Bioprospecting of an Endolichenic Fungus Phanerochaete sordida Isolated from Mangrove-Associated Lichen Bactrospora myriadea
Abstract
Bioassay-guided fractionation of the ethyl acetate extract of Phanerochaete sordida, an endolichenic fungus (ELF) isolated from the host lichen Bactrospora myriadea, collected from Negombo lagoon, Sri Lanka, led to the isolation of a bioactive compound. Following the identification of the fungus using morphological and DNA barcoding techniques, the pure compound was isolated using column chromatography, preparative TLC, and semipreparative HPLC. The structure elucidation was carried out using IR, HR-ESI-MS and 1H, 13C & 2D NMR spectroscopic methods. The in vitro bioassays conducted revealed that compound 1 has a high antioxidant activity with ABTS•+ (IC5058.91 ± 1.35 μM), moderate anti-inflammatory activity (IC50254.79 ± 1.41 μM), comparable antibacterial activity against the oral-bacterial strain Streptococcus mutans (MIC 898.79 μM and MLC 1797.58 μM), moderate tyrosinase inhibition (IC501713.69 ± 8.65 μM), and moderate cytotoxicity against oral cancer (IC5013.65 ± 0.02 μM), in comparison with respective positive controls. The in silico experiments conducted for tyrosinase inhibition and cytotoxicity using Schrödinger revealed results in line with the in vitro results, thus confirming the bioactivities. The molecule also satisfies the key features of drug likeliness according to pharmacokinetic studies.
1. Introduction
Endolichenic fungi (ELF), asymptomatic fungal associates which live in the interior of a lichen thallus, serve as an incredible source of novel bioactive compounds. While adding evidence in proving lichens to be “cradles of fungal diversification” [1], this distinct group of fungi has been found to produce compounds with antioxidant, antibacterial, anti-inflammatory, antifungal, anticancer, and numerous other potentials [2]. Subsequent to the first report of such isolated compounds in 2007 [3], more than 172 compounds have been isolated and explored from ELF while more than 99 of them had been novel molecules [4]. Experiments conducted in Sri Lanka revealed numerous such potent compounds, which include polyketides from ELF Penicillium citrinum [5] and Curvularia trifolli [6] along with more metabolites from Daldinia eschscholtzii [7] Xylaria psidii [8], and Neurospora ugadawe [9].
The ELF used in the present study, Phanerochaete sordida, was isolated from the host lichen Bactrospora myriadea, collected from the mangrove ecosystem of Negombo lagoon, Sri Lanka, in early March, 2018. This particular strain is known to degrade the lignin, cellulose, and hemicellulose [10]. Phanerochaete sordida is a member of the division basidiomycete and subjected for further studies since not many ELF from this division are reported. It was hypothesized that if a basidiomycete survives as an ELF, it should be metabolically unique. A thorough literature survey revealed that this is the first report of isolation of a bioactive compound from P. sordida.
The aim of the present study was to bioprospect and conduct compound isolation on the ethyl acetate extract of the ELF P. sordida using bioassay-guided fractionation and investigate their bioactive potentials using in vitro and in silico techniques. The compound was identified using spectroscopic data. The in vitro assays were conducted to evaluate the antioxidant, anti-inflammatory, tyrosinase inhibitory, antibacterial, and cytotoxic potentials of the compound. In silico approach was used in tyrosinase inhibitory and cytotoxic screenings, to establish the validity of the obtained in vitro results. Following the widely used trend of molecular docking, interactions of the compound with selected receptors in cancer cells and tyrosinase enzyme were assessed using Schrödinger (2021-1) software. Molecular docking has improved the drug discovery process by providing estimations of a compound’s affinity to receptors and thereby expediting the lead optimization processes [11]. Furthermore, pharmacokinetic studies were carried out to assess the perfect balance between high biological activity and low toxicity. The ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) parameters which comprehensively address the chemical drug-likeness of the compound were assessed using the freely accessible SwissADME web tool [12]. This comprehensive report contains the details of preliminary identification and screening for biological activities of the fungus combined with the isolation, identification, bioactivity screening, and computational studies of the isolated compound.
2. Materials and Methods
2.1. Ethical Statement
Permit to collect lichen samples from the mangrove forest in Negombo lagoon was obtained by the Forest Department of Sri Lanka. The samples were collected from mangrove and mangrove-associated plants in the area of “Kadolkele” under the observation of a representative of the department, without causing any disturbance to the surroundings.
2.2. Collection of Lichen Sample
Lichen host was collected on the 31st of March, 2018, at the mangrove forest of Negombo lagoon, Sri Lanka. The study site was “Kadolkele” area situated at 7.195528 N 79.84389 E GPS coordinates belonging to the Negombo lagoon area in Sri Lanka. Sample was immediately placed in an acid free paper bag, labeled appropriately, and transported to the Department of Chemistry, University of Kelaniya, and stored in 4°C to be processed for the isolation of ELF within two weeks of sampling.
