Transcriptome Profiling of Different State Callus Induced from Immature Embryo in Maize
Abstract
Embryogenic and regenerable tissue cultures are widely used in plant transformation. To dissect the molecular mechanism of embryogenesis, we used inbred line A188 as the material; the immature embryo of kernels (15 day after pollination, 15DAP) was isolated and cultured in inducing medium and subjected to RNA-Seq. The results revealed that 5,076 differentially expressed genes (DEGs) were involved in morphological and histological changes and endogenous indole-3-acetic acid (IAA) alteration. Functional analysis showed that the DEGs were related to metabolic pathways and biosynthesis of secondary metabolites. In particular, ARF16 and ARF8 genes of auxin response factors (ARF) were upregulated from EC to IDC and EC to IRC. Meanwhile, BBM2, SERK1, and SERK2 genes of the embryogenic pathway were upregulated, and WIP2 and ESR genes of the wound-inducible were upregulated from EC to IDC and EC to IRC. These changes can improve conversion efficiency from EC to IRC, which is important for elucidating the underlying molecular mechanisms of callus formation.
1. Introduction
Maize is the main feed and food crop in the world and is very important for humans and livestock. In recent years, maize has changed from a single food crop and feed crop to a cash crop and industrial raw material. The genetic improvement of food crops, including conventional technology and biotechnology, is important for the needs of 8.3 billion people [1, 2]. Currently, most of the maize genetic engineering systems still greatly depend on callus induction from young embryos (called embryonic callus), which is the prerequisite for the genetic transformation of maize inbred lines [3]. The genetic transformation of maize mainly depends on the in vitro callus formation of young embryos, which is the process of plant cells to regain totipotency and is an important process of plant cell fate transformation. Callus induction from young embryos is strongly genotype-dependent; only specific genotypes have embryogenic competence in tissue culture and are able to develop callus [4]. The study shows that the callus induction rate of maize is related to the genotype of maize. Many maize inbred lines barely induce callus formation or the callus induction rate is very low [5–7]. Therefore, this genotype-dependent culture limits the application of crop improvement [8].
Callus formation is an important factor affecting maize genetic transformation efficiency, and the regulatory molecular mechanisms of embryogenesis remain unclear. It is commonly believed that embryogenesis mainly involves lots of cell reprogramming and signal activation [9, 10]. The efficiency of embryogenesis dedifferentiation is a quantitative trait, which is controlled by additive gene effects. Pan et al. mapped five QTLs (quantitative trait loci) on chromosomes 1, 3, 7, and 8 by composite interval mapping, which explained 5.25–23.4% of the phenotypic variation [11, 12].
Inbred line A188 has been widely used in genetic improvement due to its high embryogenic efficiency and regeneration ability [13–17]. To reveal the molecular mechanisms of callus induced and/or regeneration, immature embryo of A188 was cultured in the initiation and regeneration medium, and the transcriptome on callus (at different states) of the A188 was analyzed by RNA-sequencing (RNA-Seq). We expect to find key genes of embryo-derived embryonic callus and provide a foundation for crop tissue culture.
2. Materials and Methods
2.1. Plant Material and Tissue Culture
The maize inbred line A188 was grown in an experimental field at the Huazhong Agricultural University (Wuhan, China). Ears were harvested at the 15th DAP (days after pollination), and the immature embryo was isolated and cultured in N6 medium (Table S1) at 28°C for 60 days [18, 19]. Callus can be divided into three types according to the characteristics of color, hardness, and granulation. The callus, with bright color and similar dryness, was embryogenic. The callus without complete dedifferentiation showed that most radicles and buds were present, and there were no small granular thin-wall callus cells. The other kind of callus was dark brown and could not be cultured in bands. The embryogenic callus was selected for further culture, and a large number of embryogenic calluses were transferred to the differentiation medium for differentiation. The culture conditions were 28°C and 16/8 h photoperiod, and regeneration seedlings were obtained after 30 days.
