3(f), 3(g), 3(h), 3(i), and 3(j)). This suggests that upregulation of ZDHHC9 correlates with reduced immune and stromal cell levels as well as increased tumor cell purity, potentially leading to shorter patient survival outcomes.
Regarding immune cell infiltration specifically, we observed negative correlations between ZDHHC9 expression and M2 macrophages as well as resting mast cells across various cancer types: BRCA (r = −0.227; p < 0.001; r = −0.150; p < 0.001), CESC (r = −0.148; p < 0.001; r = −0.158; p < 0.001), KIRP (r = −0 0.121; p = 0.014; r = −0.217; p < 0.001), and HNSC (r = −0137, p = 0.001; r = −0.162, p < 0.001) (Figures 3(k), 3(l), 3(m), 3(n), and 3(o)).
Furthermore, we conducted a meticulous examination of the relationship between the expression of ZDHHC9 and immune checkpoint molecules (Figure 3(p)). The comprehensive analysis and in-depth exploration of the data obtained revealed a distinct and positive correlation between the expression levels of ZDHHC9 and the majority of immune-related genes (all p values < 0.05). This finding strongly suggests that this particular gene might exert a significant influence on tumor behavior through the modulation of immune expression profiles. It implies that ZDHHC9 could potentially play a crucial role in shaping the immune response within the tumor microenvironment and thereby have implications for tumor progression and therapeutic strategies.
4.4. GSEA and Functional Enrichment Analysis of ZDHHC9
Human tumor samples from the GSEA database were meticulously classified into high and low expression groups of ZDHHC9. Subsequently, a comprehensive KEGG pathway analysis was carried out with great precision to thoroughly explore the potential pathways and genes that might have a significant influence on the tumorigenicity of four specific tumors (Figures 4(a), 4(b), 4(c), and 4(d)). The Venn diagram clearly indicated that ZDHHC9 predominantly regulates the development of BRCA, CESC, and HNSC through a set of five key pathways, namely, asthma, inflammatory bowel disease, autoimmune thyroid disease, primary immunodeficiency, and allograft rejection. Furthermore, ZDHHC9 primarily affects the tumorigenesis of KIRP via pathways closely associated with the cell cycle and thyroid hormone synthesis (Figures 4(e) and 4(f)). This detailed analysis provides valuable insights into the underlying mechanisms of tumor development and potentially paves the way for the development of more targeted therapeutic strategies.
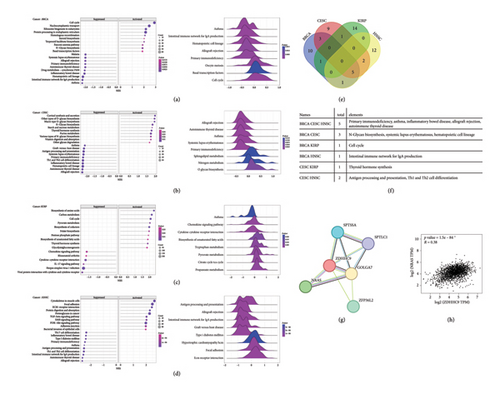
Further in-depth analysis of the PPI network was meticulously conducted in order to comprehensively elucidate the intricate molecular mechanism through which ZDHHC9 functions and operates within tumors. The PPI network uncovered a highly significant and notable interaction between ZDHHC9 and neuroblastoma RAS virus (v-ras) oncogene homologs, particularly NRAS. NRAS is an N-ras oncogene that encodes a membrane protein which holds the responsibility for the continuous shuttling process between the Golgi apparatus and the plasma membrane. This specific shuttling process is precisely regulated by the delicate balance of palmitoylation and depalmitoylation within the complex formed by ZDHHC9 and GOLGA7 (Figure 4(g)). Significantly, NRAS demonstrated a strikingly strong positive correlation with ZDHHC9 in various types of cancers such as BRCA, CESC, KIRP, and HNSC (r = 0.38, p < 0.001) (Figure 4(h)).