2.3. Lichen Identification
A portion of lichen of the particle (2 × 2 cm) was air-dried for two weeks and refrigerated. The identification of this lichen was carried out using morphological and molecular techniques at Natural History Museum, London, and photographs were taken using Olympus stereomicroscopes and Olympus compound microscopes with interference contrast, connected to a Nikon Coolpix digital camera. Tap water was utilized for section mounting for all measurements [9]. Duplicate of the specimen was deposited in the Department of Chemistry, University of Kelaniya.
2.4. Isolation of Endolichenic Fungi
Lichen thallus was washed in running tap water to remove unwanted particles and then surface sterilized using consecutive immersion in 96% ethanol for 10 s, 0.5% sodium hypochlorite for 2 min, and 70% ethanol for 2 min and dried using sterile filter paper. Small segments of the lichen thallus (approximately 3 × 3 mm) was then cut and placed on 2% malt extract agar (MEA) supplemented with 0.01% streptomycin (Sigma-Aldrich). Sealed plates were then incubated under ambient conditions (room temperature 30°C) for 14 days and observed regularly. Hyphal tips from each morphologically distinct colony was placed in new PDA plates and subjected to several subculturing to obtain a pure culture [3]. Among various fungal isolates, a sterile and fast-growing basidiomycete isolate was selected for further studies since isolation of basidiomycetes as ELF is as not as frequent as ascomycetes.
2.5. Extraction of Genomic DNA and Polymerase Chain Reaction (PCR)
The isolate was inoculated in Potato Dextrose Broth (PDB) and incubated at room temperature (30 ± 32°C) for one week. Mycelia were separated from the broth, washed with sterilized water, and remove excess water, and DNA was extracted using a slightly modified method of CTAB DNA extraction protocol [13]. Quality of extracted DNA sample was tested using 1% agarose gel electrophoresis, and sample was stored at -20°C for further utilization. The nuclear ribosomal internal transcribed spacer region (rDNA-ITS) was amplified using polymerase chain reaction (PCR); using the fungal-specific ITS5 forward and ITS4 reverse universal primers [14]. The PCR reaction procedure was conducted as described in Maduranga et al. (2018)[15] Clean PCR products were subjected to bidirectional Sanger dideoxy sequencing at Genetech Institute, Colombo, Sri Lanka. DNA sequences were manually edited using BioEdit sequence alignment editor (Version 7.2.5) and compared with the sequences available in the GenBank using Basic Local Alignment Search Tool (BLAST) to assess homology. The isolated fungus was identified comparing sequence of ITS-rDNA region with the established (voucher specimens or published) records with the highest percent similarity of the GenBank accessions. DNA sequence of the identified ELF species was deposited at the NCBI database, and an accession number was obtained [9].
2.6. Extraction and Isolation of Secondary Metabolites from the Endolichenic Fungus
The ELF was cultured on PDA plates of 150 mm diameter (100 plates) and incubated at room temperature for 2 weeks. The mycelium with the medium were cut into small pieces, extracted to ethyl acetate (6 × 500 mL), and the solvent was evaporated under reduced pressure using the Rotary Evaporator (IKA RV 10B S96, Germany). The evaporated ethyl acetate extract was then transferred to a glass vial separately, and N2 gas was passed through the sample to remove any remaining solvent. The resulting semisolid extract was stored at 0°C for further analysis [5].
2.7. Bioassay-Guided Isolation of the Compound
Since the ethyl acetate extract showed significant antioxidant, antibacterial, and anti-inflammatory activity, 5.84 g of the crude extract was partitioned with hexane, chloroform, and methanol, in order to separate nonpolar, moderately polar, and polar compounds, respectively. The chloroform fraction (4.2 g) was found to possess the highest activity in antioxidant, antibacterial, and anti-inflammatory assays. Hence, it was subjected to column chromatography using normal phase silica gel. The column (70-230 mesh, 19.0 g, 2 cm × 50 cm) was eluted with 100% chloroform containing increasing amounts of methanol and finally methanol. The collected fractions were combined according to thin layer chromatography (TLC) profiles to yield 7 major fractions (F1–F7). Of the seven fractions, F1 (38.7 mg) and F2 (314.5 mg) contained the same major compound and were subjected to column chromatography with the same polarity gradient. Column fractions obtained from F1 and F2 were combined after running the TLC and purified using preparative TLC (PTLC) to afford two fractions of the same pure compound, compound 1 (weights: 7.0 and 11.7 mg). After utilizing a portion to perform bioassays (please note that cytotoxicity assay was conducted after the final purification), the remaining of the fractions were sent to obtain spectral data. The combined fractions were further purified using preparative HPLC with a semipreparative column (Waters, Xterra Prep RP C18 column, 5 μm, 19 × 150 mm) applying the following method: (A) 1% formic acid in deionized water and (B) methanol, gradient; 95% A/5% B (0 min), 95% A/5% B (5 min), 5% A/95% B (15 min), 5% A/95% B (25 min), 95% A/5% B (28 min), and 95% A/5% B (30 min); flow rate 5 mL/min. One pure compound (compound 1, weight: 4 mg) was isolated, and the structure of the compound was determined by 1H-NMR and 13C-NMR spectroscopy (Bruker, 500 MHz in CDCl3) in combination with DQF-COSY, DEPT, HMBC, HSQC, and NOESY correlations. Molecular mass of the compound 1 was obtained using HR-ESI-MS.