2.2. RNA-Seq Library Construction and Sequencing
Total RNA was isolated from callus after morphological classification using a plant RNA kit (OMEGA) [20]. RNA quality was checked by the Bioanalyzer (2100, Agilent Technologies, Palo Alto, CA, USA). The mRNA was enriched using oligo (dT) magnetic beads [21]. The target mRNA was reversely transcribed to cDNA, phosphorylated at the 5′ end, adhered to “A” base at the 3′ end, and ligated with adapters [22, 23]. The products were amplified by two specific primers and prepared using the Illumina TruSeq Stranded Total RNA HT Library Preparation Kit (Illumina). Transcriptional sequencing was performed on the Illumina HiSeqTM 2500 by Shanghai OE Biotech Co., Ltd.
2.3. Sequencing Analysis and Differential Expression Analysis
The B73 reference genomic and annotated files were used as the database [24]. The software HTSEQ-count was used to the reads of each gene [25, 26], and the software Cufflinks was used to calculate the FPKM (fragments per kilobase per million mapped fragments) values [27].
The reads containing ploy-N and the low-quality reads of raw data were removed using Trimmomatic [28, 29]. The resulting clean reads were mapped to the B73 reference genome [26]. DEGs were identified using the DESeq according to the following criteria-fold change >2 and corrected P value < 0.05 [30–36]. All DEGs were mapped to each term in the gene ontology database (http://www.geneontology.org/), and Gene Ontology (GO) enrichment analysis was performed using WEGO 2.0 [37–39]. Pathway enrichment analysis of DEGs was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) [40]. The GO terms and pathways with P value ≤ 0.05 and FDR≤0.01 were considered to be significantly enriched in DEGs [23].
2.4. Real-Time PCR Validation
Eight DEGs were selected for qRT-PCR verification. The primers were designed using Primer Premier 5.0. Total RNA was reversely transcribed into cDNA using a cDNA synthesis kit (Thermo Fisher Science). The qRT-PCR was performed using the CFX96 Real-time system [41, 42], according to the method used by Petersen [43], and actin was used as the internal reference. The standard error among the three biological replicates was calculated.
2.5. Accession Numbers
The raw data of RNA-Seq have been submitted to https://ngdc.cncb.ac.cn/:PRJCA009242. Temporary Submission ID: subPRO013562; PRJCA009242 records will be accessible upon publication on the indicated release date.
3. Results
3.1. Callus Culture and Phenotype Identification
According to transformation of morphological feature, after three cycles of induction culture, callus was produced in most of the immature embryo, and callus obtained can be divided into three categories, according to the morphological characteristics (Figure 1). A part of the callus which only expanded, accompanied by a large number of nonremovable root bud structures, was named incomplete dedifferentiation callus (IDC). However, in the materials that produced callus, two kinds of callus exist simultaneously: callus that was yellow, loose, and small granular, and the other was the browning dead, which cannot be further cultured, which were named embryonic callus (EC) and browning dead callus (BDC), respectively. Regeneration plants were redifferentiated from the embryogenic callus, and incipient callus was produced. After 10 days of regeneration, incipient redifferentiation callus (IRC) was produced on the regeneration medium.




3.2. Statistical of Transcriptome Data
Thirty calluses with the same growth state were selected from each material, and RNA was extracted after mixing. The callus in each state was repeated twice. A total of eight libraries was established from four turntable callus for transcriptome sequencing analysis. Overall, 32.76 G of data was obtained in total, with Q30 bases distributed in 91.96–92.35%, and the average GC content was 54.41% (Table S2). The genome alignment ratio of each sample was 88.80–90.23%; after removing the low-quality tags, 35066073 (84.84%), 34749515 (83.76%), 34788501 (84.58%), 35596745 (86.45%), 35315481 (85.46%), 35923796 (86.50%), 35012769 (84.70%), and 35563113 (83.11%) clean tags were left. According to the comparison between the sequences and the exons of the reference genome, 60.78–62.77% of the sequences were completely compared to the exons, and 20.44–24.01% of the single-ended forces were compared to the exons. Reads mapped in proper pairs showed 78.70% (EC-1), 77.47% (EC-2), 77.60% (IDC-1), 79.91% (IDC-2), 78.82% (BDC-1), 80.25% (BDC-2), 78.08% (IRC-1), and 76.16% (IRC-2), separately.
