Common Expression Quantitative Trait Loci Shared by Histone Genes
Abstract
A genome-wide association study (GWAS) was conducted to examine expression quantitative trait loci (eQTLs) for histone genes. We examined common eQTLs for multiple histone genes in 373 European lymphoblastoid cell lines (LCLs). A linear regression model was employed to identify single-nucleotide polymorphisms (SNPs) associated with expression of the histone genes, and the number of eQTLs was determined by linkage disequilibrium analysis. Additional associations of the identified eQTLs with other genes were also examined. We identified 31 eQTLs for 29 histone genes through genome-wide analysis using 29 histone genes (P < 2.97 × 10−10). Among them, 12 eQTLs were associated with the expression of multiple histone genes. Transcriptome-wide association analysis using the identified eQTLs showed their associations with additional 80 genes (P < 4.75 × 10−6). In particular, expression of RPPH1, SCARNA2, and SCARNA7 genes was associated with 26, 25, and 23 eQTLs, respectively. This study suggests that histone genes shared 12 common eQTLs that might regulate cell cycle-dependent transcription of histone and other genes. Further investigations are needed to elucidate the transcriptional mechanisms of these genes.
1. Introduction
Histone mRNA transcripts and proteins are important for packing DNA into chromatin and are thus tightly regulated in most human cells [1]. In humans, the genes encoding histones are gathered on chromosomes 1 and 6. It has been suspected that the clustered structure of genes can provide a manageable unit for coordinating transcription [1]. Recently, genome-wide chromatin interaction analysis with paired-end-tag sequencing (ChIA-PET) has shown that some histone genes can share promoters [2].
While many efforts have been made to understand the mechanisms for the transcription of histone genes, they have not yet been well defined. Nuclear protein of the ataxia-telangiectasia-mutated locus (NPAT), which promotes the transcription of histone genes, is located near the Cajal body [1]. Clusters of histone genes are also located near the Cajal body [3]. The positions of histone gene clusters near the Cajal body have been observed between the restriction point (R-point) and the G1/S transition (S-point) during the cell cycle [4]. The objective of this study was to select simultaneously expressed histone genes, identify their expression quantitative trait loci (eQTLs), and examine the functions of those eQTLs.
2. Material and Methods
2.1. Subjects and Data
The subjects of this study were 373 Europeans including 95 Finnish in Finland, 94 British in England and Scotland, 93 Tuscans from Italy, and 91 Utahn residents with Northern and Western European ancestry from the CEPH collection. Their genotypic data were derived from the phase 1 dataset produced by the 1000 Genomes Project [5] (http://www.internationalgenome.org/). This study utilized genotypic data at 5,796,145 SNPs after filtering out the SNPs with minor allele frequency < 0.05, with missing rate > 0.05, or in Hardy-Weinberg disequilibrium with P < 0.001.
Transcriptional data on 10,518 human genes were obtained in lymphoblastoid cells of the subjects by the Geuvadis RNA sequencing project (http://www.geuvadis.org/web/geuvadis/rnaseq-project). The unit used for the mRNA expression level was reads per kilobase per million mapped reads (RPKM). Outliers were removed based on sample similarity, which was estimated by the Spearman rank correlation between RPKMs and the exon counts of the samples [6]. Sample swaps or contaminated samples were excluded based on allele-specific expression analysis [6]. For details on the quality control process, see t Hoen et al. [7].
2.2. Statistical Methods
We selected histone genes that were expressed simultaneously. Pairwise gene expression relationships were estimated using Pearson’s correlation coefficient (r). The significance of the correlation was determined by P < 0.05.
We investigated genome-wide associations of the expression of the selected histone genes. A regression model was employed to identify SNPs associated with expressions of histone genes using PLINK [8]. The Bonferroni correction was applied as a multiple testing, and the significance was determined by P < 2.97 × 10−10.
Linkage disequilibrium (LD) between the identified SNPs was estimated using the HaploView program [9]. The LD block was determined according to the 95% confidence interval of the D′ value for pairwise LD between the nucleotide variants with minor allele frequency > 0.05 [10].
The identified eQTLs were further analyzed for their associations with the expression of nonhistone genes throughout the genome. The Bonferroni multiple testing based on t-statistic was also applied with a significance threshold value of P = 4.75 × 10−6.
The functions of identified SNPs were examined using the Ensembl Variant Effect Predictor program [11] and RegulomeDB [12] (e.g., the motif of DNA footprinting assay, chromatin structure by DNA-seq, and protein binding by ChIP-seq).
3. Results
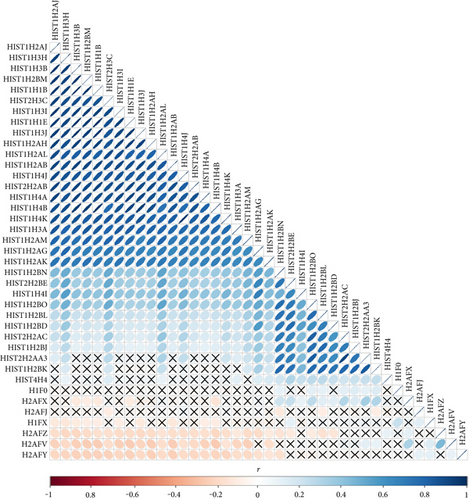
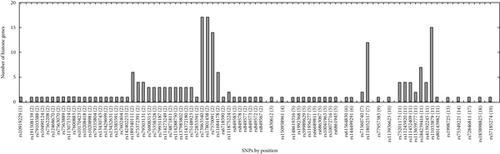
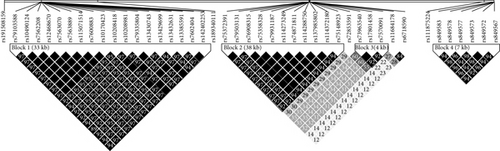
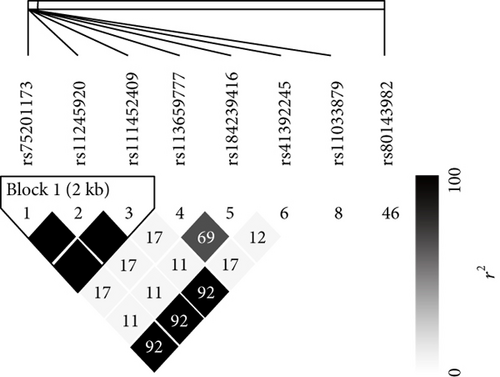
We observed numerous correlations amid the expression of the histone genes investigated in the current study (Figure 1). In particular, the expression of 29 genes showed correlations significantly (P < 0.05). Genome-wide association analysis showed that 74 SNPs were associated with the expression of the 29 histone genes (P < 2.97 × 10−10; Table 1). Among them, 26 SNPs were simultaneously associated with the expression of multiple histone genes, and 5 out of 26 SNPs were associated with the expression of more than 10 histone genes (Figure 2). Thirty-one LD blocks were constructed covering the identified SNPs (Figure 3). The eQTLs corresponded to functional sites provided by various functional search sites. The rs79335804 had the most probable function with a RegulomeDB score of 2b (Table S1 available online at https://doi.org/10.1155/2017/6202567).
