Correctness of Protein Identifications of Bacillus subtilis Proteome with the Indication on Potential False Positive Peptides Supported by Predictions of Their Retention Times
Abstract
The predictive capability of the retention time prediction model based on quantitative structure-retention relationships (QSRR) was tested. QSRR model was derived with the use of set of peptides identified with the highest scores and originated from 8 known proteins annotated as model ones. The predictive ability of the QSRR model was verified with the use of a Bacillus subtilis proteome digest after separation and identification of the peptides by LC-ESI-MS/MS. That ability was tested with three sets of testing peptides assigned to the proteins identified with different levels of confidence. First, the set of peptides identified with the highest scores achieved in the search were considered. Hence, proteins identified on the basis of more than one peptide were taken into account. Furthermore, proteins identified on the basis of just one peptide were also considered and, depending on the possessed scores, both above and below the assumed threshold, were analyzed in two separated sets. The QSRR approach was applied as the additional constraint in proteomic research verifying results of MS/MS ion search and confirming the correctness of the peptides identifications along with the indication of the potential false positives.
1. Introduction
Liquid chromatography (LC) combined with tandem mass spectrometry (MS/MS) plays an essential role in the field of protein research. In this technique, proteins and peptides are separated with the use of liquid chromatography methods and then identified by tandem mass spectrometry analysis. Thanks to high resolution, accuracy, and sensitivity of LC-MS/MS systems, equipped with sophisticated techniques of fragmentation, not only can simple proteins be directly investigated, but also research on the level of whole proteomes became possible [1]. However, proteins/peptides identification from biological matrices is still an analytical challenge because of the great complexity of the samples, enormous concentration ranges of the occurring proteins and lack of proper standards. It all makes an exact and precise peptide or protein identification and, consequently, proteome coverage limited [2].
Proteomic research requires also higher throughput of the protein identification in LC-MS/MS. Peptide identification in MS/MS is based on matching to parent ion m/z and m/z values of daughter ions. This procedure allows to assign an identification confidence for this particular peptide, which contributes independently to the overall confidence of the protein identification. One of the most commonly applied method for protein definition in complex samples relies on correlation algorithm Sequest proposed by Yates and coworkers [3–6]. This algorithm matches the investigated peptide tandem mass spectrometry data with proper data from protein database. To increase reliability of the identification, several statistic parameters have been considered. First, the difference between the normalized cross-correlation functions for the first and second ranked results (∆Cn) is applied to indicate a correctly selected peptide sequence. The other criteria are cross-correlation score between the observed peptide fragment mass spectrum and the theoretically predicted one (Xcorr), the preliminary score based on the number of ions in the MS/MS spectrum that match the experimental data (Sp), the rank of the certain match during the preliminary scoring (RSp), and the ions value (I) describing how many of the observed ions match the theoretical ions for the listed peptide. Currently, the most often applied criteria in protein study are cross-correlation score between the observed peptide fragment mass spectrum and the theoretically predicted one (Xcorr) and cross-correlation functions for the first and second ranked results (∆Cn). Washburn et al. [7] applied the following criteria of correctness of peptide identification: Xcorr above 1.9 for single charged fully tryptic peptides, over 2.2 and 3.75 for fully or partially tryptic doubly and triply charged peptides, respectively, and the ∆Cn values higher than 0.08. On the other hand, in the studies performed by Peng et al. [8] the peptides were classified as properly identified when Xcorr was, in case of fully tryptic peptides, higher than 2.0, 1.5, or 3.3 for the charge states of 1+, 2+, 3+, correspondingly, and over 3.0 (2+ charged) or 4.0 (3+ charged) considering partially tryptic peptides, when ∆Cn score was above 0.08. The relationship between application of different filtering criteria and degree of false positive identifications has also been recently demonstrated by Qian et al. [9]. There it was shown that all previously applied filtering criteria were derived using either relatively simple proteomes (e.g., the yeast proteome) or standard proteins. The degree of false positive identifications, when these criteria are extended to considerably more complex mammalian proteomes, especially human proteome, is still problematic and requires improvement of the strategies to distinguish correct from incorrect ones. Therefore, to decrease the probability of random match, which is growing up with the size of the protein database, two new sets of filtering criteria were independently developed for human cell line and human plasma samples [9]. For human cell line samples, the new criteria were as follows: Xcorr ≥ 1.5 for fully tryptic peptides and Xcorr ≥ 3.1 for partially tryptic peptides for the 1+ charge state, Xcorr ≥ 1.9 for fully tryptic peptides and Xcorr ≥ 3.8 for partially tryptic peptides for 2+ charge state, and Xcorr ≥ 2.9 for fully tryptic peptides and Xcorr ≥ 4.5 for partially tryptic peptides for the 3+ charge state. All the criteria had ∆Cn value of ≥0.1. The new criteria for peptides from human plasma samples include for the 1+ charged, Xcorr ≥ 2.0 and ≥3.0 for fully and partially tryptic peptides, respectively; for the 2+ charged, Xcorr ≥ 2.4 for fully and ≥3.5 for partially tryptic peptides, consequently; and for the 3+ charged, Xcorr ≥ 3.7 for fully and ≥4.5 for partially tryptic peptides, accordingly. The ∆Cn values were in all cases ≥0.1 as well.