3.3. Gene Expression Level Analysis among Different State Callus
According to the differences in gene expression number and gene expression value distribution in samples (Figure 2(a)), the expression value (FPKM) was divided into four intervals (Table S3). There are 10,630 genes with more than 0.5–1(FPKM), 1,701 genes with 1–10 (FPKM), and 14,499 genes with FPKM≥10 in EC1. Meanwhile, in EC2, there were 10,409 genes with expression levels between 0.5 and 1, 1,768 genes with expression levels between 1 and 10, and 15,342 genes with expression levels greater than 10, using the FPKM value as the standard. In IDC callus, the number of genes in the expression range FPKM <0.5, 0.5 < FPKM <1, 1 < FPKM <10, and FPKM≥10 was 9516 and 9441, 11123 and 10328, 1835 and 1847, and 14457 and 15315, respectively. The number of genes expressed in BDC callus was between 8662 and 9639 (FPKM <0.5), 10582 and 9856 (0.5 < FPKM <1), 1984 and 1767 (1 < FPKM <10), and 15703 and 15669 (FPKM≥10), respectively. In the differentiated callus (IRC), with the FPKM value as parameter, 9831 and 8996 genes were expressed at less than 0.5, respectively, in the two groups of materials. There were 11094 genes and 11852 genes with 0.5–1 expression levels in the two groups of materials, respectively. The number of genes with expression levels between 1 and 10 (FPKM) was between 1797 and 1835, respectively. The number of genes with expression levels greater than or equal to 10 (FPKM) was between 14209 and 14284, respectively, in two replicates.
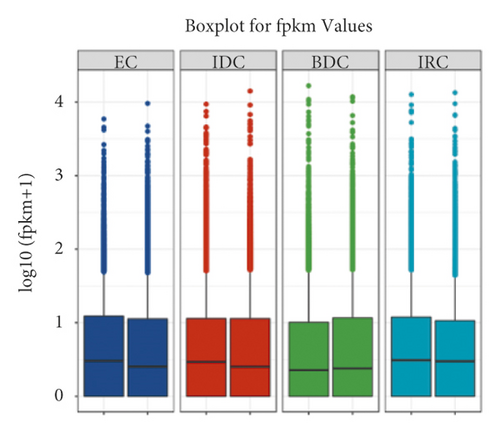
The function of highest expressed genes involves serine-type endopeptidase inhibitor activity, RNA binding, ATP binding, polysaccharide catabolic process, and DNA binding. sci1 is annotated as a response to wounding by-product subtilisin-chymotrypsin inhibitor homolog 1. Another gene related to plant defense that was among the highest expressed genes was LOC100283098, which was with an FPKM value of 11648.33 at BDC, which decreased over 2-fold to IDC (5769.86) and IRC (5122.97), respectively. The highest expressed was about 4-fold to EC with an FPKM value of 2387.30. Finally, we obtained 28,076, 28,113, 27,776, and 28,707 unique labels for four states of callus, EC, IDC, BDC, and IRC, respectively.