| SNPa | Gene | Effectb | Locationc | A1d | A2d | MAF | Genetic associations | Beta | P |
|---|---|---|---|---|---|---|---|---|---|
| rs10919229 | SELE (2Kb upstream) | 1:169735986 | T | A | 0.05 | HIST1H4B | 1.035 | 2.51E−10 | |
| rs191508159 | Intergenic | 2:35957035 | T | C | 0.06 | HIST1H4A | 0.466 | 3.41E−11 | |
| rs79103588 | Intergenic | 2:48891009 | A | T | 0.05 | HIST1H4B | 1.045 | 1.02E−10 | |
| rs10490124 | Intergenic | 2:48897335 | G | A | 0.06 | HIST1H4B | 1.023 | 1.65E−10 | |
| rs7562208 | Intergenic | 2:48898231 | C | T | 0.06 | HIST1H4B | 1.023 | 1.65E−10 | |
| rs12468670 | Intergenic | 2:48902054 | T | G | 0.06 | HIST1H4B | 1.023 | 1.65E−10 | |
| rs7563070 | Intergenic | 2:48902202 | T | C | 0.06 | HIST1H4B | 1.023 | 1.65E−10 | |
| rs7563889 | Intergenic | CTCF | 2:48902751 | T | G | 0.06 | HIST1H4B | 1.023 | 1.65E−10 |
| rs115071514 | Intergenic | 2:48902928 | T | C | 0.06 | HIST1H4B | 1.023 | 1.65E−10 | |
| rs7600883 | Intergenic | 2:48905331 | G | A | 0.06 | HIST1H4B | 1.023 | 1.65E−10 | |
| rs10170423 | Intergenic | 2:48906784 | G | A | 0.06 | HIST1H4B | 1.023 | 1.65E−10 | |
| rs10208418 | Intergenic | 2:48912244 | A | T | 0.05 | HIST1H4B | 1.079 | 3.82E−11 | |
| rs10208981 | Intergenic | 2:48912890 | G | T | 0.05 | HIST1H4B | 1.049 | 8.60E−11 | |
| rs79335804 | Intergenic | 2:48915424 | T | C | 0.05 | HIST1H4B | 1.079 | 3.82E−11 | |
| rs13430743 | Intergenic | 2:48916396 | T | G | 0.05 | HIST1H4B | 1.079 | 3.82E−11 | |
| rs13429699 | Intergenic | 2:48921530 | G | C | 0.05 | HIST1H4B | 1.049 | 8.60E−11 | |
| rs13432631 | Intergenic | 2:48921948 | A | C | 0.05 | HIST1H4B | 1.108 | 1.78E−11 | |
| rs13385591 | Intergenic | 2:48923227 | T | C | 0.05 | HIST1H4B | 1.079 | 3.82E−11 | |
| rs7602404 | Intergenic | 2:48923414 | A | G | 0.05 | HIST1H4B | 1.049 | 8.60E−11 | |
| rs142402253 | Intergenic | 2:48924921 | A | G | 0.05 | HIST1H4B | 1.049 | 8.60E−11 | |
| rs189340111 | Intergenic | 2:68450091 | G | A | 0.06 | HIST1H4B | 0.995 | 2.99E−12 | |
| HIST1H1B | 2.319 | 2.91E−11 | |||||||
| HIST1H1E | 2.044 | 6.90E−11 | |||||||
| HIST1H3J | 1.044 | 8.17E−11 | |||||||
| HIST1H2AH | 0.745 | 2.34E−10 | |||||||
| HIST1H2BM | 1.027 | 2.58E−10 | |||||||
| rs75372391 | LRP1B (intron 2) | 2:141415941 | G | A | 0.05 | HIST1H2BM | 1.268 | 6.19E−12 | |
| HIST1H4B | 1.102 | 1.39E−11 | |||||||
| HIST1H1B | 2.645 | 2.92E−11 | |||||||
| HIST1H3B | 2.016 | 2.00E−10 | |||||||
| rs79503131 | LRP1B (intron 2) | 2:141417401 | T | C | 0.05 | HIST1H2BM | 1.268 | 6.19E−12 | |
| HIST1H4B | 1.102 | 1.39E−11 | |||||||
| HIST1H1B | 2.645 | 2.92E−11 | |||||||
| HIST1H3B | 2.016 | 2.00E−10 | |||||||
| rs76908315 | LRP1B (intron 2) | 2:141428575 | G | A | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs75358328 | LRP1B (intron 2) | 2:141435763 | C | T | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs79931187 | LRP1B (intron 2) | 2:141440679 | C | T | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs114273249 | LRP1B (intron 2) | 2:141441139 | T | C | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs74871811 | LRP1B (intron 2) | 2:141441981 | T | A | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs114288750 | LRP1B (intron 2) | 2:141442588 | A | C | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs137985802 | LRP1B (intron 2) | 2:141442673 | T | C | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs114372180 | LRP1B (intron 2) | 2:141442782 | A | G | 0.05 | HIST1H2BM | 1.246 | 9.09E−12 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.590 | 4.75E−11 | |||||||
| rs75148923 | LRP1B (intron 2) | 2:141454893 | T | C | 0.05 | HIST1H2BM | 1.236 | 1.33E−11 | |
| HIST1H4B | 1.080 | 2.15E−11 | |||||||
| HIST1H1B | 2.588 | 4.96E−11 | |||||||
| rs72853591 | LRP1B (intron 2) | OC | 2:141557777 | C | T | 0.06 | HIST1H4B | 1.015 | 9.68E−11 |
| rs73963540 | LRP1B (intron 2) | 2:141710041 | T | C | 0.05 | HIST1H1B | 2.975 | 1.62E−14 | |
| HIST1H2BM | 1.369 | 3.12E−14 | |||||||
| HIST1H3B | 2.277 | 1.75E−13 | |||||||
| HIST1H4B | 1.154 | 4.90E−13 | |||||||
| HIST1H2AB | 0.310 | 8.94E−13 | |||||||
| HIST1H3J | 1.273 | 1.28E−12 | |||||||
| HIST1H1E | 2.439 | 3.33E−12 | |||||||
| HIST1H2AJ | 1.711 | 4.01E−12 | |||||||
| HIST1H2AL | 0.552 | 9.35E−12 | |||||||
| HIST2H2AB | 0.263 | 1.43E−11 | |||||||
| HIST2H3C | 0.609 | 4.10E−11 | |||||||
| HIST1H4K | 1.649 | 4.54E−11 | |||||||
| HIST1H4J | 2.204 | 4.67E−11 | |||||||
| HIST1H3H | 1.277 | 5.05E−11 | |||||||
| HIST1H3I | 0.574 | 6.66E−11 | |||||||
| HIST3H2BB | 0.268 | 6.82E−11 | |||||||
| HIST1H4A | 0.474 | 1.04E−10 | |||||||
| rs17801458 | LRP1B (intron 2) | 2:141710903 | T | C | 0.05 | HIST1H1B | 2.975 | 1.62E−14 | |
| HIST1H2BM | 1.369 | 3.12E−14 | |||||||
| HIST1H3B | 2.277 | 1.75E−13 | |||||||
| HIST1H4B | 1.154 | 4.90E−13 | |||||||
| HIST1H2AB | 0.310 | 8.94E−13 | |||||||
| HIST1H3J | 1.273 | 1.28E−12 | |||||||
| HIST1H1E | 2.439 | 3.33E−12 | |||||||
| HIST1H2AJ | 1.711 | 4.01E−12 | |||||||
| HIST1H2AL | 0.552 | 9.35E−12 | |||||||
| HIST2H2AB | 0.263 | 1.43E−11 | |||||||
| HIST2H3C | 0.609 | 4.10E−11 | |||||||
| HIST1H4K | 1.649 | 4.54E−11 | |||||||
| HIST1H4J | 2.204 | 4.67E−11 | |||||||
| HIST1H3H | 1.277 | 5.05E−11 | |||||||
| HIST1H3I | 0.574 | 6.66E−11 | |||||||
| HIST3H2BB | 0.268 | 6.82E−11 | |||||||
| HIST1H4A | 0.474 | 1.04E−10 | |||||||
| rs7570091 | LRP1B (intron 2) | 2:141714131 | G | T | 0.05 | HIST1H2BM | 1.297 | 1.36E−13 | |
| HIST1H1B | 2.787 | 1.48E−13 | |||||||
| HIST1H3B | 2.132 | 1.34E−12 | |||||||
| HIST1H2AL | 0.543 | 4.65E−12 | |||||||
| HIST1H4B | 1.074 | 4.79E−12 | |||||||
| HIST1H3J | 1.201 | 5.93E−12 | |||||||
| HIST1H2AB | 0.289 | 7.28E−12 | |||||||
| HIST1H2AJ | 1.628 | 1.12E−11 | |||||||
| HIST2H2AB | 0.256 | 1.13E−11 | |||||||
| HIST3H2BB | 0.267 | 1.89E−11 | |||||||
| HIST1H1E | 2.277 | 2.38E−11 | |||||||
| HIST2H3C | 0.583 | 7.77E−11 | |||||||
| HIST1H3I | 0.545 | 1.78E−10 | |||||||
| HIST1H3H | 1.199 | 2.26E−10 | |||||||