Nevertheless, considering the variety and dynamic range of the proteins, occurring in the different organisms, there is still a possibility of false positive or false negative identification. Growing concerns about the quality of MS data affected in various ideas to harden protein identification by using bioinformatics’ methods, for example, decoy search strategies [10] or additional information obtained during analysis, for example, peptide pI or retention time [11]. The retention time is very practical parameter in proteomics as it is easy to obtain from LC-MS data and does not require a lot of instrumental effort [2, 12]. Comparison of the experimental and predicted retention times of the occurring peptides may examine the correctness of the identification and then enable to exclude the incorrectly identified ones. However, to predict properly peptides’ retention highly accurate models should be developed. Recently, some models have been proposed which characterize quantitatively the structure of a peptide and predict its gradient RP-LC retention at given separation conditions [13, 14].
Liquid chromatography (LC) is an analytical technique which can provide a great amount of quantitative, comparable, and reproducible (retention) data for large sets of structurally diversified compounds (analytes). On the other hand, chromatographic retention time can be considered as a chemical structure dependent parameter, which is constant for given separation conditions (mobile phase composition, stationary phase, temperature, pH). Due to that, quantitative structure (chromatographic) retention relationships (QSRR) have been considered as a model approach to establish strategy of retention predictions. However, to predict properly peptides’ retention highly accurate models should be developed [15–17]. In particular, in proteomics, the structural descriptors obtained from QSRR studies can contribute to better predictions of retention times and therefore harden peptides identification.
Several previous reports [18–21] prove that retention of peptides in reversed-phase liquid chromatography (RP-LC) depends on their amino acids composition. There, the regression analysis was used to derive the regression coefficients, which represented the contribution of each amino acid in the peptide’s sequence to its retention. This approach was applied in proteomics analysis, to predict the retention times of peptides’ tryptic digests [22]. Then, it was also employed to increase the reliability of the peptides identification to check the predictive capability of artificial neural networks (ANNs) by Petritis et al. [23] or by Shinoda et al. [24], where created ANN was then applied to predict the retention times of peptides from Escherichia Coli proteome. The correlation between amino acid composition and peptide’s retention time was used as well to provide the identity information, given by the tandem mass spectrometry, of the peptides from Drosophila melanogaster proteome, to exclude the false positive identifications [25].
Recently, a QSRR model based on multiple linear regression has been proposed [26] to quantitatively characterize the structure of a peptide and to predict its gradient RP-LC retention at established separation conditions. The logarithm of the sum of gradient retention times of the amino acids composing the individual peptide, log SumAA, the logarithm of the peptide Van der Waals volume, log VDWVol, and the logarithm of its calculated n-octanol-water partition coefficient, clogP, were employed [26–29].
The aim of the study was to derive the retention time prediction model and check its predictive capability based on quantitative structure-retention relationships (QSRRs). The newly modified QSRR model was derived with the use of set of peptides identified with the highest scores and originated from eight model proteins [13, 24, 30–32]. Therefore, no synthesized peptides with known amino acid sequences were used to derive and check the model [14, 31]. Moreover, descriptors applied in the new QSRR model were obtained in the new, facilitated from practical point of view, manner. Finally, its predictive ability was supported by further investigation with the use of a Bacillus subtilis proteome digest (not like previously just applying synthesized peptides with known amino acid sequences). To demonstrate that ability three sets of testing peptides received from proteins identified with different levels of confidence were used. Moreover, the additional attempts were performed to demonstrate the utility of QSRR approach as the additional constraint confirming the correctness of the peptides identifications.
2. Material and Methods
2.1. Standards
The standard amino acids solutions were prepared by dissolving seven amino acids among twenty naturally occurring ones (isoleucine, leucine, methionine, phenylalanine, tryptophan, tyrosine, and valine, all from Fluka BioChemika, Buchs, Switzerland) in 0.1% aqueous solution of trifluoroacetic acid (TFA). Water was deionized by passing through a Direct-Q (Millipore) system (Millipore, Bedford, MA, USA). The concentrations of the samples were approximately 0.6 mg/mL.
