Investigation of the Potential Targets and Mechanism Actions of Berberine in the Treatment of Pulpitis Based on Bioinformatics Analysis
Abstract
Berberine, an active compound extracted from the Chinese herb, was reported to have an antibacterial effect and an anti-inflammatory effect and promote osteogenic differentiation of human dental pulp stem cells. However, the underlying therapeutic mechanism of berberine in pulpitis is still unknown. Here, bioinformatics analysis was performed to investigate the potential mechanism of berberine against pulpitis. First, we identified the collective targets of berberine and pulpitis from several databases using a Venny online tool. The pattern of interaction between berberine and the targeted protein was visualized by molecular docking. Moreover, we performed GSEA, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to obtain potential pathways. Cell experiments were performed to validate the bioinformatics analysis results. Prostaglandin-endoperoxide synthase 2 (PTGS2) was identified as the crucial antipulpitis target of berberine via the CytoHubba plugin. Docking analysis indicated that berberine could fit in the binding pocket of the PTGS2 protein. Additionally, berberine exerted its therapeutic effects against pulpitis via multiple pathways. Overall, berberine possessed therapeutic effects against pulpitis through the inactivation of toll-like receptor and NOD-like receptor signaling pathways. The present research proposes a novel approach to explore the therapeutic mechanism of natural products.
1. Introduction
Pulp inflammation (pulpitis) is one of the common oral diseases, which is accompanied by spontaneous or provoked pain evoked by stimulating nerve cells of the dental pulp [1, 2]. Pulpitis is caused by a sustained and severely stimulated response that may lead to inflammatory damage and apoptosis of pulp cells, resulting in long-term tissue impairments [3–5]. It has been demonstrated that pathogenic microorganism infection is closely related to the initiation and progression of pulpitis [6]. Immune responses are one of the primary factors implicated in inflammatory responses in inflamed pulp tissue. The balance between repair processes and inflammation in the host defense response could affect the degree of injury in pulpitis, in which multiple signaling pathways have participated, including toll-like receptors, nuclear factor-kappa B, and NLR family pyrin domain containing 3 (NLRP3) inflammasomes’ signaling pathways [7, 8]. Nonimmune and immune cells including endothelial cells, odontoblasts, and macrophages could identify pathogen-related molecules and trigger immune responses via the expression of toll-like receptors in pulp tissue [9–11]. The previous report indicated that nitric oxide-induced inflammatory cytokines expression is involved in nuclear factor-kappa B activation in primary cultured pulp cells [12]. Besides, astrocyte-elevated gene-1 could initiate the generation of proinflammatory cytokines via the nuclear factor-kappa B pathway in pulp cells [13]. It has revealed that lipopolysaccharide (LPS) stimulation activated the NLRP3 inflammasome pathway, thereby leading to the secretion of proinflammatory cytokines, which contributes to the immune responses in human dental pulp tissue [14]. Currently, clinical interventions for the prevention and treatment of pulpitis are limited due to the lack of understanding of the specific molecular mechanisms underlying pulpitis. Thus, clarifying potential molecular mechanisms involved in the progression of pulpitis and inflammatory responses might be useful for the prevention and treatment of this oral disease.
Berberine is an isoquinoline alkaloid extracted from popular medicinal plants, such as Chelidonium majus, Xanthoriza simplicissima, and Xanthoriza simplicissima. Recently, studies have indicated that berberine exerts a wide range of pharmacological effects, such as antiviral, antitumor, anti-inflammatory, hypolipidemic, anti-infective, neuroprotective, and hepatoprotective effects [15, 16]. Berberine has been found to inhibit interleukin 6 (IL-6) and C-C motif chemokine ligand 11 (CCL11) production via regulation of the signal transducer and activator of transcription 6 (STAT6) pathway in proinflammatory cytokine-activated human bronchial epithelial cells [17]. Berberine reportedly alleviates streptozotocin (STZ)-induced diabetic nephropathy by inhibiting the TLR4/NF-кB signaling pathway [18]. Berberine has been reported to protect the intestinal mucosal barrier via the regulation of the toll-like receptor pathway [19]. Importantly, berberine promotes odontoblast differentiation via activating the Wnt/β-catenin pathway, and it could promote human dental pulp cell osteogenic differentiation via EGFR-MAPK-Runx2 signaling pathway activation, suggesting that berberine is a potential therapeutic agent for the treatment of pulpitis [20, 21]. Moreover, our previous study revealed that berberine regulates LPS-induced inflammation in human dental pulp fibroblast via the miR-21/KBTBD7 axis [22]. A mechanistic randomized controlled trial demonstrated that berberine effectively reduces total cholesterol levels and potentially lowers LDL-c, while maintaining a good safety profile [23]. A comprehensive meta-analysis of randomized controlled trials revealed that the utilization of berberine in individuals suffering from metabolic syndrome and associated disorders exhibited a notable reduction in inflammatory markers, such as CRP, TNF-α, and IL-6 [24]. Based on these reports, we speculated that berberine might be a potential therapeutic drug for pulp inflammation via the inactivation of inflammatory-related pathways.