| rs11684178 | LRP1B (intron 2) | 2:141788340 | C | G | 0.06 | HIST1H2BM | 1.090 | 6.71E−12 | |
| HIST1H1B | 2.283 | 2.56E−11 | |||||||
| HIST1H3B | 1.778 | 6.72E−11 | |||||||
| HIST2H2AB | 0.221 | 1.04E−10 | |||||||
| HIST1H3J | 1.019 | 1.10E−10 | |||||||
| HIST1H2AL | 0.455 | 1.67E−10 | |||||||
| rs6718590 | LRP1B (intron 1) | 2:141817609 | G | C | 0.07 | HIST1H2BM | 0.968 | 1.15E−10 | |
| rs111875224 | Intergenic | 2:202309382 | A | C | 0.05 | HIST1H2AM | 0.594 | 1.24E−11 | |
| HIST1H4B | 1.023 | 8.51E−11 | |||||||
| rs849583 | NRP2 (intron 7) | 2:205737518 | A | G | 0.08 | HIST1H4A | 0.398 | 8.66E−11 | |
| rs849578# | NRP2 (intron 7) | 2:205738798 | T | G | 0.08 | HIST1H4A | 0.390 | 1.69E−10 | |
| rs849577 | NRP2 (intron 7) | 2:205739705 | A | G | 0.07 | HIST1H4A | 0.407 | 4.48E−11 | |
| rs849573 | NRP2 (intron 8) | 2:205741099 | T | C | 0.08 | HIST1H4A | 0.407 | 4.48E−11 | |
| rs849572 | NRP2 (intron 8) | 2:205741317 | A | G | 0.08 | HIST1H4A | 0.398 | 8.98E−11 | |
| rs849567 | NRP2 (intron 9) | 2:205744540 | T | C | 0.07 | HIST1H4A | 0.403 | 7.11E−11 | |
| rs830612 | Intergenic | 3:71634179 | C | T | 0.08 | HIST1H4B | 0.889 | 1.28E−10 | |
| rs150958961 | Intergenic | 4:9006930 | C | A | 0.1 | HIST1H4A | 0.341 | 8.77E−11 | |
| rs148619316 | Intergenic | 5:14080602 | T | C | 0.06 | HIST1H4B | 1.006 | 4.81E−11 | |
| rs1992368 | Intergenic | 5:14092868 | A | G | 0.06 | HIST1H4B | 0.974 | 9.25E−11 | |
| rs55989629 | Intergenic | 5:14093354 | T | C | 0.06 | HIST1H4B | 0.974 | 9.25E−11 | |
| rs11956277 | Intergenic | 5:14095038 | T | C | 0.06 | HIST1H4B | 0.974 | 9.25E−11 | |
| rs66689811 | Intergenic | 5:14095266 | T | C | 0.06 | HIST1H4B | 0.974 | 9.25E−11 | |
| rs6863067 | Intergenic | 5:14098914 | A | C | 0.06 | HIST1H4B | 0.974 | 9.25E−11 | |
| rs10041963 | Intergenic | 5:14099801 | A | G | 0.06 | HIST1H4B | 0.974 | 9.25E−11 | |
| rs10072716 | Intergenic | 5:14101222 | T | C | 0.06 | HIST1H4B | 0.974 | 9.25E−11 | |
| rs6861945 | Intergenic | 5:53398953 | A | G | 0.11 | HIST1H3J | 0.780 | 2.75E−10 | |
| rs61564830 | Intergenic | 6:129768950 | T | C | 0.06 | HIST1H3J | 1.036 | 2.62E−10 | |
| rs144499291 | UTRN (intron 22) | 6:144471095 | G | A | 0.06 | HIST1H4A | 0.461 | 2.88E−10 | |
| rs71560740 | Intergenic | 7:85896218 | C | T | 0.06 | HIST1H2AJ | 1.491 | 1.48E−10 | |
| HIST1H3A | 0.228 | 2.41E−10 | |||||||
| rs118052517 | Intergenic | 7:153287269 | C | T | 0.06 | HIST1H3J | 1.234 | 8.16E−13 | |
| HIST1H4A | 0.494 | 1.89E−12 | |||||||
| HIST1H2BM | 1.222 | 2.40E−12 | |||||||
| HIST1H2AJ | 1.648 | 3.64E−12 | |||||||
| HIST1H3B | 2.052 | 6.16E−12 | |||||||
| HIST1H1B | 2.540 | 1.39E−11 | |||||||
| HIST1H1E | 2.269 | 1.78E−11 | |||||||
| HIST1H2AB | 0.270 | 1.16E−10 | |||||||
| HIST1H4K | 1.532 | 2.14E−10 | |||||||
| HIST1H4J | 2.039 | 2.62E−10 | |||||||
| HIST1H3I | 0.534 | 2.77E−10 | |||||||
| HIST1H3H | 1.180 | 2.95E−10 | |||||||
| rs77957383 | Intergenic | 9:1715766 | T | C | 0.07 | HIST1H4B | 0.894 | 1.85E−10 | |
| rs113936625 | Intergenic | 10:111124417 | C | G | 0.05 | HIST1H4B | 1.056 | 2.76E−10 | |
| rs75201173 | Intergenic | CTCF | 11:1062030 | C | A | 0.07 | HIST1H2BM | 1.092 | 1.45E−11 |
| HIST2H3C | 0.529 | 1.22E−10 | |||||||
| HIST1H1B | 2.227 | 1.75E−10 | |||||||
| HIST1H3B | 1.766 | 1.94E−10 | |||||||
| rs11245920 | Intergenic | CTCF | 11:1062519 | C | T | 0.07 | HIST1H2BM | 1.092 | 1.45E−11 |
| HIST2H3C | 0.529 | 1.22E−10 | |||||||
| HIST1H1B | 2.227 | 1.75E−10 | |||||||
| HIST1H3B | 1.766 | 1.94E−10 | |||||||
| rs111452409 | Intergenic | 11:1064929 | G | A | 0.07 | HIST1H2BM | 1.092 | 1.45E−11 | |
| HIST2H3C | 0.529 | 1.22E−10 | |||||||
| HIST1H1B | 2.227 | 1.75E−10 | |||||||
| HIST1H3B | 1.766 | 1.94E−10 | |||||||
| rs113659777 | MUC2 (intron 2) | 11:1088149 | A | G | 0.07 | HIST1H2BM | 1.101 | 2.89E−11 | |
| HIST1H4B | 0.951 | 8.17E−11 | |||||||
| rs184239416 | MUC2 (intron 2) | 11:1088154 | G | A | 0.06 | HIST1H2BM | 1.262 | 1.56E−12 | |
| HIST1H4B | 1.077 | 9.13E−12 | |||||||
| HIST1H1B | 2.566 | 2.84E−11 | |||||||
| HIST1H3J | 1.174 | 3.88E−11 | |||||||
| HIST1H3B | 1.979 | 1.17E−10 | |||||||
| HIST1H1E | 2.206 | 2.06E−10 | |||||||
| HIST3H2BB | 0.257 | 2.37E−10 | |||||||
| rs41392245 | MUC2 (intron 2) | 11:1089095 | T | C | 0.07 | HIST1H2BM | 1.068 | 4.19E−11 | |
| HIST2H3C | 0.528 | 1.34E−10 | |||||||
| HIST1H3B | 1.761 | 2.23E−10 | |||||||
| HIST1H1B | 2.201 | 2.90E−10 | |||||||
| rs11033879 | MMP26 (intron 1) | 11:4786997 | G | A | 0.05 | HIST1H2AJ | 1.711 | 2.53E−13 | |
| HIST1H1E | 2.374 | 9.71E−13 | |||||||
| HIST1H3B | 2.102 | 9.80E−13 | |||||||
| HIST2H2AB | 0.263 | 1.12E−12 | |||||||
| HIST1H1B | 2.635 | 1.18E−12 | |||||||
| HIST1H4A | 0.490 | 1.62E−12 | |||||||
| HIST1H2BM | 1.195 | 4.53E−12 | |||||||
| HIST2H3C | 0.594 | 1.15E−11 | |||||||
| HIST1H3J | 1.148 | 2.08E−11 | |||||||
| HIST1H4K | 1.593 | 2.16E−11 | |||||||
| HIST1H2AB | 0.275 | 3.14E−11 | |||||||
| HIST1H4J | 2.095 | 4.85E−11 | |||||||
| HIST1H3I | 0.544 | 7.46E−11 | |||||||
| HIST1H3H | 1.190 | 1.28E−10 | |||||||
| HIST1H2AH | 0.793 | 2.70E−10 | |||||||
| rs80143982 | Intergenic | 11:110990257 | A | G | 0.05 | HIST1H4A | 0.482 | 2.65E−10 | |
| rs75349221 | Intergenic | 14:44176974 | T | A | 0.05 | HIST1H4A | 0.471 | 2.75E−10 | |
| rs72846811 | RBFOX3 (intron 3) | 17:79231229 | A | G | 0.06 | HIST1H4A | 0.489 | 1.90E−11 | |
| rs183898825 | TAF4B (intron 9) | 18:26303160 | C | A | 0.05 | HIST1H4A | 0.466 | 2.60E−10 | |
| rs57185274 | CACNA1A (intron 3) | 19:13406972 | A | T | 0.05 | HIST1H4B | 1.034 | 2.04E−10 | |
- aSNP in bold indicates the representative variant in each linkage disequilibrium block. SNP with superscript “#” was associated with autism in a Chinese population [15]. bThe effects were predicted using Ensembl Variant Effect Predictor program [11]. OC: open chromatin region; CTCF: CTCF binding site. cChromosome: base pair position from GRCh38.p2. dA1: minor allele; A2: major allele. SNP: single-nucleotide polymorphism; MAF: minor allele frequency.