2.8. Radical Scavenging Ability by (2,2 ′-Azino-Bis(3-Ethylbenzothiazoline-6-Sulfonic Acid)) (ABTS·+) Method
2.9. Anti-Inflammatory Assay (HRBC Stabilization Method)
The anti-inflammatory assay was conducted using human red blood cell stabilization method which uses the heat induced hemolysis principle [17]. Test sample and the positive control aspirin (1 mg of each) were dissolved in 0.5 mL of DMSO (dimethyl sulfoxide) and diluted up to 5 mL by addition of normal saline. A half dilution series was prepared with a control, and the reaction mixture was prepared with 2.5 mL of the solution and 0.25 mL of 10% v/v RBC suspension. All the centrifuge tubes containing reaction mixtures were incubated in a water bath at 56°C for 30 min after which the tubes were cooled under running tap water. The reaction mixture was centrifuged at 3000 rpm for 10 min to obtain a clear liquid phase, and the absorbance of the supernatants was measured at 560 nm using Microplate Reader (Biotek, USA) by using aliquots of 200 μL for each well in a 96 well-plate from each mixture. The test was performed in triplicates, and the percentage stability was calculated using Equation (1). IC50 values were calculated using the statistical software GraphPad Prism 7.04.
2.10. Tyrosinase Inhibitory Assay
2.11. Antibacterial Assay (Agar Well Diffusion Method)
Antibacterial assay against the anaerobic bacterial strain Streptococcus mutans (ATCC 700610) was conducted using agar well diffusion method [19]. An aliquot of overnight cultures in Trypticase soy broth (supplemented with blood with lysed RBC and grown under anaerobic conditions in a jar containing anaerobic sachets at 37°C) was diluted hundred times, and a quantity of 100 μL was spread evenly on the surface of the Trypticase soy agar (supplemented with lysed RBC) plates prepared. Wells with a diameter of 0.5 cm were made on the plates using a cork borer. Sample and chlorhexidine (0.2%) (positive control) were dissolved in DMSO to meet 5 mg/mL concentration for initial screening. In order to determine minimum inhibitory concentration (MIC) and minimum lethal concentration (MLC) of the compound 1, a concentration series of 1000, 750, 500, 250, 100, 50, and 10 μg/mL was utilized. The wells were filled with 100 μL of the test solutions while keeping a negative control of DMSO. After an incubation of 24 hours at room temperature under anerobic conditions, the diameters of observable inhibition zones were measured in mm [20].
2.12. Cytotoxicity Assay (Alamar Blue Method)
2.13. In Silico Studies
Protein sequences were retrieved from Universal Protein Resource (UniProtKB) database, and homology modelling was used accordingly to select and retrieve protein structures representing CAL-27 (oral cancer) cell lines and tyrosinase enzyme. Rho-associated protein kinase (ROCK 1), programmed cell death 6-interacting protein (ALIX) [22], and tumor susceptibility gene 101 protein (TSG101) [23] were selected as receptors for CAL-27 cell. Molecular details were obtained from UniProtKB database with UniProt ID Q13464, Q8WUM4, and Q99816, respectively. The protein structures were obtained from RCSB (Research Collaboratory for Structural Bioinformatics), PDB (Protein Data Bank) with ID 2ETR, 3C3R, and 3OBQ having 2.60 Å, 2.02 Å, and 1.40 Å resolutions, respectively. For tyrosinase enzyme, the enzyme itself was selected as the suitable receptor in line with UniProt ID P1469, and structure was obtained from ModBase. Please note that the structures retrieved for cell lines and tyrosinase enzyme were from human sources.