Network pharmacology is a novel method for drug development and pharmacological mechanistic research, which combines multidirection pharmacology, bioinformatics, system biology, and interdisciplinary theories to screen therapeutic targets quickly, economically, and efficiently [25]. In recent years, molecular docking is a well-established computational method based on the structure of molecules, which is extensively utilized in the field of drug discovery [26, 27]. In the present study, computational tools and public databases were used to screen relevant therapeutic targets of berberine in the treatment of pulpitis. Subsequently, molecular docking was performed to validate whether berberine exerts an antipulpitis effect by binding to PTGS2 protein. Finally, the potential mechanisms were systematically predicted based on the results of enrichment analyses. The flowchart of the present study is presented in Figure 1. We aimed to investigate the potential targets and mechanism of berberine against pulpitis, and our findings provided the theoretical basis for the investigation of molecular mechanisms of berberine in the treatment of pulpitis.

2. Materials and Methods
2.1. Collection of Putative Targets of Pulpitis and Berberine
We collected the microarray dataset (GSE77459) from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/) with the keyword “pulpitis.” The selection criteria for the datasets involved in the present study are as follows: the species is “Homo sapiens,” the data type is “expression profile microarray,” and GEO2R analysis can be performed. The GSE77459 dataset contains 6 inflamed pulps from patients with irreversible pulpitis and 6 pulp tissue samples from healthy donors. The raw expression data were processed using Bioconductor Packages, which included steps such as creating an expression matrix and matching probes to gene symbols. To ensure accuracy, a manufacturer-provided annotation file was used to convert probes to gene symbols. Additionally, duplicate probe sets were eliminated by calculating the median expression value of all corresponding probes. Probes that did not have corresponding gene symbols were also excluded. The microarray datasets were preprocessed and normalized using the affy package in the R environment. Subsequently, the R package was used to screen differentially expressed genes (DEGs) between the healthy donors and inflamed pulps. ∣log fold change∣≥ 1 and p < 0.05 as the cutoff criteria were considered statistically significant. The chemical structure of berberine was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and inputted into the SwissTargetPrediction database (https://www.swisstargetprediction.ch/) to obtain berberine-related genes [28]. Moreover, we searched the keyword “berberine” in the comparative toxicogenomics database (CTD) (https://ctdbase.org/) to collect berberine-related genes. Then, the berberine-associated targets were obtained by combining these targets derived from the two databases. We used a Venny online tool to visualize the intersection targets of berberine-related targets and DEGs. The volcano map and heat map of DEGs were visualized using a bioinformatics platform. In addition, the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of berberine were obtained from the pkCSM (https://biosig.unimelb.edu.au/).
2.2. Construction of the Integrated Network in Antipulpitis Targets of Berberine
The shared targets of berberine and pulpitis were imported to the String database (https://www.string-db.org/) with a confidence score greater than 0.9 to generate the protein-protein interaction (PPI) network construction [29]. Then, the PPI network was visualized using Cytoscape software, and the CytoHubba plugin was used to screen hub genes based on 10 algorithms [30]. Besides, the common genes of berberine and DEGs were grouped into two clusters using the molecular complex detection (MCODE) algorithm in Metascape [31].
2.3. Molecular Docking Validation
We performed molecular docking to interpret the potential binding mode of berberine to a macromolecular receptor. The hub gene (PTGS2) was selected for molecular docking analysis. The 3D conformations of proteins with a crystal resolution <3 Å, as measured by X crystal diffraction, were chosen. The 3D structure of the PTGS2 protein (PDB ID: 5IKR with resolution 2.342 Å) was downloaded from the PDB (https://www.rcsb.org) database [32]. The PyMOL software package was applied to remove the solvent and organic of the protein conformations. For the processing of ligand molecules, we applied ChemDraw 19.0 software to draw the molecular structure of the small molecule compound, imported the format files into Chem3D 19.0 software, performed the program to optimise the energy, and saved them in the PDB format. The PDB format file was then imported into AutoDockTools 1.5.6 software, which was applied to adjust the charge of the ligand and saved as a PDBQT file. Then, AutoDock software (1.5.6) was used to perform molecular docking analysis. The detailed procedure for molecular docking is as follows: the PDBQT file of the receptor macromolecule and ligand was opened in AutoDockTools 1.5.6 software. To identify the active binding site for molecular docking, we utilized the coordinates of the cocrystallized ligand. Based on the active pocket of the target protein, we then set the grid-box coordinates and size. The grid-box parameters were set as follows: spacing of 0.503 Å, x-dimension of 80 points, y-dimension of 80 points, and z-dimension of 80 points. Docking was performed with the Vina force field, using the Lamarckian GA (4.2) algorithm. In this study, a total of 100 GA runs were conducted while keeping other docking criteria fixed at default settings. The result of molecular docking was visualized by using PyMOL software.
2.4. Enrichment Analysis of Crucial Targets
The Metascape online platform was used to perform (gene ontology) GO enrichment analyses including biological process (BP), cellular component (CC), and molecular function (MF) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Based on p < 0.05 and a high count, the top 10 BP, CC, MF, and KEGG pathways were chosen to visualize circle, bubble, and heat graphs using the bioinformatics platform (https://www.bioinformatics.com.cn/). Besides, the Gene Set Enrichment Analysis (GSEA) was used to identify the enriched pathways in patients with pulpitis; p.adjust <0.05 and false discovery rate (FDR) <0.25 were considered as the inclusion criteria.