Transcriptome-wide association analysis revealed 80 additional genes associated with the 31 identified eQTLs (P < 4.75 × 10−6; Table 2). The genes encoding ribonuclease P RNA component H1 (RPPH1) and some small Cajal body-specific RNAs (scaRNAs), in particular, were associated with more than half of the eQTLs (>15 eQTLs; Table 2).
| eQTL | Gene | P | Gene | P | Gene | P | Gene | P |
|---|---|---|---|---|---|---|---|---|
| rs10919229 | HIST1H4B (6) | 2.51E−10 | HIST1H3J (6) | 3.40E−08 | HIST1H3H (6) | 2.02E−07 | SASH3 (X) | 2.89E−06 |
| HIST1H2BM (6) | 4.38E−09 | HIST1H3B (6) | 4.64E−08 | HIST1H1E (6) | 2.50E−07 | HIST1H2AH (6) | 4.61E−06 | |
| HIST1H1B (6) | 1.81E−08 | RPPH1 (14) | 5.82E−08 | HIST1H2AB (6) | 2.54E−07 | |||
| SCARNA13 (14) | 3.38E−08 | SCARNA12 (12) | 1.22E−07 | HIST1H2AM (6) | 9.76E−07 | |||
| rs191508159 | SNORA53 (12) | 4.38E−13 | SCARNA2 (1) | 5.28E−08 | HIST2H3C (1) | 1.41E−07 | HIST1H3H (6) | 5.46E−07 |
| HIST1H4A (6) | 3.41E−11 | HIST1H1E (6) | 7.02E−08 | HIST1H2AL (6) | 1.99E−07 | G3BP2 (4) | 5.51E−07 | |
| HIST1H2AJ (6) | 5.72E−09 | HIST2H2AB (1) | 7.35E−08 | HIST1H1B (6) | 2.67E−07 | HIST1H2AH (6) | 3.21E−06 | |
| HIST1H3J (6) | 3.33E−08 | HIST1H2AB (6) | 1.02E−07 | HIST1H4K (6) | 2.99E−07 | HIST1H3A (6) | 4.06E−06 | |
| HIST1H4J (6) | 4.63E−08 | HIST1H3B (6) | 1.14E−07 | HIST1H3I (6) | 5.12E−07 | HIST1H2BM (6) | 4.73E−06 | |
| rs13432631 | HIST1H4B (6) | 1.78E−11 | HIST1H3B (6) | 1.13E−08 | HIST1H2AJ (6) | 1.86E−07 | HIST1H2AH (6) | 6.84E−07 |
| SCARNA12 (12) | 1.70E−10 | HIST1H2AB (6) | 2.83E−08 | HIST3H2BB (1) | 1.91E−07 | RPPH1 (14) | 7.69E−07 | |
| HIST1H2BM (6) | 1.95E−09 | HIST1H3I (6) | 1.01E−07 | HIST1H3H (6) | 2.56E−07 | HIST1H3A (6) | 2.10E−06 | |
| HIST1H1B (6) | 3.33E−09 | SCARNA13 (14) | 1.23E−07 | HIST1H4K (6) | 3.45E−07 | HIST1H4J (6) | 2.48E−06 | |
| HIST1H3J (6) | 1.11E−08 | HIST1H1E (6) | 1.74E−07 | HIST2H2AB (1) | 6.28E−07 | HIST1H4A (6) | 2.81E−06 | |
| rs189340111 | HIST1H4B (6) | 2.99E−12 | HIST1H3I (6) | 1.73E−09 | HIST1H4J (6) | 3.30E−08 | SCARNA2 (1) | 4.27E−07 |
| HIST1H1B (6) | 2.91E−11 | HIST2H2AB (1) | 1.78E−09 | HIST1H3A (6) | 3.60E−08 | PIGF (2) | 6.15E−07 | |
| HIST1H1E (6) | 6.90E−11 | HIST1H2AJ (6) | 2.58E−09 | HIST1H4A (6) | 7.70E−08 | SCARNA13 (14) | 1.55E−06 | |
| HIST1H3J (6) | 8.17E−11 | HIST1H3H (6) | 3.93E−09 | HIST1H2AM (6) | 1.00E−07 | RPPH1 (14) | 1.91E−06 | |
| HIST1H2AH (6) | 2.34E−10 | HIST1H4K (6) | 9.71E−09 | SCARNA12 (12) | 1.05E−07 | HIST1H2AL (6) | 2.40E−06 | |
| HIST1H2BM (6) | 2.58E−10 | HIST1H2AB (6) | 1.81E−08 | SCARNA7 (3) | 1.81E−07 | |||
| HIST1H3B (6) | 4.62E−10 | HIST3H2BB (1) | 3.14E−08 | HIST2H3C (1) | 2.90E−07 | |||
| rs75372391 | HIST1H2BM (6) | 6.19E−12 | HIST1H3I (6) | 5.56E−09 | HIST1H4A (6) | 3.57E−08 | HIST1H2AG (6) | 1.77E−07 |
| HIST1H4B (6) | 1.39E−11 | HIST1H2AJ (6) | 6.33E−09 | HIST1H2AH (6) | 5.13E−08 | HIST1H2AK (6) | 6.45E−07 | |
| HIST1H1B (6) | 2.92E−11 | HIST1H2AB (6) | 7.88E−09 | HIST1H2AM (6) | 5.45E−08 | SCARNA7 (3) | 1.95E−06 | |
| RPPH1 (14) | 1.53E−10 | HIST3H2BB (1) | 9.94E−09 | HIST1H4J (6) | 5.90E−08 | HIST1H3A (6) | 2.31E−06 | |
| HIST1H3B (6) | 2.00E−10 | HIST1H3H (6) | 1.10E−08 | HIST2H3C (1) | 9.35E−08 | SCARNA2 (1) | 2.85E−06 | |
| HIST1H1E (6) | 6.26E−10 | HIST2H2AB (1) | 2.33E−08 | SCARNA12 (12) | 9.42E−08 | |||
| HIST1H3J (6) | 1.15E−09 | HIST1H4K (6) | 3.21E−08 | HIST1H2AL (6) | 1.09E−07 | |||
| rs72853591 | HIST1H4B (6) | 9.68E−11 | SCARNA13 (14) | 1.03E−08 | SCARNA12 (12) | 2.12E−07 | HIST1H2AH (6) | 2.69E−06 |
| HIST1H1B (6) | 5.88E−10 | HIST1H3J (6) | 1.17E−08 | HIST2H2AB (1) | 4.57E−07 | HIST1H2AL (6) | 2.72E−06 | |
| HIST1H2BM (6) | 6.02E−10 | HIST1H1E (6) | 1.88E−08 | HIST1H4A (6) | 1.16E−06 | HIST1H4J (6) | 3.52E−06 | |
| RPPH1 (14) | 1.96E−09 | HIST1H3H (6) | 1.56E−07 | HIST1H4K (6) | 1.57E−06 | HIST1H3A (6) | 4.33E−06 | |
| HIST1H3B (6) | 4.11E−09 | HIST1H2AJ (6) | 1.74E−07 | SNORD17 (20) | 1.66E−06 | HIST2H3C (1) | 4.67E−06 | |
| HIST1H3I (6) | 8.43E−09 | HIST1H2AB (6) | 1.84E−07 | SCARNA2 (1) | 2.24E−06 | |||
| rs17801458 | RPPH1 (14) | 1.31E−15 | HIST1H2AL (6) | 9.35E−12 | SCARNA2 (1) | 9.35E−10 | HIST1H2AK (6) | 1.75E−06 |
| HIST1H1B (6) | 1.62E−14 | HIST2H2AB (1) | 1.43E−11 | HIST1H3A (6) | 5.23E−09 | VGLL4 (3) | 1.88E−06 | |
| HIST1H2BM (6) | 3.12E−14 | HIST2H3C (1) | 4.10E−11 | HIST1H2AG (6) | 6.96E−09 | SNORA53 (12) | 2.09E−06 | |
| HIST1H3B (6) | 1.75E−13 | HIST1H4K (6) | 4.54E−11 | HIST1H2AH (6) | 1.34E−08 | WASF2 (1) | 3.49E−06 | |
| HIST1H4B (6) | 4.90E−13 | HIST1H4J (6) | 4.67E−11 | SNORD17 (20) | 2.36E−08 | UBE2R2 (9) | 4.47E−06 | |
| HIST1H2AB (6) | 8.94E−13 | HIST1H3H (6) | 5.05E−11 | SCARNA12 (12) | 2.60E−08 | |||
| HIST1H3J (6) | 1.28E−12 | HIST1H3I (6) | 6.66E−11 | SCARNA7 (3) | 1.91E−07 | |||
| HIST1H1E (6) | 3.33E−12 | HIST3H2BB (1) | 6.82E−11 | SCARNA13 (14) | 4.53E−07 | |||
| HIST1H2AJ (6) | 4.01E−12 | HIST1H4A (6) | 1.04E−10 | HIST1H2AM (6) | 1.16E−06 | |||
| rs11684178 | HIST1H2BM (6) | 6.71E−12 | HIST1H1E (6) | 3.59E−10 | HIST1H3A (6) | 2.78E−09 | NOL11 (17) | 4.61E−07 |
| RPPH1 (14) | 7.92E−12 | HIST1H4J (6) | 3.64E−10 | HIST2H3C (1) | 3.55E−09 | SCARNA12 (12) | 8.16E−07 | |
| HIST1H1B (6) | 2.56E−11 | HIST1H4B (6) | 3.69E−10 | HIST3H2BB (1) | 9.84E−09 | SCARNA13 (14) | 1.09E−06 | |
| HIST1H3B (6) | 6.72E−11 | HIST1H2AB (6) | 4.40E−10 | HIST1H4A (6) | 2.03E−08 | DDX17 (22) | 1.32E−06 | |