Binding pocket prediction was conducted utilizing CASTp and Ghecom for 2ETR, 3C3R, 3OBQ, and tyrosinase. The compound was docked against all selected proteins using the docking software Schrödinger (2021-1). Grids were generated for each of the protein with following amino acids: (a) 2ETR: Met 156, Ile 82, Phe 368, Leu 205, Val 90, Asp 202, Tyr 155, Met 153, Glu 154, Ala 103, Ala 205, Asp 216, and Asn 203; (b) 3C3R: Glu 137, Asp141, Ala 151, Tyr 153, Leu 201, Thr 204, Asp 206, Pro 238, Ile 251, Asn 255, Val 297, Phe 300, and Lys 303; (c) 3OBQ: Thr 58, Arg64, Asn 66, Tyr 68, Asn 69, Lys 98, Thr 92, Met 95, Phe 142, and Ser143; (d) tyrosinase: His 19, Gln 68, Val 74, Asp 76, Phe 84, Gly 154, Met 159, Tyr 181, Leu 188, Arg 196, His 202, Leu 288, Thr 309, Ala 357, His 363, Ile 396, Pro417, and His 420. Resultant docking complex was evaluated for ligand efficiency, binding energy scores, and intermolecular H-bonds.
The drug likeliness and pharmacokinetic profile of the compound were studied using SwissADME tool.
3. Results and Discussion
3.1. Identification and Isolation of Endolichenic Fungus
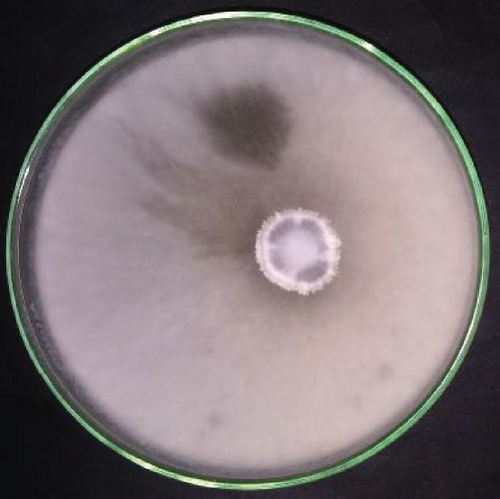
The lichen was collected from the mangrove plant Rhizophora mucronata, a true mangrove species among one of the frequently found plant species in the mangrove ecosystem of Negombo lagoon, Sri Lanka [24]. The lichen host of the ELF isolate was identified using morphological and molecular techniques as Bactrospora myriadea. The obtained ITS sequence of the ELF sample showed 98.5% sequence homology to Phanerochaete sordida sequence submitted by Kamei et al. [25] (accession number AB210078.1) in GenBank. The obtained sequence was deposited under the accession number MT507808 in the GenBank. The isolate showed rapid growth on PDA, covering a 150 mm diameter petri-dish within two weeks. The colony colour was white to grey by the second week after incubation at the room temperature (Figure 1).
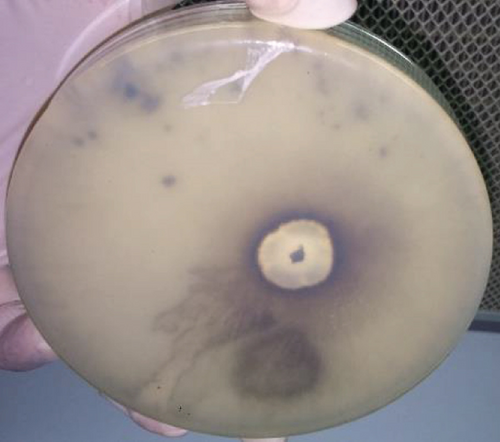
3.2. In Vitro Assays
As per bioassay-guided fractionation, the in vitro assays were first conducted (except anticancer assay) to the ethyl acetate crude extract followed by hexane, chloroform, and methanol partitions. According to the results, the chloroform partition was subjected to further isolation owing to its highest activity compared to the others. The comparison of results is presented in Table 1.
ABTS assay Activity at highest concentration of 50 μg/mL (%) |
HRBC stabilization assay IC50 value (μg/mL) |
Tyrosinase inhibitory assay Activity at highest concentration of 666.67 μg/mL (%) |
Antibacterial assay against S. mutans Inhibition zone diameter (mm) |
|
|---|---|---|---|---|
| Crude extract | 34.13 ± 0.62 | 124.00 ± 0.45 | 12.58 ± 0.37 | 16 ± 0.15 |
| Hexane partition | 24.57 ± 0.22 | 170.27 ± 3.02 | 22.54 ± 0.97 | — |
| Chloroform partition | 43.17 ± 0.55 | 88.62 ± 0.76 | 42.42 ± 0.50 | 11 ± 0.04 |
| Methanol partition | 13.95 ± 3.76 | Max < 50% | 9.91 ± 1.28 | — |
- Three replicates were used in each assay, and the values are given with ±standard error. Max < 50% indicates that activity showed by the maximum dosage did not surpass 50%.
Bioassay-guided fractionation of the chloroform fraction using column chromatography, PTLC, and preparative HPLC yielded pure compound 1, and the bioactivity of the compound 1 was evaluated using the in vitro bioassays.