2.5. Human Dental Pulp Stem Cell Culture and Treatment
Human dental pulp stem cells were incubated in alpha modification of Eagle’s medium containing 10 U/mL penicillin, 10 mg/mL streptomycin, and 20% fetal bovine serum and maintained in an incubator supplemented with 5% CO2 at 37°C. Berberine (purity >98%) and LPS (Escherichia coli) were obtained from Sigma-Aldrich and dissolved in DMSO. Cells from the sixth passage were used in the next experiments. Human dental pulp stem cells were treated or not treated with berberine (10 or 20 μM) in the presence of LPS (1 μg/ml) for 24 h. The pulpitis model induced by LPS was established based on a previous study [33].
2.6. Determination of Proinflammatory Cytokines
The cell culture media were collected and centrifuged at 1000 × g for 20 min. Then, the supernatant was collected, and the level of IL-6 was measured using ELISA kits as recommended by the manufacturer’s protocols (Elabscience Biotechnology, Wuhan, China).
2.7. Quantitative Polymerase Chain Reaction (qPCR)
The total RNA was extracted from human dental pulp stem cells using TRIzol (Invitrogen, USA) based on the protocols recommended by the manufacturer. 1 μg of total RNA was reverse-transcribed by a First Strand cDNA Synthesis Kit (Invitrogen, USA). qPCR was carried out using the SYBR Green PCR master mix reagent (Takara) in a thermocycler instrument (Applied Biosystems, USA). Relative quantification of target gene expression was measured by the 2−ΔΔCt method, and GAPDH was used as the reference gene.
2.8. Statistical Analysis
The means ± SD were used to present all data. GraphPad Prism software was employed for conducting all statistical analyses. One-way analysis of variance followed by Dunnett’s multiple comparison test was utilized to compare the statistical differences between two groups. The statistical difference was considered significant at a p value of less than 0.05.
3. Results
3.1. Prediction of ADMET Properties of Berberine
In the present study, we used pkCSM to estimate the ADMET properties of berberine. As shown in Table 1, the human intestinal absorption of berberine is 97.14, indicating that it showed good absorption. The blood-brain barrier (BBB) permeability (LogBB) was used to assess the brain distribution of berberine. The result of BBB permeability is 0.198, indicating that berberine could permeate the BBB. In addition, berberine may not inhibit the activity of CYP2C19 and CYP2C9 enzymes, indicating that it may not impact the clearance and metabolism of the drug substrates of CYP2C19 and CYP2C9. The renal total clearance is 1.27, and berberine is not likely to be organic cation transporter 2 (OCT2) substrate. Furthermore, the toxicity data indicated that berberine exhibited a promising safety profile.
| Property | Model name | Predicted value | Unit |
|---|---|---|---|
| Absorption | Intestinal absorption (human) | 97.147 | Numeric (% absorbed) |
| Distribution | BBB permeability | 0.198 | Numeric (log BB) |
| Metabolism | CYP2C19 inhibitor | No | Categorical (yes/no) |
| CYP2C9 inhibitor | No | Categorical (yes/no) | |
| Excretion | Total clearance | 1.27 | Numeric (log ml/min/kg) |
| Renal OCT2 substrate | No | Categorical (yes/no) | |
| Toxicity | Oral rat acute toxicity (LD50) | 2.571 | Numeric (mol/kg) |
| Oral rat chronic toxicity (LOAEL) | 1.89 | Numeric (log mg/kg·bw/day) | |
| Skin sensitisation | No | Categorical (yes/no) | |
| Max. tolerated dose (human) | 0.144 | Numeric (log mg/kg/day) | |
3.2. Prediction of Potential Targets Involved in the Antipulpitis Effect of Berberine
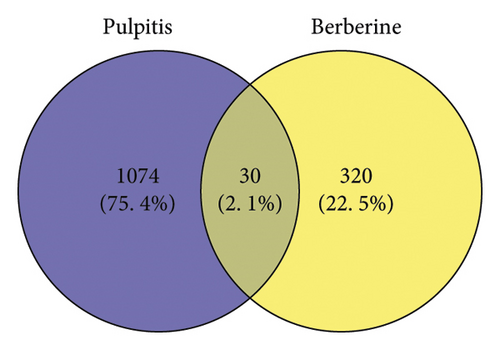
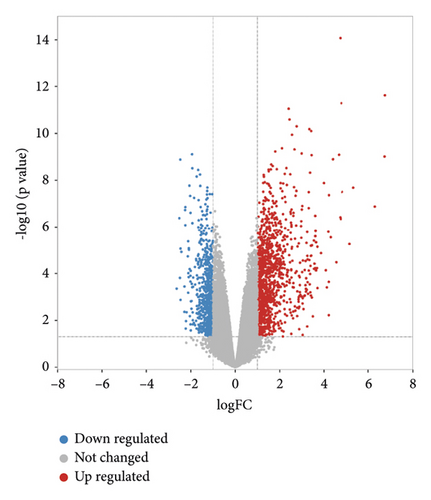
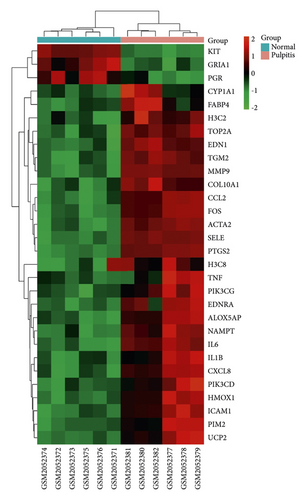
As shown in Figure 2(a), we identified 350 berberine-related genes from CTD and SwissTargetPrediction databases. A total of 1104 pulpitis-related DGEs were collected from the GSE77459 dataset, which included 325 downregulated genes and 779 upregulated genes, and the volcano plot of DEGs is presented in Figure 2(b). Then, 30 intersection genes were chosen as potential targets of berberine in the treatment of pulpitis. The heat diagram of 30 overlapping targets is presented in Figure 2(c).