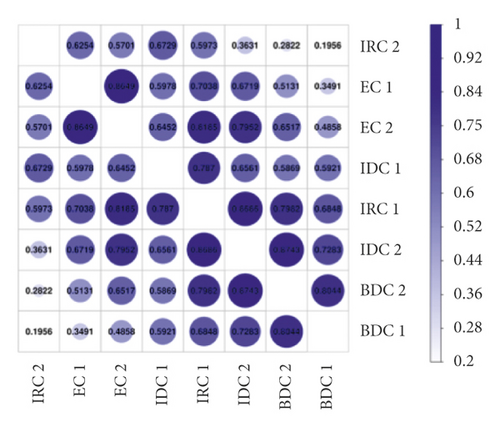
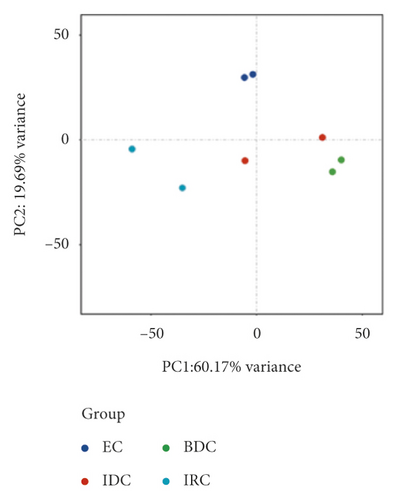
Meanwhile, transcripts in the different samples were analyzed to perform a correlation analysis (Figure 2(b)). The correlation coefficients between two replicates of the same material were 0.8649 (EC 1 vs EC 2), 0.6561 (IDC 1 vs IDC 2), 0.8044 (BDC 1 vs BDC 2), and 0.5973 (IRC 1 vs IRC 2), respectively. The PCA results of the 8 samples showed a clear separation between the different stages of callus (Figure 2(c)). Additionally, the replicates of each treatment clustered together.
The trend of gene expression, the stability, and data reliability of transcriptome analysis by correlation analysis and PCA analysis were analyzed. Correlation analysis results also showed a similar trend, that is, there was a significant correlation between the same type of tissue samples, while the correlation between different tissues decreased, which was consistent with research expectations and consistent with the basis of this study. Finally, PCA analysis also showed a trend consistent with correlation analysis, with significant clustering among other materials except IDC. In general, the consistent results of the above three analyses indicated that the RNA-Seq data of all samples were credible and the differences between materials were significant, which was suitable for expression analysis of callus in different states.
3.4. Identification of Differentially Expressed Genes (DEGs)
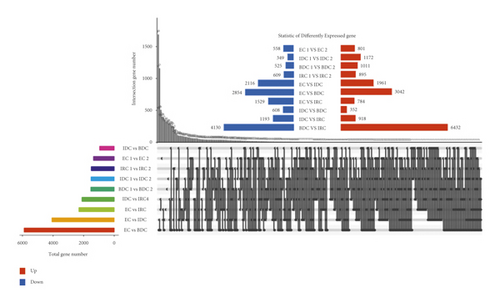
The number of DEGs between each pair of compared groups was analyzed using NOISeq [44, 45]. Venn diagrams show genes expressed consistently and differentially between repeats of different materials (Figure 3). As shown in the figure, the two groups of materials with the most DEGs were IRC and BDC, with 10,562 genes. EC and BDC followed, each with 5896 DEGs. The third largest group was 4077 DEGs in EC and IDC. Then, EC and IRC were with 2,313 DEGs. The least difference was expressed between IDC and BDC (960). The number of DEGs between different groups was 4077 genes between IDC and EC, 5896 genes between BDC and EC, 960 genes between BDC and IDC, and 2313 genes between IRC and EC.