3.3. Structure Elucidation
Bioassay-guided fractionation of the ethyl acetate extract of P. sordida yielded the compound as a white-coloured solid. The compound gave [M + H]+ ion in the high-resolution electrospray ionization mass spectrometry (HR-ESI-MS) at m/z 279.1603, which was consistent with a molecular formula of C16H22O4 (calcd.278.15186), requiring 6 sites of unsaturation. The absorption peaks appearing in the IR spectrum occurring at 3343 cm-1 (broad peak) and 1620 cm-1 (towards the lower end of carbonyl stretch due to the attached aromatic group) suggested the presence of hydroxyl and carbonyl groups, respectively. UV spectrum showed maximum absorption bands at 220 and 260 nm confirming the presence of a carbonyl group. With the attained information, a thorough search was done using SciFinder and Dictionary of Natural Products (DNP) using species and genus specific filters. Since no matches were found, structure elucidation was conducted using NMR data.
The aromatic region of the 1H NMR spectrum exhibited two signals at δΗ 6.23(d, 2 Hz) and δΗ 6.28 (d, 2 Hz), and this spin system consisting of two 1H doublets was shown to be due to metacoupled protons with 2.2 Hz. The 13C NMR signals at δC 101.37, 105.56, 110.80, 149.48, 160.08, and 165.34 completed the carbon skeleton of the aromatic ring. The carbon signal at δC 75.17 was indicative of a carbon connected to an oxygen with a single bond. The D2O experiment carried out revealed significant lowering of intensity of the 1H peaks at δΗ 5.53 and 12.00, affirming the protons as of hydroxyl groups. An analysis of the spectra 13C NMR, DEPT, and HSQC revealed one methyl carbon (δC 20.11), seven methylene carbons (δC 21.16, 24.12, 24.68, 27.24, 30.78, 31.08, and 33.54), and one sp3 methine carbon (δC 75.17) with the olefinic carbons in the aromatic ring mentioned above (δC 101.37 and 110.80). Four out of the five quaternary carbons were embedded in the aromatic ring at δC 105.56, 149.48, 160.08, and 165.34 with latter two being more deshielded due to each being bonded to an oxygen atom. An analysis of 1H-1H correlations observed in the DQF-COSY spectrum revealed the presence of CH3 CH CH2 and CH2 CH2 CH2 isolated spin systems. The 1H-1H correlations observed in the DQF-COSY and HSQC further indicated that two Hs of carbons at δC 33.54 (δH 2.50 and 3.28) and δC 31.08 (δH 1.79 and 1.92) were nonequivalent methylene protons. In HMBC spectrum, the aromatic proton at δH 6.28 showed correlations with δC 105.56, 110.80, 160.08, and 165.34 while the proton at δH 6.23 showed correlations to the aromatic carbons at δC 101.37 and 105.56 as well as to the benzylic carbon at δC 33.54. The 13C NMR spectrum of compound 1 when analyzed with the help of the HMBC and DEPT spectra showed the presence of a caboxylic carbonyl (δ 171.9), and it further indicates the correlation of the proton in carboxyl group at δH 12.00 with δC 101.37, 105.56, and 165.34. The HMBC correlations were observed from δH 1.36 to δC 75.17; δH 1.79 to δC 20.11; δH 1.92 to δC 24.12, and 75.17 with δH 1.48 long range coupling to δC 20.11. The NOESY correlations depict that the protons at δH 1.79, 5.53, and 12.00 are in close proximity to each other in spacial arrangement of the molecule. The structure of the compound 1 was thus elucidated as 12,14-dihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-benzo[c][1]oxacyclododecin-1-one.
The 1H and 13C NMR data pertaining to the structure elucidation is presented in Table 2, and the structure is shown in Figure 2.
| Position | δ 13C | δ 1H (multiplicity, nH, J/Hz) | DEPT |
|---|---|---|---|
| 1 | 20.11 | 1.36 (d, 3H, 5) | CH3 |
| 2 | 21.16 | 1.48 (m, 2H) | CH2 |
| 3 | 24.12 | 1.48 (m, 2H) | CH2 |
| 4 | 24.68 | 1.61 (m, 2H) | CH2 |
| 5 | 27.24 | 1.55 (m, 2H) | CH2 |
| 6 | 30.78 | 1.61 (m, 2H) | CH2 |
| 7 | 31.08 |
|
CH2 |
| 8 | 33.54 |
|
CH2 |
| 9 | 75.17 | 5.17 (m, H) | CH |
| 10 | 101.37 | 6.28 (d, H, 2) | CH |
| 11 | 105.56 (quaternary) | ||
| 12 | 110.80 | 6.23 (d, H, 2) | CH |
| 13 | 149.48 (quaternary) | ||
| 14 | 160.08 (quaternary) | ||
| 15 | 165.34 (quaternary) | ||
| 16 | 171.90 (quaternary) | 12.00 (OH) |
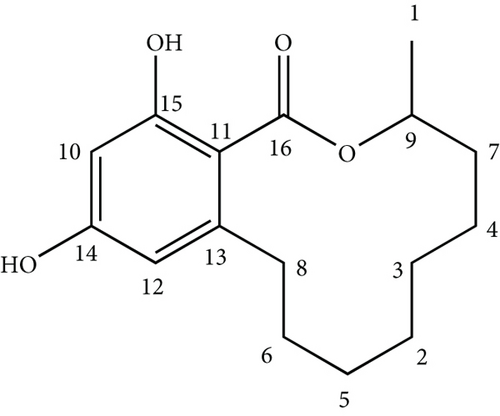
Subsequent search in SciFinder of the elucidated structure revealed that this compound has previously been isolated firstly from the fungus Lasiodiploidia theobromae [26]. It is a member of the large group of resorcinylic acid lactones and is known as (R)-de-O-methyllasiodiplodin. This is the first time the compound has been isolated from the genus Phanerochaete until date.