3.3. Identification of Hub Genes and Molecular Docking Study
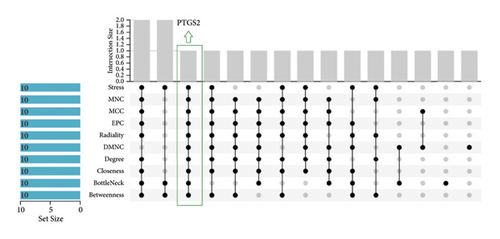
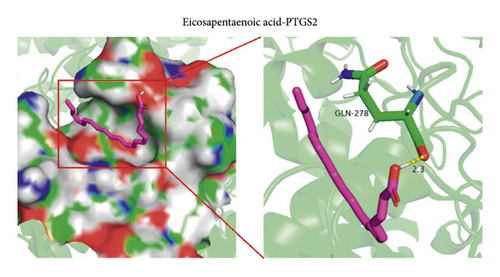
PTGS2 was identified as a hub gene based on the overlapped parameters of the top 10 genes in 10 algorithms (including stress, MNC, MCC, EPC, radiality, DMNC, degree, closeness, bottleneck, and betweenness) (Figure 3(a)). Then, molecular docking analysis was performed on berberine with the PTGS2 gene to validate the potential target of berberine against pulpitis. The results showed that the berberine binds to PTGS2 with a binding energy of −9.0 kcal/mol. Eicosapentaenoic acid (EA) is a typical PTGS2 enzyme inhibitor, so it was used as a control compound in the present study. EA binds to PTGS2 with a binding energy of −5.79 kcal/mol. Berberine formed hydrophobic interactions with TRP-387 and ASN-382 residues; EA formed hydrophobic interactions with GLN-278 residue. These findings implied that berberine showed a strong binding affinity for PTGS2 protein. The binding modes and interactions between the PTGS2 receptor and small molecule ligands are presented in Figures 3(b) and 3(c).

3.4. PPI Network from 30 Overlapping Genes
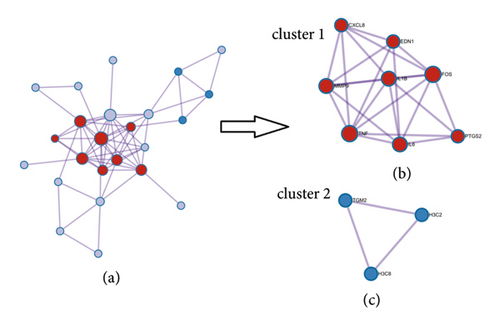
In the present study, 30 overlapping genes were inputted into the Metascape platform to construct the PPI network. As shown in Figure 4(a), this network contained 24 nodes and 63 edges. All targets were ranked in descending order of degree value, and the top 10 genes are presented in Table 1. Besides, the PPI network was grouped into two clusters using the MCODE algorithm. As shown in Figures 4(b) and 4(c), cluster 1 contained several inflammation-related genes, such as CXCL8, TNF, IL1B, and IL-6. Cluster 2 contained three genes (TGM2, H3C2, and H3C8).