The solutions of standard proteins annotated as eight model proteins (about 3 mg/mL) were as follows: bovine serum albumin (BSA), chicken egg ovalbumin (CEO), bovine milk lactoglobulin (BML), bovine milk β-casein (BMC), bovine myoglobin (BM), human serum albumin (HSA) and ribonuclease B (RibB) from Sigma-Aldrich (Steinheim, Germany), and insulin-like growth factor-binding protein 1 (IGFBP-1), which was purified from human amniotic fluid following a previously reported procedure [33]. They were obtained by dissolving the lyophilized standard proteins in deionized water and then treated as shown below in digestion protocol.
2.2. Bacillus subtilis Sample Preparation
2.2.1. Growth Conditions
Bacillus subtilis strains were grown in nutrient broth (NB) supplemented with 0.2% KCl, 0.05% MgSO4 (final concentration) and antibiotics, if appropriate with shaking at 37°C.
2.2.2. Spore Purification
As described before [33] forty-eight-hour cultures in nutrient broth were pelleted (10000 ×g, 10 minutes) and washed three times with 1/4 volume of cold water. The pellet was resuspended in 1/5 of the initial volume of cold MQ water and incubated overnight at 4°C. On subsequent days the suspension was centrifuged (20000 ×g, 20 min, 4°C). The pellet was resuspended in fresh cold MQ water. This procedure was repeated for 5 to 10 days. Purified spores were kept in water suspension at 4°C in the dark. Once per week the spore were centrifuged and suspended in fresh water to avoid spontaneous germination.
2.2.3. Protein Extraction
The spore pellet (approximately 20 mg spores) was resuspended in 1 mL of extraction buffer (50 mM Tris-HCl, pH = 7.8; 2% SDS; 10% glycerol; 0,2 M DTT) and boiled for 5 min and vortexed for 30 seconds. These steps were repeated twice. Unlysed spores and spore debris were removed by centrifugation at 12,000 ×g for 5 min at 4°C. The supernatant was precipitated with acidified acetone/methanol mixture. To one volume of protein solution four volumes of cold precipitation reagent were added and kept on at −20°C. Precipitate was spun down at 15,000 × g, at 4°C and supernatant was discharged an samples were drained, then resuspended in water, and stored at −80°C. Concentration of proteins was determined with the use of Bradford assay kit (Bio-Rad Laboratories) and it equalled 1.2–1.5 mg/mL.
2.3. Digestion Protocol
To 1 mL of each protein (BSA, CEO, BML, BMC, BM, HAS, RibB, and IGFBP-1) sample (~3 mg/mL), 300 μL of DTT (dithiothreitol) (Sigma-Aldrich, Steinheim, Germany) (100 mM, freshly prepared in 100 mM ammonium bicarbonate buffer, pH 8.5) were added. The samples were kept in 60°C for 30 min, to allow reduction of the disulfide bridges. Then 50 μg of trypsin was added (ratio 1 : 50 E/S) to each sample. Samples were digested for 12 hours (overnight digestion) at 37°C. After that 0.1 mL of TFA was added to each sample to stop the digestion. Obtained standard solutions concentrations were about 50 pmol/μL.
To 1 mL of Bacillus subtilis spore cells lizates (1.2–1.5 mg/mL), 150 μL of DTT (Sigma-Aldrich, Steinheim, Germany) (100 mM, freshly prepared in 100 mM ammonium bicarbonate buffer, pH 8.5) were added. The samples were kept in 60°C for 30 min, to allow reduction of the disulfide bridges. Then 25 μg of trypsin was added (ratio 1 : 50 E/S) to each sample. Samples were digested for 12 hours (overnight digestion) at 37°C. After that 0.05 mL of TFA was added to each sample to stop the digestion. Obtained standard solutions concentrations were about 50 pmol/μL.
Tryptic digests were stored at −20°C (if frozen in this reaction mixture the disulfide bonds would not reoxidase). The LC-ESI-MS/MS analyses were performed in three weeks at the latest (the shelf life of such frozen solution is couple of months) (http://www.thermo.com/).
2.4. LC Conditions
The chromatographic analysis was performed on C-18 analytical column: XTerra MS C18 3.5 μm (2.1 × 100 mm) column (Waters, Milford, MA, USA).