| HIST2H2AB (1) | 1.04E−10 | HIST1H2AH (6) | 7.01E−10 | HIST1H4K (6) | 3.26E−08 | MRPS24 (7) | 3.22E−06 | |
| HIST1H3J (6) | 1.10E−10 | HIST1H2AJ (6) | 8.58E−10 | SCARNA7 (3) | 3.37E−08 | FASTKD2 (2) | 3.42E−06 | |
| HIST1H2AL (6) | 1.67E−10 | HIST1H3H (6) | 1.56E−09 | HIST1H2AG (6) | 3.80E−08 | SNORD17 (20) | 3.99E−06 | |
| HIST1H3I (6) | 3.41E−10 | SCARNA2 (1) | 2.47E−09 | HIST1H2AK (6) | 7.75E−08 | EFTUD2 (17) | 4.04E−06 | |
| rs6718590 | HIST1H2BM (6) | 1.15E−10 | HIST1H1E (6) | 2.68E−09 | HIST1H3H (6) | 1.57E−08 | NOL11 (17) | 4.12E−07 |
| RPPH1 (14) | 1.92E−10 | HIST1H3I (6) | 3.57E−09 | HIST1H3A (6) | 1.98E−08 | SCARNA12 (12) | 1.10E−06 | |
| HIST1H1B (6) | 3.63E−10 | HIST1H4B (6) | 4.90E−09 | SCARNA2 (1) | 2.30E−08 | APPBP2 (17) | 1.77E−06 | |
| HIST1H2AL (6) | 4.03E−10 | HIST1H2AB (6) | 6.23E−09 | HIST1H2AG (6) | 8.36E−08 | ATP5A1 (18) | 3.68E−06 | |
| HIST2H2AB (1) | 5.83E−10 | HIST1H2AH (6) | 6.89E−09 | HIST1H4A (6) | 1.92E−07 | PITPNB (22) | 3.92E−06 | |
| HIST1H3J (6) | 9.78E−10 | HIST1H4J (6) | 9.48E−09 | SCARNA7 (3) | 2.51E−07 | |||
| HIST3H2BB (1) | 1.30E−09 | HIST2H3C (1) | 1.01E−08 | HIST1H4K (6) | 3.19E−07 | |||
| HIST1H3B (6) | 1.74E−09 | HIST1H2AJ (6) | 1.10E−08 | HIST1H2AK (6) | 3.22E−07 | |||
| rs111875224 | HIST1H2AM (6) | 1.24E−11 | SCARNA7 (3) | 2.79E−08 | HIST1H3H (6) | 2.63E−07 | GTF2E2 (8) | 2.28E−06 |
| HIST1H4B (6) | 8.51E−11 | HIST3H2BB (1) | 3.71E−08 | SCARNA2 (1) | 3.89E−07 | C18orf32 (18) | 2.68E−06 | |
| HIST1H1B (6) | 7.63E−10 | HIST1H3J (6) | 9.07E−08 | HIST1H2AJ (6) | 4.51E−07 | CD63 (12) | 2.80E−06 | |
| HIST1H1E (6) | 1.11E−09 | SCARNA12 (12) | 1.02E−07 | SCARNA13 (14) | 4.99E−07 | SERINC1 (6) | 3.24E−06 | |
| HIST1H2BM (6) | 1.41E−09 | HIST1H3I (6) | 1.14E−07 | HIST1H3A (6) | 9.59E−07 | |||
| HIST1H3B (6) | 1.41E−08 | HIST1H2AB (6) | 1.32E−07 | HIST1H2AL (6) | 1.60E−06 | |||
| HIST1H2AH (6) | 2.27E−08 | HIST2H2AB (1) | 1.38E−07 | RPPH1 (14) | 1.87E−06 | |||
| rs849577 | HIST1H4A (6) | 4.48E−11 | HIST1H1E (6) | 3.42E−08 | HIST1H3I (6) | 4.33E−07 | HIST1H3A (6) | 2.03E−06 |
| SNORA53 (12) | 3.16E−10 | HIST2H2AB (1) | 4.00E−08 | HIST1H1B (6) | 5.65E−07 | HIST1H3H (6) | 2.06E−06 | |
| HIST1H4J (6) | 8.79E−10 | HIST1H3B (6) | 8.53E−08 | HIST1H3J (6) | 6.70E−07 | RPPH1 (14) | 3.22E−06 | |
| SCARNA2 (1) | 1.23E−09 | HIST1H2AL (6) | 1.35E−07 | HIST1H2AH (6) | 6.75E−07 | HIST1H2BM (6) | 4.42E−06 | |
| HIST1H4K (6) | 2.46E−09 | HIST2H3C (1) | 2.09E−07 | SCARNA7 (3) | 1.71E−06 | |||
| HIST1H2AJ (6) | 1.12E−08 | HIST1H2AB (6) | 3.52E−07 | SNORD17 (20) | 1.77E−06 | |||
| rs830612 | HIST1H4B (6) | 1.28E−10 | HIST1H3B (6) | 1.16E−08 | SNORD17 (20) | 1.52E−07 | RPPH1 (14) | 6.10E−07 |
| HIST1H1B (6) | 6.19E−10 | HIST1H3H (6) | 1.67E−08 | SASH3 (X) | 1.74E−07 | HIST1H2AJ (6) | 6.67E−07 | |
| HIST1H2BM (6) | 2.66E−09 | HIST1H1E (6) | 2.00E−08 | HIST1H2AH (6) | 2.54E−07 | SCARNA7 (3) | 1.56E−06 | |
| HIST1H3J (6) | 1.05E−08 | HIST1H3I (6) | 4.17E−08 | HIST1H3A (6) | 3.80E−07 | HIST1H2AB (6) | 3.15E−06 | |
| rs150958961 | HIST1H4A (6) | 8.77E−11 | HIST1H1B (6) | 4.03E−09 | HIST1H4K (6) | 3.84E−08 | HIST2H2AB (1) | 4.70E−07 |
| HIST1H3J (6) | 3.41E−10 | SCARNA2 (1) | 6.96E−09 | HIST1H3H (6) | 4.81E−08 | SCARNA7 (3) | 5.49E−07 | |
| HIST1H2AJ (6) | 3.78E−10 | HIST1H2AH (6) | 1.72E−08 | HIST1H3I (6) | 1.01E−07 | HIST1H3A (6) | 1.04E−06 | |
| HIST1H2AB (6) | 3.81E−10 | HIST1H2AL (6) | 1.86E−08 | HIST1H4B (6) | 1.38E−07 | SNORA57 (11) | 1.66E−06 | |
| HIST1H3B (6) | 4.54E−10 | HIST1H2BM (6) | 2.30E−08 | HIST3H2BB (1) | 1.62E−07 | HIST1H2AG (6) | 2.46E−06 | |
| RPPH1 (14) | 7.11E−10 | HIST1H4J (6) | 2.31E−08 | SNORA53 (12) | 4.22E−07 | |||
| HIST1H1E (6) | 1.80E−09 | HIST2H3C (1) | 2.57E−08 | SNORD17 (20) | 4.60E−07 | |||
| rs148619316 | HIST1H4B (6) | 4.81E−11 | HIST1H3H (6) | 6.81E−08 | HIST1H2AJ (6) | 1.23E−06 | PPM1G (2) | 2.97E−06 |
| RPPH1 (14) | 3.50E−09 | HIST1H1E (6) | 1.11E−07 | HIST1H3I (6) | 1.33E−06 | THRAP3 (1) | 3.14E−06 | |
| HIST1H1B (6) | 4.92E−09 | HIST1H2AH (6) | 1.66E−07 | SNORD17 (20) | 1.52E−06 | SUPT5H (19) | 3.91E−06 | |
| HIST1H3J (6) | 5.46E−09 | DDX28 (16) | 5.22E−07 | SCARNA12 (12) | 1.68E−06 | ZNF207 (17) | 4.48E−06 | |
| HIST1H3B (6) | 9.95E−09 | HIST1H2AB (6) | 6.80E−07 | HIST1H2AK (6) | 1.69E−06 | |||
| HIST1H2BM (6) | 1.70E−08 | HIST2H2AB (1) | 1.14E−06 | C20orf11 (20) | 2.61E−06 | |||
| SASH3 (X) | 3.00E−08 | HIST3H2BB (1) | 1.22E−06 | SCARNA7 (3) | 2.67E−06 | |||
| rs6861945 | HIST1H3J (6) | 2.75E−10 | HIST1H4J (6) | 1.67E−08 | HIST1H3I (6) | 7.68E−08 | HIST1H2AK (6) | 1.18E−06 |
| HIST1H3B (6) | 4.30E−09 | HIST1H1E (6) | 2.30E−08 | HIST1H2AB (6) | 1.38E−07 | HIST2H2AB (1) | 1.32E−06 | |
| HIST1H4B (6) | 4.44E−09 | HIST1H4K (6) | 3.40E−08 | SCARNA2 (1) | 1.55E−07 | SCARNA7 (3) | 2.25E−06 | |
| HIST1H2BM (6) | 7.98E−09 | HIST1H2AH (6) | 3.78E−08 | HIST1H4A (6) | 1.55E−07 | SNORD17 (20) | 2.57E−06 | |
| HIST1H1B (6) | 1.04E−08 | HIST1H3H (6) | 4.52E−08 | SASH3 (X) | 4.33E−07 | HIST1H2AL (6) | 4.34E−06 | |
| RPPH1 (14) | 1.11E−08 | HIST1H2AJ (6) | 5.09E−08 | HIST1H3A (6) | 8.63E−07 | |||
| rs61564830 | HIST1H3J (6) | 2.62E−10 | SCARNA2 (1) | 5.85E−09 | HIST1H3I (6) | 3.37E−08 | HIST1H2AB (6) | 2.54E−06 |
| HIST1H2AH (6) | 8.25E−10 | HIST1H1B (6) | 6.35E−09 | HIST1H2BM (6) | 3.45E−08 | HIST1H2AL (6) | 2.85E−06 | |
| HIST1H1E (6) | 9.74E−10 | HIST1H2AJ (6) | 1.00E−08 | SCARNA13 (14) | 1.68E−07 | UBE2R2 (9) | 4.53E−06 | |
| HIST1H3B (6) | 1.50E−09 | SNORD17 (20) | 1.23E−08 | RPPH1 (14) | 3.47E−07 | |||