3.5. Functional Analysis of DEGs of Callus Induction
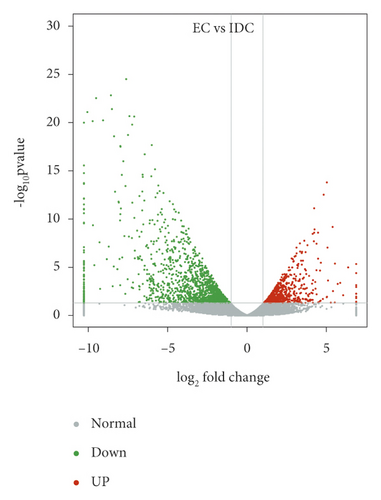
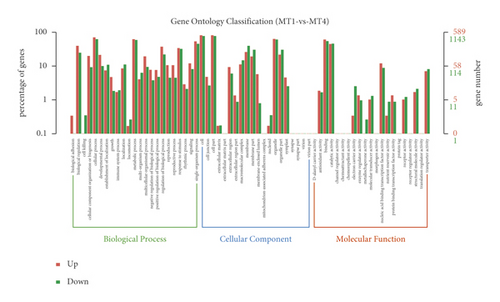
DEGs were analyzed by Geno Ontology (GO) functional classification in IDC and BDC. These DEGs were grouped into the categories of BP (biological process, 23 GOs), CC (cellular component, 20 GOs), and MF (molecular function, 21 GOs) (Figure 4). For induced callus of different states, 1341 upregulated genes (Figure 4(a)) in IDC were significantly enriched in 49 ontologies and 1603 downregulated genes (Figure 4(a)) were significantly enriched in 52 ontologies (Figure 4(d)). The most abundant ontologies include biological regulation (GO:0065007), cellular process (GO:0009987), metabolic process (GO:0008152), and regulation of biological process (GO:0050791), single-organism process (GO:0044702), and binding (GO:0005488). Compared with EC, significantly enriched genes in BDC included 2118 upregulated genes and 2210 downregulated genes (Figure 4(b)), including 51 ontologies and 52 ontologies (Figure 4(d)), respectively. Ontology with a large number of enriched genes was highly consistent with EC vs IDC. Genes significantly differentially expressed between IDC and BDC were enriched in 47 ontologies and 50 ontologies (Figure 4(c) and Figure 4(d)). However, the number of enriched genes was significantly less than that between IDC and BDC and EC, respectively. The largest ontologies of the gene enrichment include the cellular process (GO:0009987), metabolic process (GO:0008152), and single-organism process (GO:0044702).