3.5. Enrichment Analyses of Crucial Targets from Cluster 1
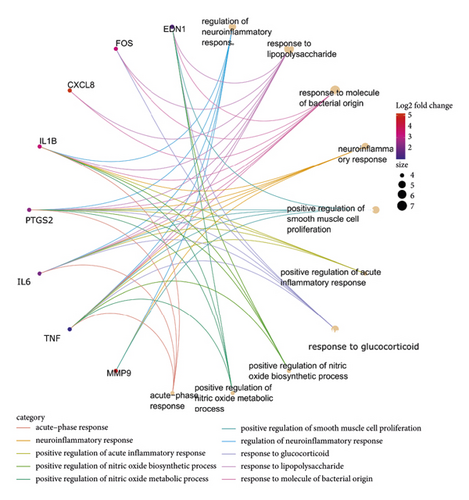
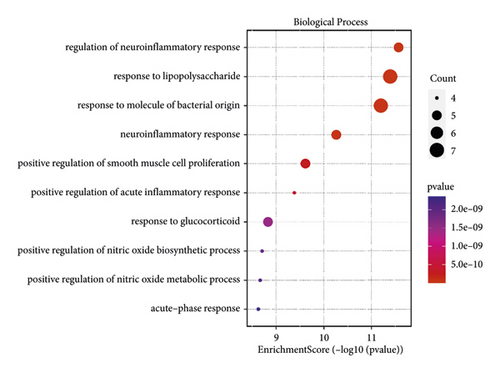
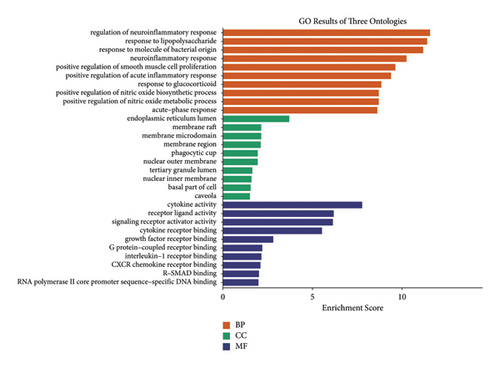
The bioinformatics platform was used for generating enriched terms in the BP, CC, and MF categories. The top 10 biological processes are shown in Figure 5 and Table 2. GO-BP enrichment analysis revealed that the genes of cluster 1 were implicated in multiple biological processes, including regulation of neuroinflammatory response, response to lipopolysaccharide, response to molecule of bacterial origin, neuroinflammatory response, and positive regulation of smooth muscle cell proliferation. CC enrichment analysis indicated that these crucial targets were mainly related to the endoplasmic reticulum lumen, membrane raft, membrane microdomain, and membrane region (Figure 6). MF enrichment analysis revealed that these crucial targets were mainly enriched in cytokine activity, receptor-ligand activity, signaling receptor activator activity, cytokine receptor binding, growth factor receptor binding, and others (Figure 5).
| ID | Description | p value | Genes | Count |
|---|---|---|---|---|
| GO:0150077 | Regulation of neuroinflammatory response | 2.69E − 12 | MMP9, TNF, IL-6, PTGS2, IL1B | 5 |
| GO:0032496 | Response to lipopolysaccharide | 4.04E − 12 | CXCL8, TNF, IL-6, PTGS2, FOS, IL1B, EDN1 | 7 |
| GO:0002237 | Response to molecule of bacterial origin | 6.33E − 12 | CXCL8, TNF, IL-6, PTGS2, FOS, IL1B, EDN1 | 7 |
| GO:0150076 | Neuroinflammatory response | 5.51E − 11 | MMP9, TNF, IL-6, PTGS2, IL1B | 5 |
| GO:0048661 | Positive regulation of smooth muscle cell proliferation | 2.43E − 10 | MMP9, TNF, IL-6, PTGS2, EDN1 | 5 |
| GO:0002675 | Positive regulation of acute inflammatory response | 4.15E − 10 | TNF, IL-6, PTGS2, IL1B | 4 |
| GO:0051384 | Response to glucocorticoid | 1.47E − 09 | TNF, IL-6, PTGS2, FOS, EDN1 | 5 |
| GO:0045429 | Positive regulation of nitric oxide biosynthetic process | 1.96E − 09 | TNF, PTGS2, IL1B, EDN1 | 4 |
| GO:1904407 | Positive regulation of nitric oxide metabolic process | 2.15E − 09 | TNF, PTGS2, IL1B, EDN1 | 4 |
| GO:0006953 | Acute-phase response | 2.35E − 09 | TNF, IL-6, PTGS2, IL1B | 4 |
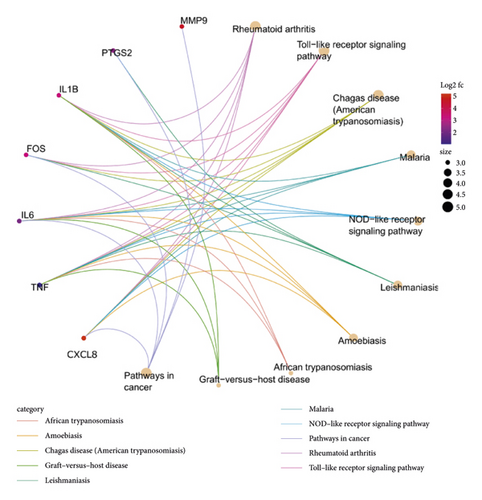
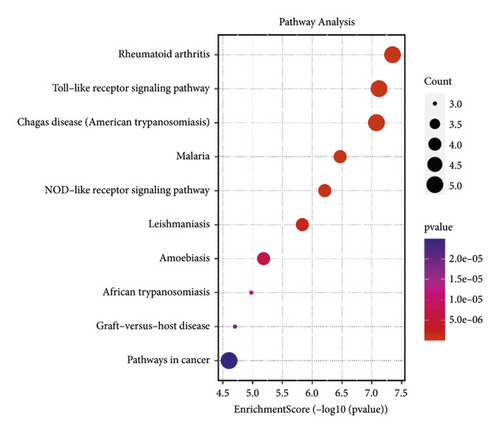
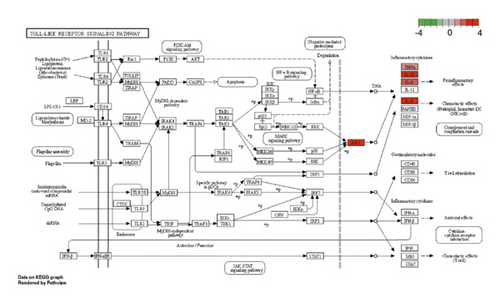
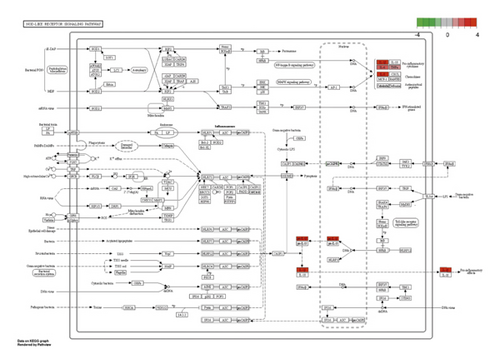
Besides, the top 10 KEGG pathways are shown in Figure 7 and Table 3. KEGG enrichment analysis revealed that these crucial genes were involved in multiple pathways, including rheumatoid arthritis, toll-like receptor signaling pathway (Figure 8), Chagas disease (American trypanosomiasis), malaria, NOD-like receptor signaling pathway (Figure 9), and leishmaniasis.