| HIST1H3H (6) | 1.58E−09 | SCARNA7 (3) | 2.46E−08 | HIST2H2AB (1) | 2.23E−06 | |||
| HIST1H4B (6) | 2.05E−09 | HIST1H4A (6) | 2.75E−08 | MRPS31 (13) | 2.36E−06 | |||
| rs144499291 | HIST1H4A (6) | 2.88E−10 | HIST1H2AK (6) | 4.16E−09 | HIST1H4K (6) | 9.65E−08 | FAM122A (9) | 2.05E−06 |
| HIST1H1B (6) | 4.57E−10 | HIST1H3H (6) | 5.46E−09 | KPNB1 (17) | 1.48E−07 | RPPH1 (14) | 3.45E−06 | |
| HIST1H2AJ (6) | 7.99E−10 | HIST1H3I (6) | 1.06E−08 | SNORD17 (20) | 1.49E−07 | HIST3H2BB (1) | 4.06E−06 | |
| HIST1H1E (6) | 9.23E−10 | HIST1H2AB (6) | 1.12E−08 | HIST2H3C (1) | 2.10E−07 | |||
| HIST1H2BM (6) | 1.15E−09 | HIST2H2AB (1) | 3.99E−08 | HIST1H4J (6) | 2.73E−07 | |||
| HIST1H3J (6) | 2.45E−09 | HIST1H4B (6) | 4.58E−08 | SCARNA7 (3) | 2.83E−07 | |||
| HIST1H3B (6) | 2.85E−09 | SCARNA2 (1) | 5.61E−08 | HIST1H2AL (6) | 4.25E−07 | |||
| HIST1H2AH (6) | 4.05E−09 | SNORA53 (12) | 9.55E−08 | HIST1H3A (6) | 5.03E−07 | |||
| rs71560740 | HIST1H2AJ (6) | 1.48E−10 | HIST1H3I (6) | 6.75E−09 | RPPH1 (14) | 1.71E−08 | HIST1H2AH (6) | 4.57E−07 |
| HIST1H3A (6) | 2.41E−10 | HIST2H2AB (1) | 9.23E−09 | SNORA53 (12) | 2.32E−08 | HIST1H3H (6) | 9.55E−07 | |
| HIST1H2AB (6) | 8.54E−10 | HIST1H4J (6) | 9.80E−09 | HIST1H3J (6) | 2.54E−08 | TDRD12 (19) | 3.26E−06 | |
| HIST2H3C (1) | 1.32E−09 | HIST1H2BM (6) | 1.13E−08 | HIST1H1E (6) | 3.18E−08 | HIST1H2AM (6) | 3.74E−06 | |
| HIST1H3B (6) | 3.46E−09 | HIST1H4A (6) | 1.27E−08 | HIST1H2AL (6) | 7.54E−08 | HIST1H4B (6) | 3.78E−06 | |
| HIST1H1B (6) | 6.55E−09 | HIST1H4K (6) | 1.29E−08 | SCARNA2 (1) | 2.35E−07 | |||
| rs118052517 | HIST1H3J (6) | 8.16E−13 | HIST1H4K (6) | 2.14E−10 | HIST1H4B (6) | 2.72E−09 | HIST3H2BB (1) | 3.49E−08 |
| HIST1H4A (6) | 1.89E−12 | SCARNA2 (1) | 2.30E−10 | HIST1H2AH (6) | 2.81E−09 | LOXL4 (10) | 1.35E−06 | |
| HIST1H2BM (6) | 2.40E−12 | HIST1H4J (6) | 2.62E−10 | HIST2H3C (1) | 6.12E−09 | KRT8 (12) | 1.53E−06 | |
| HIST1H2AJ (6) | 3.64E−12 | HIST1H3I (6) | 2.77E−10 | SNORA53 (12) | 8.62E−09 | HIST1H2AM (6) | 1.63E−06 | |
| HIST1H3B (6) | 6.16E−12 | HIST1H3H (6) | 2.95E−10 | HIST1H3A (6) | 1.04E−08 | TRNP1 (1) | 1.82E−06 | |
| HIST1H1B (6) | 1.39E−11 | RPPH1 (14) | 1.53E−09 | HIST1H2AK (6) | 1.27E−08 | SCUBE2 (11) | 4.20E−06 | |
| HIST1H1E (6) | 1.78E−11 | HIST1H2AL (6) | 1.73E−09 | SNORD17 (20) | 1.61E−08 | |||
| HIST1H2AB (6) | 1.16E−10 | HIST2H2AB (1) | 2.34E−09 | SCARNA7 (3) | 3.31E−08 | |||
| rs77957383 | HIST1H4B (6) | 1.85E−10 | HIST3H2BB (1) | 4.38E−09 | SCARNA12 (12) | 1.82E−07 | CDC5L (6) | 3.27E−06 |
| RPPH1 (14) | 3.19E−10 | HIST1H1E (6) | 2.17E−08 | SCARNA2 (1) | 2.84E−07 | HIST2H3C (1) | 4.43E−06 | |
| HIST1H2BM (6) | 4.46E−10 | HIST2H2AB (1) | 2.20E−08 | HIST1H3H (6) | 7.97E−07 | |||
| HIST1H3B (6) | 1.04E−09 | HIST1H2AJ (6) | 2.95E−08 | HIST1H2AK (6) | 1.03E−06 | |||
| HIST1H1B (6) | 1.38E−09 | SCARNA7 (3) | 3.95E−08 | HIST1H4A (6) | 1.61E−06 | |||
| HIST1H3J (6) | 3.57E−09 | HIST1H3I (6) | 4.73E−08 | SCARNA13 (14) | 2.67E−06 | |||
| HIST1H2AB (6) | 3.98E−09 | HIST1H2AH (6) | 1.49E−07 | HIST1H2AL (6) | 3.22E−06 | |||
| rs113936625 | HIST1H4B (6) | 2.76E−10 | SCARNA13 (14) | 1.12E−07 | HIST1H3H (6) | 3.64E−07 | RPPH1 (14) | 2.97E−06 |
| HIST1H2BM (6) | 2.29E−09 | SCARNA12 (12) | 1.20E−07 | HIST1H2AJ (6) | 9.36E−07 | HIST1H2AL (6) | 3.00E−06 | |
| HIST1H1B (6) | 8.82E−09 | HIST1H3B (6) | 1.36E−07 | HIST2H2AB (1) | 1.25E−06 | HIST1H3A (6) | 3.44E−06 | |
| HIST1H2AM (6) | 1.42E−08 | HIST1H2AB (6) | 1.79E−07 | HIST1H3I (6) | 1.41E−06 | SNORD17 (20) | 3.63E−06 | |
| HIST1H3J (6) | 3.32E−08 | HIST1H1E (6) | 2.11E−07 | HIST1H2AH (6) | 2.45E−06 | |||
| rs75201173 | HIST1H2BM (6) | 1.45E−11 | HIST1H2AK (6) | 2.05E−09 | STYX (14) | 4.33E−07 | HMHA1 (19) | 2.49E−06 |
| RPPH1 (14) | 6.79E−11 | HIST1H2AB (6) | 3.62E−09 | PRKAR1A (17) | 4.54E−07 | OSBPL11 (3) | 2.57E−06 | |
| HIST2H3C (1) | 1.22E−10 | SCARNA2 (1) | 1.15E−08 | SMAD2 (18) | 5.85E−07 | FBXL12 (19) | 2.73E−06 | |
| HIST1H1B (6) | 1.75E−10 | HIST1H2AL (6) | 1.56E−08 | EFTUD2 (17) | 6.59E−07 | FNBP1 (9) | 2.92E−06 | |
| HIST1H3B (6) | 1.94E−10 | HIST2H2AB (1) | 3.33E−08 | ZFP91 (11) | 8.19E−07 | SLC44A2 (19) | 3.26E−06 | |
| HIST1H4B (6) | 3.04E−10 | HIST1H3I (6) | 5.13E−08 | GBF1 (10) | 1.03E−06 | SCARNA12 (12) | 3.62E−06 | |
| HIST1H3H (6) | 6.76E−10 | HIST1H2AH (6) | 5.81E−08 | APBA3 (19) | 1.15E−06 | SCARNA13 (14) | 4.24E−06 | |
| HIST1H2AJ (6) | 6.95E−10 | HIST1H2AG (6) | 7.13E−08 | COX15 (10) | 1.90E−06 | ACLY (17) | 4.49E−06 | |
| SCARNA7 (3) | 1.45E−09 | HIST1H4K (6) | 1.46E−07 | SERBP1 (1) | 2.00E−06 | |||
| HIST1H2AM (6) | 1.61E−09 | HIST3H2BB (1) | 1.64E−07 | HIST1H2BN (6) | 2.09E−06 | |||
| HIST1H1E (6) | 1.79E−09 | HIST1H4A (6) | 3.81E−07 | HIST1H3A (6) | 2.19E−06 | |||
| HIST1H3J (6) | 1.92E−09 | HIST1H4J (6) | 4.30E−07 | HIST1H2BO (6) | 2.22E−06 | |||
| rs113659777 | HIST1H2BM (6) | 2.89E−11 | HIST1H1E (6) | 1.72E−09 | SNORD17 (20) | 1.14E−07 | SCARNA12 (12) | 1.62E−06 |
| RPPH1 (14) | 7.27E−11 | HIST1H4K (6) | 3.31E−09 | HIST3H2BB (1) | 1.27E−07 | HTATSF1 (X) | 2.97E−06 | |
| HIST1H4B (6) | 8.17E−11 | HIST1H2AJ (6) | 5.31E−09 | HIST1H2AK (6) | 1.40E−07 | AGGF1 (5) | 3.52E−06 | |
| HIST1H1B (6) | 3.27E−10 | HIST1H2AB (6) | 7.80E−09 | SCARNA13 (14) | 2.84E−07 | MTFMT (15) | 3.89E−06 | |
| HIST1H3B (6) | 6.97E−10 | HIST1H2AL (6) | 1.05E−08 | HIST1H4A (6) | 3.39E−07 | |||
| HIST2H2AB (1) | 9.75E−10 | SCARNA2 (1) | 1.72E−08 | HIST1H3I (6) | 3.63E−07 | |||