This macrolide is previously known to be a nonsteroidal mineralocorticoid receptor antagonist and to possess a variety of bioactivities [27].
Here, we are presenting and discussing activities which have not been reported before for this compound. Activity comparison of the compound 1 with relevant positive controls for all the in vitro assays is presented in Table 3.
| In vitro assay | Activity of the compound 1 | Activity of the positive control |
|---|---|---|
| ABTS assay | IC5058.91 ± 1.35 μM | IC5055.20 ± 4.04 μM |
| HRBC stabilization assay | IC50254.79 ± 1.41 μM | IC50134.16 ± 1.33 μM |
| Tyrosinase inhibitory assay | IC501713.69 ± 8.65 μM | IC50148.43 ± 13.04 μM |
| Antibacterial assay | MIC 898.79 μM, MLC 1797.58 μM | MIC 98.92 μM, MLC 197.85 μM |
| Anticancer assay against oral cancer | IC5013.65 ± 0.02 μM | IC5001.20 ± 0.03 μM |
- Three replicates were used in each assay and the values are given with ±standard error.
According to literature, the compound is known to reduce DPPH radical [28]. In this study, radical scavenging activity was assessed using the in vitro assay stated under methods against the radical monocation ABTS•+. As shown in Table 3, the close IC50 value shown by the compound 1 (IC5058.91 ± 1.35 μM) to that of the positive control BHT (IC5055.20 ± 4.04 μM) against ABTS•+ radical monocation displays promising antioxidant potential of the compound 1.
Compound 1 has been reported to inhibit the production of nitric oxide [29] and prostaglandin [30] which act as inflammatory mediators. Supportingly, in this study, the anti-inflammatory activity is assessed by the ability to stabilize the lysosomal membrane through HRBC stabilization assay. The activity shown by the compound 1 (IC50254.79 ± 1.41 μM) is slightly comparable to the positive control aspirin (IC50134.16 ± 1.33 μM). It could be assumed that binding mechanism of the compound 1 is not similar to that of aspirin (acetylsalicylic acid) though further studies are required to confirm its validity.
Inhibition of tyrosinase enzyme is the most convenient way of controlling melanogenesis, since it catalyzes the first and only rate-limiting step in the process. Hence, the demand for tyrosinase inhibitors has increased to solve the issue of hyperpigmentation [31]. This compound showed moderate activity compared to the commercially available inhibitor kojic acid. Further, in silico analysis was conducted to assess the binding of the compound to the enzyme, which will be discussed later in the manuscript.
The oral-bacterial strain Streptococcus mutans, known for its pathogenicity of dental caries, is now even proved to be a potential risk factor in causing systemic diseases such as ulcerative colitis [32]. In an era where the antibiotic resistance has become a major issue, even in oral health, emergence of compounds with promising antibacterial activity is vital [33]. The agar well diffusion method carried out to assess the antibacterial activity of the compound 1 with a series of concentrations as stated under methods section revealed that the minimum inhibitory concentration (MIC) and minimum lethal concentration (MLC) values of the compound 1 were 898.79 μM and 1797.58 μM, respectively. These MIC and MLC values were higher than that of the commercially available antibiotic chlorohexidine (MIC 98.92 μM, MLC 197.85 μM).
The burden of cancer and mortality is increasing at an alarming rate as per the global cancer statistics. Only in 2020, 19.3 million new cancer cases and almost 10.0 million cancer deaths have been reported worldwide. Among many others, tongue squamous cell carcinoma (tongue cancer) has been found to be one of the most common malignancies seen in the oral maxillofacial region [34]. In order to determine the anticancer potency of the compound 1 against oral cancer, the cell line CAL-27 (oral cancer) was used in Alamar Blue cell-culture assay. It seems to possess comparable activity against CAL-27 (IC5013.65 ± 0.02 μM) cancerous cell line. The cytotoxicity on normal cells was assessed by using the normal breast cell line Mcf10a, and it was found to be nontoxic up to a concentration of 100 μM (Please note that maximum percentage of methanol used was 0.2 percent, which did not show any toxicity as per the vehicle control fluorescence observed.). Moreover, in silico studies were conducted to determine interactions of the compound 1 with the receptors of this particular cell line and are discussed later in the manuscript.