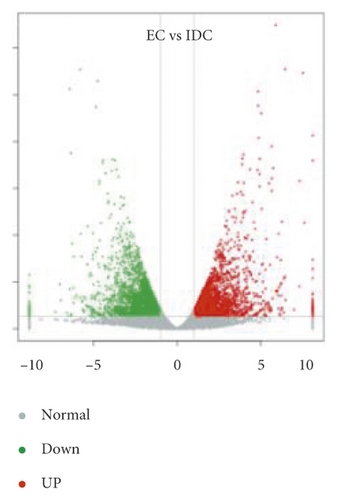
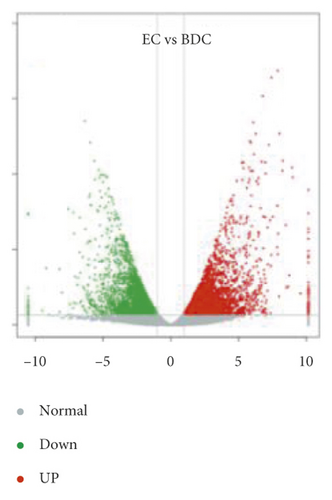
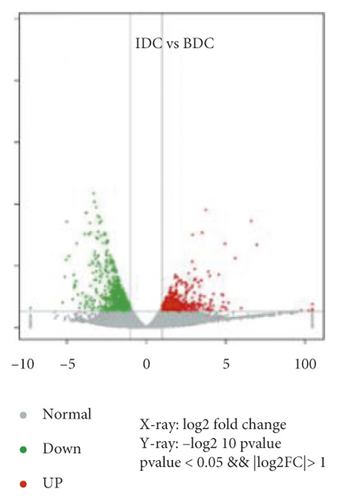

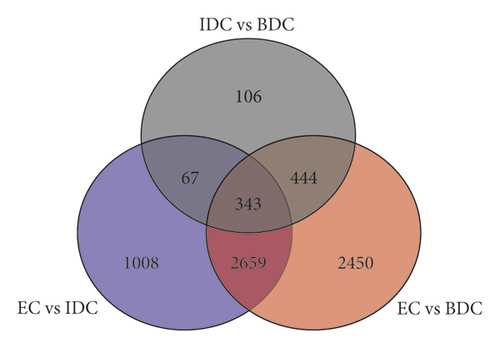
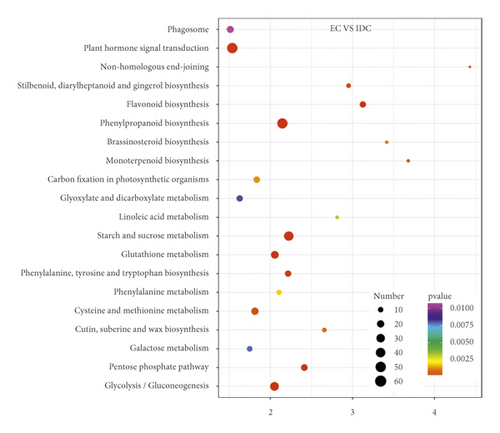
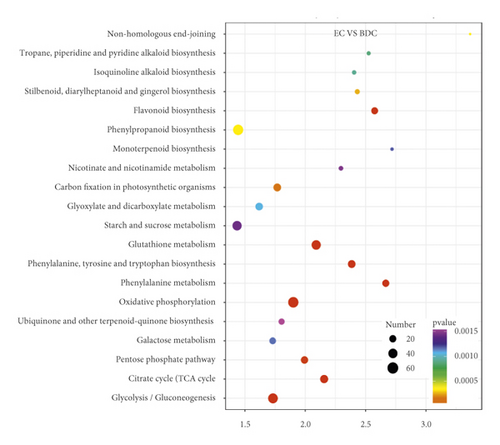
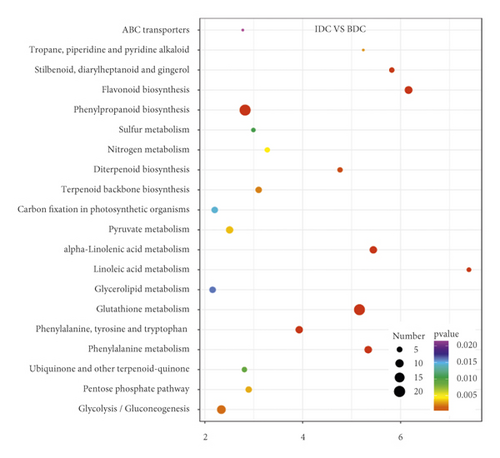
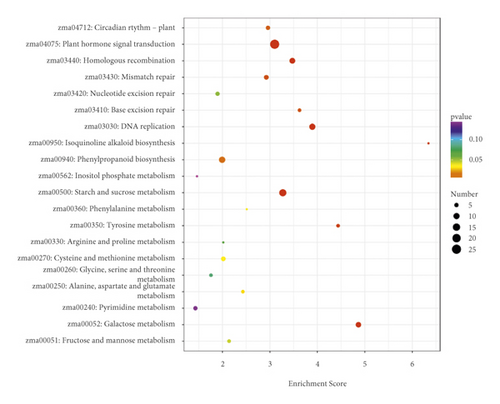
Overlap of DEGs in tissues under three different induced states is shown in Figure 5(a). There are 3002 overlaps of DEGs between EC vs IDC and EC vs BDC and 410 overlaps between EC vs IDC and IDC vs BDC. There were 787 genes that overlapped between EC vs BDC and IDC vs BDC. There were 343 DEGs in all three tissue materials. The expression levels of 343 genes are shown in Figure 5(e) and varied in different materials. In general, most of the genes maintained a low expression level in EC, and there was a certain upregulated expression level in IDC, while most of the genes in BDC showed a significant upregulated expression trend. Pathway analysis showed that DEGs were annotated into 20 KEGG pathways for each group comparison shown in Figures 5(b)–5(d). Pathways with the highest enrichment in EC vs IDC have plant hormone signal transduction, phenylpropanoid biosynthesis, starch and sucrose metabolism, and glycolysis gluconeogenesis. Between EC and BDC, pathways with the highest enrichment are glutathione metabolism and oxidative phosphorylation. Phenylpropanoid biosynthesis and glutathione metabolism are the most enriched pathways in IDC vs BDC.