| HIST1H2AM (6) | 9.86E−10 | HIST1H4J (6) | 4.06E−08 | HIST1H2AG (6) | 4.70E−07 | |||
| HIST1H3J (6) | 1.08E−09 | HIST1H2AH (6) | 9.03E−08 | HIST1H3A (6) | 4.83E−07 | |||
| HIST1H3H (6) | 1.37E−09 | HIST2H3C (1) | 9.54E−08 | SCARNA7 (3) | 1.13E−06 | |||
| rs184239416 | HIST1H2BM (6) | 1.56E−12 | HIST2H2AB (1) | 2.30E−09 | SCARNA12 (12) | 9.43E−08 | HIST2H3C (1) | 1.42E−06 |
| RPPH1 (14) | 2.76E−12 | HIST1H3H (6) | 2.98E−09 | HIST1H4A (6) | 9.92E−08 | GNAI3 (1) | 1.60E−06 | |
| HIST1H4B (6) | 9.13E−12 | HIST1H2AL (6) | 3.09E−09 | SCARNA13 (14) | 1.11E−07 | MAPRE1 (20) | 1.73E−06 | |
| HIST1H1B (6) | 2.84E−11 | HIST1H2AJ (6) | 3.83E−09 | HIST1H3I (6) | 1.46E−07 | RIOK3 (18) | 2.38E−06 | |
| HIST1H3J (6) | 3.88E−11 | HIST1H2AM (6) | 8.62E−09 | CAPZA1 (1) | 1.84E−07 | HIST1H2AK (6) | 2.55E−06 | |
| HIST1H3B (6) | 1.17E−10 | HIST1H2AB (6) | 1.50E−08 | HIST1H2AG (6) | 2.41E−07 | AGGF1 (5) | 2.75E−06 | |
| HIST1H1E (6) | 2.06E−10 | SCARNA2 (1) | 3.83E−08 | HIST1H3A (6) | 2.62E−07 | ARL6IP6 (2) | 3.00E−06 | |
| HIST3H2BB (1) | 2.37E−10 | HIST1H2AH (6) | 6.24E−08 | HIST1H4K (6) | 4.45E−07 | |||
| SNORD17 (20) | 1.19E−09 | SCARNA7 (3) | 6.81E−08 | HIST1H4J (6) | 4.76E−07 | |||
| rs41392245 | HIST1H2BM (6) | 4.19E−11 | HIST1H1E (6) | 3.18E−09 | HIST1H4K (6) | 2.14E−07 | HMHA1 (19) | 2.79E−06 |
| RPPH1 (14) | 1.07E−10 | HIST1H2AB (6) | 4.10E−09 | PRKAR1A (17) | 3.46E−07 | FBXL12 (19) | 3.05E−06 | |
| HIST2H3C (1) | 1.34E−10 | HIST1H2AK (6) | 1.08E−08 | ZFP91 (11) | 3.49E−07 | SNORD17 (20) | 3.06E−06 | |
| HIST1H3B (6) | 2.23E−10 | SCARNA2 (1) | 1.47E−08 | SCARNA12 (12) | 4.21E−07 | APBA3 (19) | 3.07E−06 | |
| HIST1H1B (6) | 2.90E−10 | HIST1H2AL (6) | 1.89E−08 | STYX (14) | 7.62E−07 | HIST1H2BN (6) | 3.26E−06 | |
| HIST1H4B (6) | 3.59E−10 | HIST2H2AB (1) | 2.27E−08 | HIST1H4J (6) | 7.71E−07 | RDH5 (12) | 3.79E−06 | |
| HIST1H3H (6) | 1.08E−09 | HIST1H2AG (6) | 2.99E−08 | HIST1H4A (6) | 9.00E−07 | ACLY (17) | 4.07E−06 | |
| HIST1H2AJ (6) | 1.10E−09 | HIST1H2AH (6) | 4.46E−08 | HIST1H2BO (6) | 9.37E−07 | ZNF622 (5) | 4.27E−06 | |
| SCARNA7 (3) | 1.16E−09 | HIST1H3I (6) | 7.59E−08 | SMAD2 (18) | 1.58E−06 | |||
| HIST1H2AM (6) | 2.39E−09 | HIST3H2BB (1) | 1.42E−07 | HIST1H3A (6) | 1.82E−06 | |||
| HIST1H3J (6) | 2.76E−09 | COX15 (10) | 1.96E−07 | EFTUD2 (17) | 2.57E−06 | |||
| rs11033879 | SNORA53 (12) | 2.29E−13 | HIST1H3J (6) | 2.08E−11 | HIST1H3A (6) | 4.97E−10 | HIST1H2AG (6) | 2.23E−07 |
| HIST1H2AJ (6) | 2.53E−13 | HIST1H4K (6) | 2.16E−11 | RPPH1 (14) | 1.18E−09 | OBFC2B (12) | 1.22E−06 | |
| HIST1H1E (6) | 9.71E−13 | HIST1H2AB (6) | 3.14E−11 | HIST3H2BB (1) | 1.51E−09 | RUNDC1 (17) | 1.82E−06 | |
| HIST1H3B (6) | 9.80E−13 | HIST1H4J (6) | 4.85E−11 | HIST1H4B (6) | 2.07E−09 | NHS (X) | 1.92E−06 | |
| HIST2H2AB (1) | 1.12E−12 | HIST1H3I (6) | 7.46E−11 | HIST1H2AK (6) | 6.39E−09 | SVIL (10) | 4.02E−06 | |
| HIST1H1B (6) | 1.18E−12 | SCARNA2 (1) | 1.10E−10 | HIST1H2AL (6) | 1.64E−08 | |||
| HIST1H4A (6) | 1.62E−12 | HIST1H3H (6) | 1.28E−10 | SCARNA7 (3) | 3.12E−08 | |||
| HIST1H2BM (6) | 4.53E−12 | HIST1H2AH (6) | 2.70E−10 | FN1 (2) | 3.67E−08 | |||
| HIST2H3C (1) | 1.15E−11 | SNORD17 (20) | 4.02E−10 | HIST1H2AM (6) | 1.68E−07 | |||
| rs80143982 | HIST1H4A (6) | 2.65E−10 | NOTCH2NL (1) | 2.32E−07 | HIST1H3I (6) | 1.73E−06 | ZNF207 (17) | 4.57E−06 |
| HIST3H2BB (1) | 3.31E−10 | RNF26 (11) | 3.73E−07 | HIST1H2AJ (6) | 2.16E−06 | |||
| SNORA53 (12) | 2.42E−08 | SCARNA2 (1) | 6.10E−07 | SNORD17 (20) | 2.25E−06 | |||
| HIST1H1E (6) | 3.15E−08 | HIST2H3C (1) | 1.51E−06 | MRFAP1L1 (4) | 4.02E−06 | |||
| rs75349221 | HIST1H4A (6) | 2.75E−10 | HIST1H2BM (6) | 8.36E−09 | HIST1H3H (6) | 1.11E−07 | HIST1H2AB (6) | 1.91E−06 |
| HIST1H1E (6) | 4.32E−10 | HIST1H3I (6) | 8.42E−09 | SCARNA2 (1) | 1.45E−07 | NDUFA8 (9) | 2.23E−06 | |
| HIST1H1B (6) | 7.02E−10 | SNORD17 (20) | 1.32E−08 | HIST1H4K (6) | 1.59E−07 | NDUFS1 (2) | 2.90E−06 | |
| HIST1H3B (6) | 1.26E−09 | SNORA53 (12) | 1.96E−08 | HIST1H4J (6) | 2.64E−07 | RPPH1 (14) | 2.95E−06 | |
| HIST1H2AJ (6) | 4.14E−09 | HIST1H4B (6) | 2.78E−08 | UBAP1 (9) | 3.35E−07 | TEX261 (2) | 3.35E−06 | |
| SCARNA7 (3) | 7.10E−09 | HIST2H2AB (1) | 5.63E−08 | HIST2H3C (1) | 1.36E−06 | RUNDC1 (17) | 3.63E−06 | |
| HIST1H3J (6) | 8.33E−09 | HIST1H2AH (6) | 8.54E−08 | SCARNA13 (14) | 1.61E−06 | HIST1H2AL (6) | 4.38E−06 | |
| rs72846811 | HIST1H4A (6) | 1.90E−11 | HIST1H3I (6) | 1.06E−07 | SCARNA7 (3) | 3.49E−07 | HIST1H3A (6) | 2.08E−06 |
| SNORA53 (12) | 1.90E−10 | SCARNA2 (1) | 1.22E−07 | HIST1H3J (6) | 5.24E−07 | HIST1H2BM (6) | 2.77E−06 | |
| HIST1H1E (6) | 8.56E−09 | HIST1H2AH (6) | 2.32E−07 | HIST2H3C (1) | 1.12E−06 | HIST2H2AB (1) | 2.79E−06 | |
| HIST1H2AJ (6) | 7.92E−08 | HIST1H3B (6) | 2.44E−07 | HIST1H1B (6) | 1.24E−06 | |||
| rs183898825 | SNORA53 (12) | 9.65E−11 | HIST1H4J (6) | 2.52E−08 | HIST1H1E (6) | 2.74E−07 | HIST1H2AH (6) | 7.54E−07 |
| HIST1H4A (6) | 2.60E−10 | HIST1H2AJ (6) | 4.29E−08 | HIST2H2AB (1) | 4.28E−07 | SNORA70 (X) | 8.38E−07 | |
| HIST1H4K (6) | 7.93E−09 | HIST2H3C (1) | 8.18E−08 | HIST1H3B (6) | 4.88E−07 | HIST1H1B (6) | 1.58E−06 | |
| HIST1H3I (6) | 1.64E−08 | SCARNA7 (3) | 1.21E−07 | HIST1H3H (6) | 4.96E−07 | HIST1H2AB (6) | 2.66E−06 | |
| SCARNA2 (1) | 1.69E−08 | SNORD17 (20) | 1.78E−07 | HIST1H3A (6) | 5.65E−07 | HIST1H3J (6) | 3.18E−06 | |
| rs57185274 | HIST1H4B (6) | 2.04E−10 | HIST1H3J (6) | 4.54E−08 | HIST1H2AB (6) | 5.47E−07 | EPCAM (2) | 2.65E−06 |