3.4. In Silico Assays
Molecular docking is one of the most widely used techniques for in silico drug design and drug discovery which involves structure-based virtual screening for identification of particular target protein [35]. The compound was docked against selected proteins using Schrödinger docking module (2021-1). The binding energy of the compound with different receptors is depicted in Table 4, and the 2D interactions are illustrated in Figure 3.
| Sr. no | Protein | PDB ID | Binding energy (kcal/Mol) |
|---|---|---|---|
| 1 | Rho-associated protein kinase 1 | 2ETR | -5.642 |
| 2 | ALIX | 3C3R | -4.878 |
| 3 | TSG101 | 3OBQ | -4.641 |
| 4 | Tyrosinase | Structure obtained from ModBase | -5.234 |
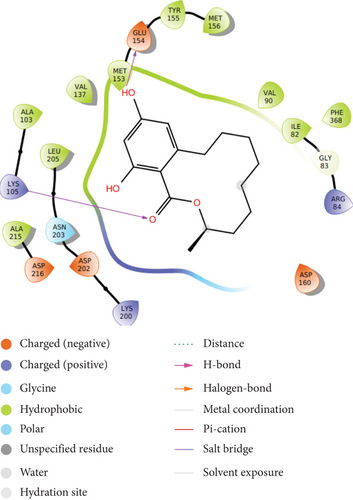
It is observed that the compound shows highest binding efficiency with the receptor 2ETR with binding energy -5.642 kcal/mol. The molecule shows hydrogen bonding at two points: carbonyl oxygen with LYS 105 and one phenolic hydroxyl group with GLU 154. Furthermore, hydrophobic interactions can be observed with amino acids LEU 205, VAL 137, MET 153, ILE 82, PHE 368, TYR 155, and ALA 215. Some positive-charged interactions with LYS 105, LYS 200, and ARG 84 as well as some negative charged interactions with ASP 160, ASP 202, and ASP 216 can also be observed. A polar interaction is shown with the amino acid ASN 203 (Figure 3(a)).
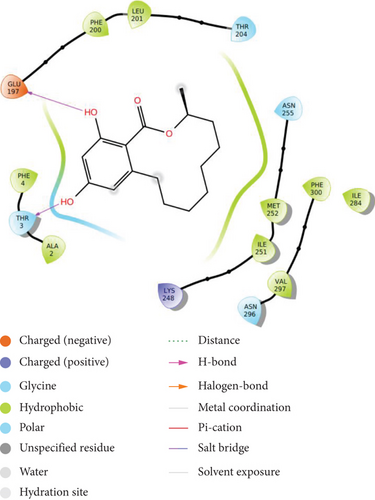
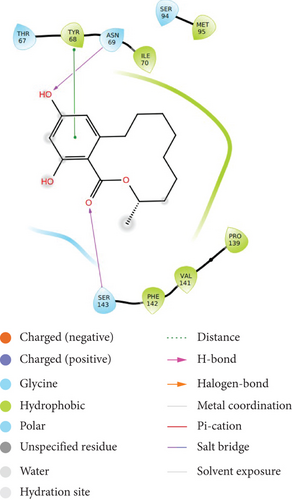
The compound shows moderate binding efficiencies with the receptors 3C3R and 3OBQ as well. In the case of 3C3R, hydrogen bonding can be observed between the two phenolic hydroxyl groups and amino acids GLU 197 and THR 3. Hydrophobic interactions with PHE 4, ALA 2, LEU 201, MET 252, ILE 251, and VAL 297 as well as the polar interactions with THR 3 and ASN 255 could be noted (Figure 3(b)). Considering the receptor 3OBQ, apart from the hydrogen bonds shown between the carbonyl oxygen and SER 143 and one phenyl hydroxyl and TYR 68, pi-pi stacking could also be observed between the aromatic ring and the amino acid TYR 68 (Figure 3(c)).
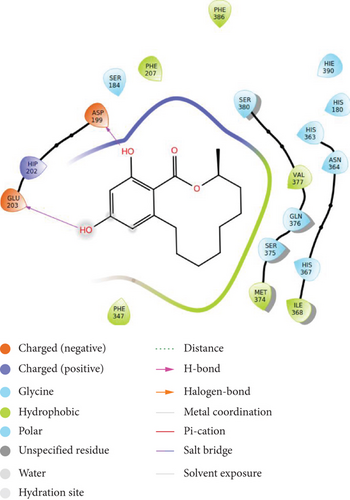
The molecule shows conventional H-bonding with the amino acid ASP 199 and GLU 203, while showing hydrophobic interactions with PHE 207, PHE 386, PHE 347, MET 274, and ILE 368 of tyrosinase enzyme as depicted in Figure 3(d). In addition to this bonding ability shown by the compound, its skin permeability (-4.48 cm/s) falling between the accepted range of -9.7 cm/s and -3.5 cm/s shows good prospect of being used in skin care industry.