4.5. ZDHHC9 Is Significantly Upregulated in Cervical Cancer Cells, Affecting the Migration and Invasion Ability of Tumor Cells
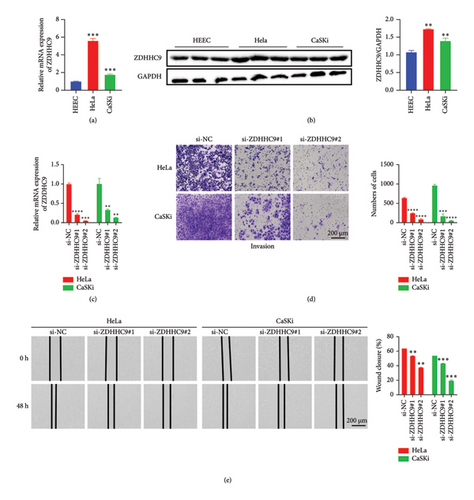
In vitro cell experiments were meticulously conducted to comprehensively evaluate the expression of the ZDHHC9 gene in tumor cells in contrast to normal epithelial cells. The outcomes of these experiments clearly indicated that both the mRNA and protein levels of the ZDHHC9 gene were conspicuously and significantly upregulated in HeLa and CaSKi cells when compared with normal HEEC cells (Figures 5(a) and 5(b)). After the successful knockdown of ZDHHC9, which was verified and confirmed (Figure 5(c)), we attentively observed a strikingly marked impairment in the migration and invasion capabilities of tumor cells. Specifically, the number of cells in the si-ZDHHC9 group within the transwell assays was significantly and notably reduced (Figure 5(d)), and the wound closure rates for cells in the si-ZDHHC9 group were conspicuously and significantly diminished during the wound-healing experiments (Figure 5(e)). These revealing findings strongly suggest that ZDHHC9 might potentially play an extremely critical role in regulating the migration and invasion processes of tumor cells.
5. Discussion
The protein encoded by ZDHHC9 functions as a palmitoyltransferase, mediating the process of protein palmitoylation. Palmitoylation is a reversible post-translational lipid modification that involves the attachment of long-chain fatty acids, such as palmitic acid, to cysteine residues. This modification plays a crucial role in facilitating vesicular transport and subcellular localization of modified proteins [12]. Notably, ZDHHC9 expression is upregulated in colon cancer, particularly within microsatellite stable (MSS) tumors [13]. To the best of our knowledge, as of now, there is a distinct and notable lack of comprehensive and in-depth literature specifically addressing the potential prognostic implications and the intricate biological functions of ZDHHC9 in pan-cancer analyses. In this study, we deliberately and purposefully utilized the most advanced and cutting-edge bioinformatics tools available to conduct a meticulous and systematic analysis of ZDHHC9 at the pan-cancer level. Our primary and key focus was precisely on thoroughly investigating the diverse expression patterns of ZDHHC9, its profound prognostic significance, and the associated immunoinfiltration and gene enrichment within the complex and heterogeneous tumor tissues. We aimed to provide a detailed and comprehensive understanding of these aspects to contribute to the existing knowledge in the field of cancer research. Analysis utilizing TCGA and GTEx databases revealed elevated expression levels of ZDHHC9 in BRCA, CESC, HNSC, and KIRP. Furthermore, comprehensive Kaplan–Meier survival analysis indicated that high expression levels of ZDHHC9 in BRCA, CESC, HNSC, and KIRP were significantly correlated with poor prognosis outcomes. The expression pattern of the ZDHHC9 protein was investigated using data from HPA database. It was observed that the ZDHHC9 protein predominantly localized to the endoplasmic reticulum and Golgi apparatus within the cytoplasm. Furthermore, its expression levels in tumor tissues were significantly elevated compared to those found in normal tissues. Consequently, abnormal expression levels of ZDHHC9 may serve as a valuable prognostic indicator for specific types of tumors.