| ID | Description | p value | GeneID | Count |
|---|---|---|---|---|
| hsa05323 | Rheumatoid arthritis | 4.48E − 08 | CXCL8, TNF, IL-6, FOS, IL1B | 5 |
| hsa04620 | Toll-like receptor signaling pathway | 7.56E − 08 | CXCL8, TNF, IL-6, FOS, IL1B | 5 |
| hsa05142 | Chagas disease (American trypanosomiasis) | 8.34E − 08 | CXCL8, TNF, IL-6, FOS, IL1B | 5 |
| hsa05144 | Malaria | 3.39E − 07 | CXCL8, TNF, IL-6, IL1B | 4 |
| hsa04621 | NOD-like receptor signaling pathway | 6.15E − 07 | CXCL8, TNF, IL-6, IL1B | 4 |
| hsa05140 | Leishmaniasis | 1.46E − 06 | TNF, PTGS2, FOS, IL1B | 4 |
| hsa05146 | Amoebiasis | 6.55E − 06 | CXCL8, TNF, IL-6, IL1B | 4 |
| hsa05143 | African trypanosomiasis | 1.05E − 05 | TNF, IL-6, IL1B | 3 |
| hsa05332 | Graft-versus-host disease | 1.98E − 05 | TNF, IL-6, IL1B | 3 |
| hsa05200 | Pathways in cancer | 2.49E − 05 | CXCL8, MMP9, IL-6, PTGS2, FOS | 5 |
3.6. GSEA
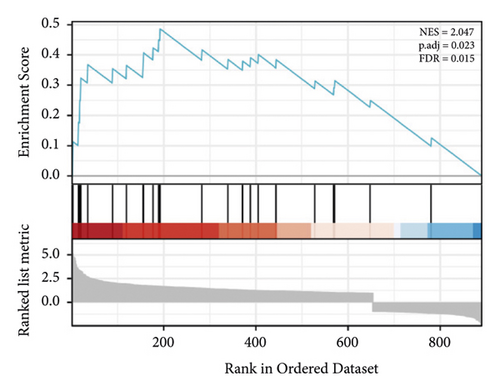
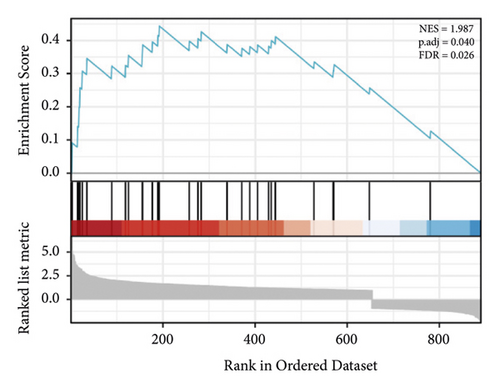
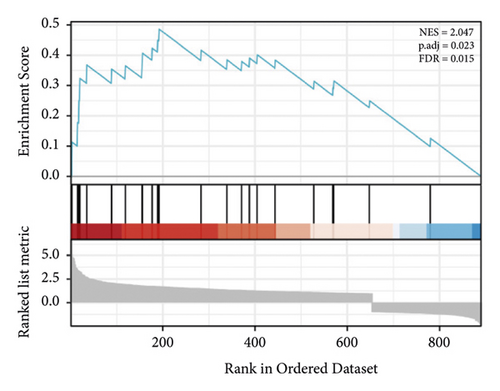
GSEA of DEGs was carried out to further assess the related pathways in the development of pulpitis. As shown in Figure 10, our results showed that the DEGs were significantly enriched in the toll-like receptor signaling pathway and regulation of the toll-like receptor signaling pathway, which is consistent with the KEGG results.