Since the compound shows high binding efficiency for receptor, it can be further studied for this receptor inhibition using various in vitro and in vivo assays.
To further establish the lead likeliness, the physiochemical properties and pharmacokinetic properties of the compound were evaluated through in silico using the comprehensive tool Swiss ADME (Swiss Institute of Bioinformatics) as depicted in Table 5.
| Physicochemical properties | Pharmacokinetic properties | Drug likeliness |
|---|---|---|
|
|
Lipinski—yes (0 violations) |
|
|
Ghose—yes |
|
|
Veber—yes |
|
|
Egan—yes |
|
|
Muegge—yes |
|
|
Bioavailability score—0.55 |
|
|
|
|
|
|
|
|
|
|
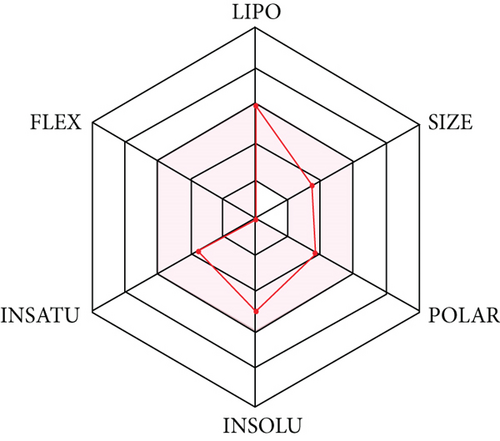
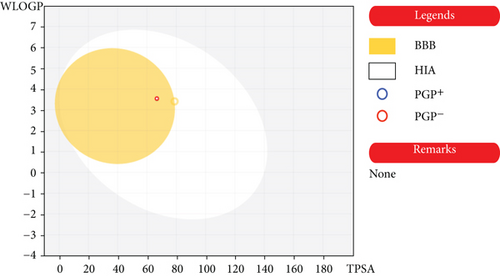
Molecule followed different rules like Lipinski, Ghose et al. [36, 37], Veber et al. [38], Egan et al. [39, 40], and Muegge et al. [41] put forth for drug likeliness evaluation with a bioavailability score of 0.55. The compound has topological polar surface area (TPSA) values below 140 Ǻ and can be characterized to have a substantial permeability in the cellular plasma membrane. The compound shows high gastro intestinal absorption with in silico absorption percentage of 89.23. The in silico absorption percentage was calculated using formula percentage absorption = 109–(0.345 × TPSA) [42]. The prediction shows it is not P-glycoprotien (P-gp) substrate, making it more suitable as an anticancer agent. The compound occupies the optimal area in SwissADME radar plot (Figure 4). As per the BOILED EGG diagram (Figure 5), the compound has high intestinal absorption as well as is blood brain barrier permeable.
4. Conclusion
The ELF Phanerochaete sordida was isolated from the lichen Bactrospora myriadea collected from Negombo lagoon, Sri Lanka. Structure of the isolated compound was elucidated as 12,14-dihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-benzo[c][1]oxacyclododecin-1-one (molecular formula: C16H22O4) using 1H, 13C NMR, DEPT, DQF-COSY, HMBC, HSQC, NOESY, and HR-ESI-MS spectral data. It exhibited high antioxidant activity against ABTS•+ in the in vitro assay conducted. It also displayed comparable anti-inflammatory, antibacterial activity with moderate tyrosinase inhibition and anticancer activity against oral (CAL-27) cancer cell line. The in silico studies done using Schrödinger (2021-1) confirmed the tyrosinase inhibitory and anticancer activity exhibited in in vitro assays with similar capacities. The molecule satisfies the key features of drug likeliness like H-bonding, molecular weight, and partition coefficient. The computational pharmacokinetics showed good oral bioavailability and blood-brain permeability as per the BOILED EGG diagram. As the molecule is not a P-gp (plasma glycoprotein) substrate, it can be considered as a good anticancer agent. This study confirms that this compound could be used as a prospective scaffold in the pharmaceutical industry in novel ways other than the already reported ones.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
The authors thank Ms. Patricia Wolseley, Algae, Fungi and Plants Division, Department of Life Sciences, The Natural History Museum, London, for the support rendered in lichen identification and Prof. Mala Amarasinghe, Department of Plant and Molecular Biology, University of Kelaniya, for helping in identification of mangrove host plants. Financial support is provided by the Ministry of Science, Technology and Research, Sri Lanka (grant no: MSTR/TRD/AGR/03/02/07).
Open Research
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.