MSI is associated with an elevated risk of cancer characterized by specific clinicopathological features, and TMB is a useful biomarker for immune checkpoint blocking (ICB) selection in certain cancer types [14, 15]. Immune cell infiltration can accurately predict the prognosis and chemotherapy effect of breast cancer patients [16]. In most cancer histologies, a higher somatic TMB and MSI are associated with enhanced efficacy of immunotherapy and improved OS rates. To accurately and comprehensively assess the potential role of ZDHHC9 in clinical immunotherapy, we meticulously conducted an in-depth analysis of the correlation between the expression level of ZDHHC9 and various crucial factors, such as TMB, MSI, and a wide range of immune checkpoint markers. The findings revealed a positive correlation between ZDHHC9 and TMB in BRCA, as well as MSI in both BRCA and HNSC. These findings suggest that elevated expression levels of ZDHHC9 in BRCA are linked to poor prognostic outcomes, whereas high MSI levels indicate promising prospects for immunotherapy in BRCA that warrant further investigation. Furthermore, a significant correlation was observed between ZDHHC9 and immune checkpoint genes across various tumors including BRCA, CESC, HNSC, and KIRP. This suggests a robust association between ZDHHC9 expression and tumor immunotherapy. Additionally, during the analysis of the tumor microenvironment correlations, negative associations were found between ZDHHC9 expression levels and both immune as well as stromal cell densities in BRCA, CESC, KIRP, and HNSC. These results indicate that the upregulation of ZDHHC9 is associated with decreased levels of immune and stromal cells, along with an increased purity of tumor cells, which may contribute to reduced patient survival times. In terms of immune cell infiltration, ZDHHC9 expression was significantly correlated with various types of immune cells, particularly M2 macrophages and resting mast cells across multiple tumor types. Specifically, notable correlations were observed in BRCA, CESC, KIRP, and HNSC. Additionally, a significant relationship between ZDHHC9 expression and immune checkpoint genes was identified in various tumors including BRCA, CESC, HNSC, and KIRP. These findings suggest a strong association between ZDHHC9 expression and tumor immunotherapy efficacy.
KEGG analysis indicated that ZDHHC9 primarily regulates tumorigenesis in BRCA, CESC, and HNSC through five key pathways: asthma, inflammatory bowel disease, autoimmune thyroid disease, primary immunodeficiency, and allograft rejection. Additionally, ZDHHC9 influences KIRP tumorigenesis mainly via the cell cycle and thyroid hormone synthesis pathways. Further PPI analysis revealed a significant interaction between ZDHHC9 and NRAS. In BRCA, CESC KIRP, and HNSC tissues, NRAS exhibited a strong positive correlation with ZDHHC9 (r = 0.38; p < 0.001). The inactivation of palmitoylacyltransferase Zdhhc9 mitigates the leukemogenic potential of oncogenic NRAS [17]. In the study conducted by Yan et al., NRAS expression was significantly upregulated in LUAD tissues and was associated with poor prognosis [18]. We hypothesize that the ZDHHC9-GOLGA7 complex regulates NRAS through palmitoylation and depalmitoylation mechanisms, thereby promoting cancer cell proliferation and metastasis. In experiments involving cervical cancer cells, we observed that both mRNA and protein levels of the ZDHHC9 gene were significantly elevated in HeLa and CaSKi cells compared to normal HEEC cells. Moreover, successful knockout of ZDHHC9 impaired the migratory and invasive capabilities of tumor cells. These findings suggest that ZDHHC9 may serve as a critical factor in the progression of cervical cancer.
The bioinformatics analysis in this study provides valuable insights into the role of ZDHHC9 in malignant tumors and lays a foundation for future research in this field. However, it still has some limitations. Further in vitro or in vivo studies are required to validate our findings and enhance the effectiveness of treatment. Moreover, ZDHHC9 expression is correlated with immune response and clinical survival in human malignancies; however, it remains uncertain whether ZDHHC9 influences clinical survival through the modulation of immune pathways.
6. Conclusion
ZDHHC9 appears abnormally in 34 tumors and is associated with poor prognosis in BRCA, CESC, KIRP, and HNSC. Clinical tumor prognosis assessment of ZDHHC9 in BRCA, CESC, KIRP, and HNSC revealed the potential of ZDHHC9 to have a good immunotherapeutic effect in these tumors. In addition, the correlation analysis of immune microenvironment indexes was also carried out. ZDHHC9 may interact with NRAS to influence tumor prognosis, suggesting that ZDHHC9 is a valuable target for potential clinical treatment prognosis of BRCA, CESC, KIRP, and HNSC.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding
This research did not receive funding from any sources.
Acknowledgments
The authors thank other esteemed colleagues in the Department of Obstetrics and Gynecology for their valuable support and helpful discussion.
Supporting Information
Figure S1: ZDHHC9 as a prognostic indicator in pan-cancer. (a, b): Forest plots illustrating the results of Cox regression analyses regarding ZDHHC9 expression and survival outcomes (disease-free interval and progression-free interval).
Open Research
Data Availability Statement
The datasets that were utilized and/or analyzed during the current study are obtainable from the corresponding author upon reasonable request. These datasets have been carefully curated and processed to ensure their accuracy and reliability for the purpose of this study. The corresponding author is willing to provide access to them to facilitate further research and exploration in the related field.