3.6. Functional Analysis of DEGs of Callus Regeneration
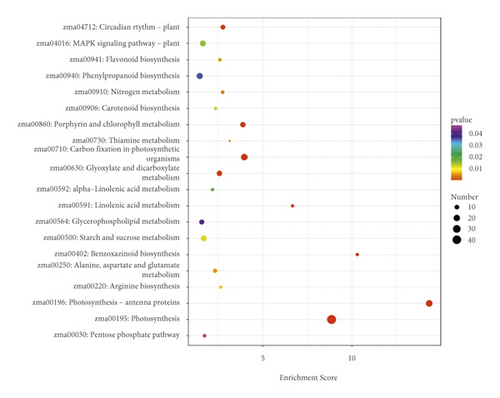
A total of 2313 DEGs was identified between the EC and IRC tissues. GO classification analysis was related to BP, CC, and MF (Figure 6). Cellular process (GO:0009987), metabolic process (GO:0008152), single-organism process (GO:0044702), response to stimulus (GO:0051869), and biological regulation (GO:0065007) were the top five classes in the BP. Cells (GO:0005623), cell parts (GO:0044464), organelles (GO:0043226), and membranes (GO:0016020) were the top four classes in the CC. Binding (GO:0005488) and catalytic activity (GO:0003824) were the top two classes in MF. Pathway analysis showed that DEGs were annotated into 20 pathways (Figure 6). The plant hormone signal transduction and starch and sucrose metabolism were the top pathways of upregulated genes, and photosynthesis and carbon fixation in photosynthetic organisms were the down pathways of upregulated genes. For the GO analysis and KEGG enrichment, similar classes and trends were detected for newly detected transcripts.
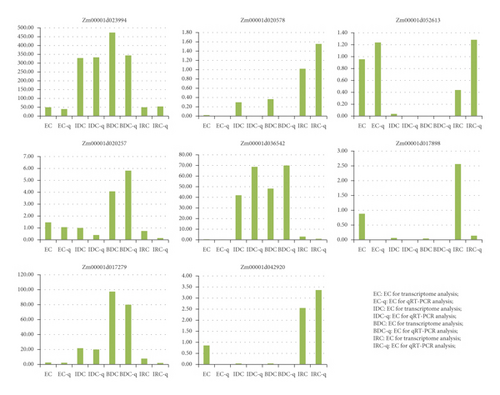
3.7. qRT-PCR Validation
qRT-PCR was performed in order to verify the expression profile obtained by transcriptome analysis (Figure 7). A total of eight DEGs, reported to be related to callus induction, was compared with qRT-PCR and transcriptomes in the study. Overall, six DEGs showed the same trend in transcriptomes and qRT-PCR. The coincidence rate of RNA-Seq and qRT-PCR was 88.89%, indicating that RNA-Seq had high accuracy and the identified pathways and candidate genes were reliable.
4. Discussion
Maize is one of the most important crops in the world and plays an important role in agricultural production and economic life. From the establishment of DNA recombination technology in the 1970s to the emergence of the world’s first transgenic plant-transgenic tobacco in 1983, the development of plant transgenic technology is changing rapidly [46]. With the development of the somatic cell regeneration system of corn and the development of transgenic technology [47, 48], based on the technology of genetically modified maize, genetic improvement technology has made a huge breakthrough, breaking reproductive isolation between species and directional import of the exogenous gene into the maize genome, so as to overcome the genetic improvement of specific traits the plight of insufficient resources [49–53]. At the same time, maize transgenic technology is still in the stage of development and improvement, especially the overall domestic maize transgenic technology, which is still relatively backward, with difficult transformation operation, low transformation efficiency, limited source of transformed recipient materials, and less independently developed carrier system, which have a great impact on and limit the research and application of maize transgenic [49, 54, 55]. The establishment of a suitable transformation receptor system is a key link in maize genetic transformation. Effective somatic cell reproduction and regeneration system is one of the preconditions for plant genetic transformation, which is related to whether suitable transformation receptor materials can be provided and whether normal transgenic plants can be successfully regenerated after transformation. The establishment of a suitable transformation receptor system is a key link in maize genetic transformation [56, 57]. Effective somatic cell reproduction and regeneration system is one of the preconditions for plant genetic transformation, which is related to whether suitable transformation receptor materials can be provided and whether normal transgenic plants can be successfully regenerated after transformation [58–61].