| HIST1H2BM (6) | 3.32E−09 | HIST1H3B (6) | 7.51E−08 | HIST1H3H (6) | 6.20E−07 | HIST2H3C (1) | 2.79E−06 | |
| HIST1H2AM (6) | 5.05E−09 | SCARNA12 (12) | 2.88E−07 | HIST1H2AJ (6) | 7.13E−07 | FNBP1 (9) | 3.03E−06 | |
| HIST1H1B (6) | 7.66E−09 | SCARNA13 (14) | 4.61E−07 | HIST2H2AB (1) | 9.68E−07 | HIST3H2BB (1) | 3.24E−06 | |
| HIST1H1E (6) | 2.02E−08 | HIST1H3I (6) | 5.12E−07 | HIST1H2AH (6) | 1.69E−06 | ZNF524 (19) | 4.57E−06 | |
- Figure in parentheses indicates chromosome number.
4. Discussion
We analyzed the eQTLs for simultaneously expressed histone genes. We found significant correlations amid the expression of 29 histone genes, which were all clustered in chromosome 1 or 6. This clustered structure of the genes may serve to control simultaneous transcription, and this is supported by the observation that the expression of other histone genes not located on chromosome 1 or 6, including H1FX and H2A family members, was not correlated with those of the 29 selected genes. Furthermore, correlation estimates showed two subgroups nested within the large group (one with 21 genes and the other with 10 genes; with strong correlation coefficients of r > 0.7), which likely provide a manageable unit for coordinating transcription.
The genome-wide eQTL analysis revealed that 12 loci were associated with the expression of multiple histone genes. The eQTLs were located on chromosomes 2, 7, and 11. Since 29 histone genes were all located on chromosome 1 or 6, we suspect that the identified eQTLs were transacting. This suggests that many histone genes are simultaneously transcribed by remote regulators.
Functional analysis of the identified eQTLs suggests that they are very important for transcription. For example, rs79335804, an SNP within an eQTL on chromosome 2, was the binding motif for Kruppel-like factor 4 (Klf4) protein in various cells including LCLs. Klf4 was associated with chromosomal aberrations and can prevent cell proliferation by acting as a transcription factor [13]. The aberrant chromatin formation could be caused by overproduction of a histone dimer set (H2A-H2B or H3-H4) [14]. Thus, we suspect that there is an association between the chromosomal aberrations by Klf4 and histone gene mRNA expression. Rs849578 within another eQTL on chromosome 2 was associated with autism in the Chinese Han population [15]. It is located in an intron of neuropilin 2 (NRP2) which may be an effector of apoptosis, proliferation, and neuronal development [16]. Histones are known to be related to developmental regulation [17], but additional study is required to elucidate underlying mechanism of the relationship between histones and NRP2.
Transcriptome-wide association analysis revealed that many nonhistone genes were also associated with the identified eQTLs. In particular, some genes were associated with 23 or more eQTLs. One was RPPH1, an RNA component of RNase P, which may assist in the cell cycle-dependent transcription of ribosomal RNAs (rRNAs) by associating with chromatin [18]. The expression of rRNAs increased from G1 to S and peaked at G2 [18]. The transcription of histone genes rapidly increased before the S phase of the cell cycle and decreased shortly thereafter [1]. Thus, many eQLTs identified in this study might be involved in the cell cycle-dependent expression of both RPPH1 and histone genes. Such a regulation of the eQTLs would be one of the key factors to solve their underlying mechanisms. The others identified with many eQTLs were the genes encoding scaRNAs (SCARNA2 and SCARNA7) located in the Cajal body, similar to the pre-mRNAs of histones, which move to the Cajal body for mRNA processing [3, 19].
Interestingly, many genes controlled by the same eQTLs as those for histones do not have polyadenylated structures [20]. In particular, the genes associated with more than 10 eQTLs were all nonpolyadenylated. They were snoRNAs, scaRNAs, and RPPH1. Considering that histones are also nonpolyadenylated, this may help us to understand the transcriptional regulation of histone genes by these eQTLs.
The expressions of histone genes play an important role in controlling chromatin accessibility [21]. Improper expression of histone genes has been associated with tumorigenesis [22–24]. Expression of NPAT, a transcriptional activator for histone genes, is also associated with human tumorigenesis [25]. The influence of histone genes on tumor developments might be supported by the eQTLs identified in the current study, because some of the eQTLs were located within anticipated tumor suppressor genes such as low-density lipoprotein receptor-related protein (LRP1B) and utrophin (UTRN) [26, 27].
In conclusion, we identified 31 eQTLs for histone genes. The eQTLs were also associated with nonhistone genes that exhibited both a cell cycle-dependent expression and a nonpolyadenylated RNA structure. Further investigations are required to understand the mechanisms regulating the transcription of the histone and nonhistone genes identified in this study and to appreciate their influence on cancer and other diseases. Moreover, identification of eQTLs using disease-specific cell types would provide resolute mechanisms by diseases.
Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
This work was funded by the National Research Foundation of Korea, the Ministry of Education, Science, and Technology (Grant no. NRF-2012M3A9D1054705).