3.7. Validation of Network Pharmacology Results by Cell Experiments
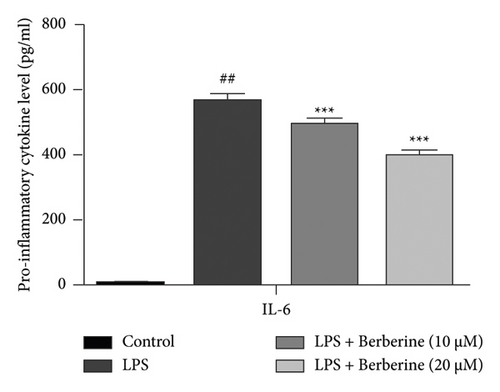
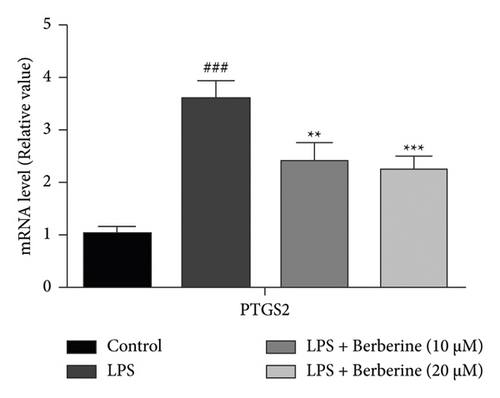
As shown in Figure 11(a), compared with the control group, the level of IL-6 was significantly increased, which was repressed by treatment with berberine. In addition, compared with the control group, the expression level of PTGS2 was significantly upregulated, which was downregulated by treatment with berberine (Figure 11(b)). These findings revealed that the activated inflammatory response in LPS stimulation of dental pulp stem cells was mitigated by berberine.
4. Discussion
Pulpitis is an oral disease mainly caused by a pathogenic microorganism infection of the pulp space, but its exact pathologic mechanisms remain unknown [6]. Berberine has been reported to promote odontoblast differentiation in human dental pulp cells, suggesting its potential therapeutic effects against pulpitis [21]. However, the therapeutic mechanism of berberine in the treatment of pulpitis remains unknown. In the field of natural product development and utilization, network pharmacology is increasingly being applied to repurpose approved drugs and to develop new therapeutic drugs [34]. In the present study, network pharmacology was applied to identify the pharmacological mechanism of berberine against pulpitis.
First, we identified 30 potential targets of berberine by combining several databases’ predication. Subsequently, a core subnetwork including 8 potential targets was identified by MCODE analysis. GO and KEGG enrichment analyses indicated that berberine exerts an antipulpitis effect via regulating multiple pathways, including regulation of neuroinflammatory response, toll-like receptor signaling pathway, and NOD-like receptor signaling pathway. In addition, the findings of molecular docking revealed that there existed hydrogen bondings between the berberine and the PTGS2 protein, which further confirmed our results from bioinformatics analysis.
Inflammation is one of the main drivers of various diseases [35]. Pulpitis is a chronic inflammatory disease, characterized by the aggregation of proinflammatory mediators, including tumor necrosis factor-α (TNF-α), IL-6, C-X-C motif chemokine ligand 8 (CXCL8), IL-1, and IL-1β [36, 37]. It has been demonstrated that the overexpression of proinflammatory cytokines could affect the differentiation of odontoblasts, thus influencing the formation of restorative dentin [8, 38]. Besides, these cytokines were considered diagnostic markers of irreversible pulpitis [39]. The activation of the toll-like receptor 2 (TLR2) signaling pathway is involved in the upregulation of IL-6, CXCL8, and TLR2 in odontoblast-like cells stimulated with lipoteichoic acid [40]. Berberine has been revealed to exert its antimicrobial and anti-inflammatory activities in the treatment of chronic diseases [41]. Importantly, berberine was reported to promote odontoblast differentiation via activating the Wnt/β-catenin pathway [20]. Interestingly, these reports supported our putative therapeutic effects that berberine could regulate inflammatory responses.
The molecular docking method is employed to examine the molecular interactions between small molecules and the binding site of the target protein. Molecular docking was performed to assess the binding modes and interactions between the core target protein (PTGS2) and berberine. The binding energy was −9.0 kcal/mol, and berberine formed hydrophobic interactions with TRP-387 and ASN-382 residues. These findings implied that berberine showed a strong binding affinity for PTGS2 protein. PTGS2, also known as COX-2, plays a vital role in the regulation of inflammation [42]. The positioning of the p-methoxy group in naproxen is directed towards the apex of the COX active site, establishing van der Waals interactions with TRP-387 and TYR-385 [43]. A recent study showed that berberine had anti-ischemia-reperfusion effects via inhibiting the PTGS2/MAPK pathway [44]. Berberine exerted the antiangiogenic effect via the inhibition of COX-2 expression and proinflammatory cytokines [45]. In line with those previous results, our findings suggested that berberine might contribute to the therapeutic effect against pulpitis by inhibiting the activity of PTGS2. However, cell or animal studies are needed to confirm the results.