Embryonic callus is induced by explants and can be regenerated by organogenesis and embryogenesis. Callus is a mass of parenchymal cells which can divide and proliferate continuously under hormone stimulation [62–65]. Embryonic callus can be cultured on a large scale in vitro, and it can divide continuously and remain undifferentiated in the proper medium under dark culture conditions. Studies have shown that induction of maize embryogenic callus is significantly limited by genotype, as well as affected by explant type, culture conditions, and exogenous hormones [8, 66]. The analysis of genetic mechanisms controlling embryonic callus cells is very important for understanding basic processes involved in plant tissue culture [67]. The research on the induction ability of embryonic callus has gradually become one of the focuses of researchers; embryogenesis-related genes have been authenticated in Arabidopsis [68]. To expound the molecular mechanism of somatic embryogenesis in maize, a large number of forward or reverse genetic studies have been carried out [69–71]. Previous studies showed that the callus induction ability of maize immature embryos was controlled by quantitative trait genes, and a series of QTLs loci were also identified in different maize populations [72].
RNA-Seq analyses are important for gene expression levels between different conditions [73]. In this study, transcriptome analysis was used to analyze the differential expression of callus in different induction states. The number of DEGs between EC and IDC was 4077, 5896 DEGs between EC and BDC, and then EC and IRC, with 2,313 DEGs. In general, DEGs in EC and IDC are mainly concentrated in cell composition and biological processes, while DEGs in EC and BDC are mainly concentrated on the biological process, cellular process, and secondary metabolic process. In EC and IRC, DEGs are mainly concentrated on the biological processes, metabolic processes, and energy metabolism.
Embryogenesis is affected by many regulatory factors in maize. Auxin plays an important role in callus formation induced by embryogenesis [74] and activates the expression of downstream transcription factors, by mediating ARFs (auxin response factors) and inducing E2Fα (E2F transcription factor) to promote the formation of callus [75]. ARF16 was upregulated from EC to IDC and ARF8 was upregulated from EC to IRC in our study. Meanwhile, baby boom (BBM), SERK1, and SERK2 involved in the embryogenic pathway. Our results demonstrated that BBM2, SERK1, and SERK2 were upregulated from EC to IDC and EC to IRC. Mechanical damage has been recognized as a common stimulus of callus induction. WIP2 and ESR were described as one of the wound-inducible genes, which were upregulated from EC to IDC and EC to IRC. These changes can improve conversion efficiency from EC to IRC, although the functions of these genes need to be further studied (Figure 8), which are important for elucidating the underlying molecular mechanisms of callus formation.

5. Conclusion
In this study, transcriptome analysis was used to analyze the differential expression of callus in different induction states. DEGs in EC and IDC are mainly concentrated in cell composition and biological process, while DEGs in EC and BDC are mainly concentrated on the biological process, cellular process, and secondary metabolic process. In EC and IRC, DEGs are mainly concentrated on the biological processes, metabolic processes, and energy metabolism. In particular, ARF16 was upregulated from EC to IDC and ARF8 was upregulated from EC to IRC. BBM2, SERK1, and SERK2 were upregulated from EC to IDC and EC to IRC. These changes can improve conversion efficiency from EC to IRC, which is important for elucidating the underlying molecular mechanisms of callus formation [76].
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was supported by the research project of the Science and Technology Department in the Guizhou Province (S20201Y050) and the special project of Animal and Plant Breeding in the Guizhou Province Agriculture department (B2018024).
Open Research
Data Availability
The data used to support the results of this study are available from the corresponding author upon request.