Toll-like receptors (TLRs) are important receptors regulating the innate immune response. Activation of TLRs is mainly related to the induction of cell-mediated immune responses and inflammatory processes via the generation of immune messenger molecules [46]. It has been demonstrated that the expression levels of toll-like receptor 4 (TLR4) and TLR2 are related to the early stage of pulpitis induced by pathogenic microorganism infection [11]. Activation of a TLR4 signaling pathway in trigeminal ganglion has been reported to be involved in the nociception associated with acute pulpitis in the rat [10]. The neural TLR2 plays an important role in pulpitis and suggesting inactivation of the TLRs signaling pathway may be a promising therapy in the treatment of pulpal inflammation [47, 48]. The anti-inflammatory effects of S14G-humanin were reported to protect LPS-stimulated human dental pulp cells via inactivating the TLR4/MyD88/NF-κB signaling pathway [49]. Phoenixin-20 was shown to inhibit LPS-induced inflammation via downregulation of TLR4 expression in dental pulp cells [50]. NOD-like receptors represent a wide range of complex signal regulators that are implicated in cellular senescence, tumorigenesis, and inflammatory responses [51, 52]. NLRP3 is one of the most characteristic members of the NOD-like receptor family. Therefore, TLRs and NOD-like receptors might be potential therapeutic targets for the prevention and treatment of oral diseases, such as pulpitis [53]. Berberine treatment downregulated TLR4 expression in necrotizing enterocolitis [54]. Berberine attenuated intestinal mucosal barrier injury through the TLRs pathway [19]. Berberine exerted an inhibitory effect on NLRP3 inflammasome activation in nonalcoholic steatohepatitis [55]. Berberine exhibited potential therapeutic effects via targeting the endothelial NLRP3 inflammasome in inflammatory vascular damage [56]. In line with these reports, our results of bioinformatics analysis indicated that berberine may exert a therapeutic effect on pulpitis via the inactivation of TLRs and NOD-like receptor signaling pathways.
The main limitation of the current study is that the results of the analyses were based on existing public databases. Therefore, our conclusions need to be verified using cell and animal experiments. In future research, we will explore the following topics: the cellular and molecular mechanism of berberine against pulpitis, the appropriate dose of berberine to improve pulpitis, and whether it is suitable for long-term maintenance treatment of pulpitis. We hope that our findings will shed light on the potential mechanism of action of berberine against pulpitis and provide a theoretical basis for the application of berberine in clinical practice.
5. Conclusion
In summary, this study employed bioinformatics analysis to explore the potential mechanism of berberine in the treatment of pulpitis. Our findings showed that berberine may play a significant role in the treatment of pulpitis by influencing key biological targets, such as PTGS2. Additionally, our molecular docking analysis indicated that berberine has the potential to effectively interact with this target. Berberine also could exert antipulpitis effects via the regulation of TLRs and NOD-like receptor signaling pathways. These results provided initial insights into the working of berberine and opened up new possibilities for future research on its mechanism in pulpitis treatment. Moreover, these findings lay the groundwork for the clinical application of berberine in pulpitis treatment. However, further pharmacological experiments are required in subsequent stages to validate the primary regulatory target of berberine in pulpitis.
Abbreviations
-
- GSEA:
-
- Gene Set Enrichment Analysis
-
- GO:
-
- Gene Ontology
-
- KEGG:
-
- Kyoto Encyclopedia of Genes and Genomes
-
- PTGS2:
-
- Prostaglandin-endoperoxide synthase 2
-
- NLRP3:
-
- NLR family pyrin domain containing 3
-
- LPS:
-
- Lipopolysaccharide
-
- IL-6:
-
- Interleukin 6
-
- CCL11:
-
- C-C motif chemokine ligand 11
-
- STAT6:
-
- Signal transducer and activator of transcription 6
-
- STZ:
-
- Streptozotocin
-
- GEO:
-
- Gene Expression Omnibus
-
- DEGs:
-
- Differentially expressed genes
-
- CTD:
-
- Comparative toxicogenomics database
-
- PPI:
-
- Protein-protein interaction
-
- MCODE:
-
- Molecular complex detection
-
- BP:
-
- Biological process
-
- CC:
-
- Cellular component
-
- MF:
-
- Molecular function
-
- FDR:
-
- False discovery rate
-
- TNF-α:
-
- Tumor necrosis factor-α
-
- CXCL8:
-
- C-X-C motif chemokine ligand 8
-
- TLR2:
-
- Toll-like receptor 2
-
- TLRs:
-
- Toll-like receptors
-
- TLR4:
-
- Toll-like receptor 4.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors’ Contributions
Bingchang Xin and Jiaying Wang designed the project plan and wrote the main manuscript text. Juan Zhou, Ran Li, and Ke Sun prepared figures and performed bioinformatics analysis. Jia Song performed cell experiments. Jin Zhang edited the article. All the authors reviewed the manuscript.
Acknowledgments
This work was sponsored by Qingdao Key Health Discipline Development Fund (2022-2024), Qingdao Clinical Research Center for Oral Diseases (22-3-7-lczx-7-nsh), Qingdao Outstanding Health Professional Development Fund, and Qingdao Medical and Health Research Project (2021-WJZD138).
Open Research
Data Availability
All data can be obtained from the website we provide and are available from the corresponding author upon request. The datasets analyzed during the present study are available in the Gene Expression Omnibus (GEO) database repository (GSE77459, https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE77459), which was used to identify the differentially expressed genes.