The mobile phase consisted of two solvents (A and B) mixed on-line. Solvent A was 0.1% aqueous (water was MS-grade) solution of trifluoroacetic acid (TFA) (Sigma-Aldrich, Steinheim, Germany) and solvent B was acetonitrile (ACN) (MS-grade, Sigma-Aldrich, Steinheim, Germany) containing 0.1% TFA. The applied linear gradient time was 90 min, from 0% B to 60% B. The flow rate was 200 μL/min. The injection volume was 10 μL. The LC-MS apparatus was equipped with thermostated column oven and surveyor autosampler controlled at 20°C (Thermo Finnigan, San Jose, CA, USA), a quaternary gradient Surveyor MS pump (Thermo Finnigan, San Jose, CA, USA) with a diode array detection (DAD) system, and LTQ linear ion trap MS system with ESI ion source controlled by Xcalibur software 1.4 (Thermo Finnigan, San Jose, CA, USA).
2.5. MS Conditions
The MS/MS analysis was performed on Finnigan LTQ instrument (Thermo Finnigan, San Jose, CA, USA). Mass spectra were generated in positive ion mode under constant instrumental conditions: source voltage 4.62 kV, capillary voltage 40.97 V, sheath gas flow rate 39.99 (arbitrary units), auxiliary gas flow 10 (arbitrary units), sweep gas flow 0.95 (arbitrary units), capillary temperature 219.96°C, and tube lens voltage 250.43 V. MS/MS spectra, obtained by CID (collision-induced dissociation) in the linear ion trap, were performed with an isolation width 3Da (m/z); the activation amplitude was 35% of ejection RF amplitude that corresponds to 1.58 V.
2.6. Protein Identification
The experimental retention times of the peptides (tR exp ) were determined at peak intensity maximum. The m/z values measured manually for the most intense peaks in acquired MS/MS spectra were automatically searched against the protein database (*fasta) using the Sequest Algorithm, incorporated into Bioworks 3.0 (Thermo Finningan, San Jose, CA, USA). The *fasta format for each protein was downloaded from Expasy (http://www.expasy.org/sprot/). During the interpretation of the results obtained after the correlation analysis done on the experimental and the predicted retention times of peptides, the exemplary filtering criteria applied in the studies were the same as those discussed previously, proposed by Washburn et al. [7]. The spectra for singly charged peptides with a cross-correlation score to a tryptic peptide (Xcorrs) greater than 1.9, the spectra for doubly charged tryptic peptides with Xcorrs of at least 2.2, and the spectra for triply charged tryptic peptides with Xcorrs above 3.75 were accepted as correctly identified according to Sequest software. For all the spectra analyzed, ∆Cn values were above 0.08.
2.7. QSRR Analysis
Multiple regression equations for model set of peptides based on the experimental retention times were derived by employing Microsoft Excel software (Microsoft Co., Redmond, WA, USA) and Statistica (StatSoft, Tulsa, OK, USA) run on a personal computer. Regression coefficients (±standard deviations), multiple correlation coefficients, R, standard errors of estimate, s, significance levels of each term and of the whole equations, p, and values of the F-test of significance, F, were calculated.
The structural descriptors of the analyzed standard amino acids and peptides from investigated, standard proteins and Bacillus subtilis cells were calculated. First of all, in contrary to the previous models [26–29], where just log SumAA was calculated by simple addition of component amino acids retention (taking into account all 20 naturally occurring amino acids), the novel QSRR peptide descriptor log Sum (k+1)AA was used. The retention factor (k) was introduced, because it is more similar for different related systems than tR as it compensates for some physical differences between columns. Descriptor log Sum (k+1)AA was calculated applying retention data for just only 7, the most retained amino acids (isoleucine, leucine, methionine, phenylalanine, tryptophan, tyrosine, and valine). The other 13 amino acids are hardly retained; therefore their presence in peptide’s sequence does not influence significantly its retention. For these 13 amino acids fixed values were ascribed (k = 0) and one was added to avoid zero in the calculation of the logarithm, according to the procedure elaborated and evaluated elsewhere [34]. On the other hand, searching for the most accurate the logarithm of its calculated n-octanol-water partition coefficient, clog P, values, different calculation methods were tested (data not shown). Briefly, to obtain clog P values HyperChem 7.5 professional software for personal computers (HyperCube, Waterloo, Canada) with the extension ChemPlus, Dragon professional 5.0 software (Milano Chemometrics and QSAR Research Group—Talete, Milano, Italy), and on-line available ALOGPS 2.1 software (http://www.vcclab.org/) were obtained. Finally, to derive the appropriate QSRR model, clog P values average, log P module in ALOGPS 2.1 software was used to determine that QSRR descriptor.
The general QSRR equation has the following form:
3. Results and Discussion
3.1. Derivation and Validation of QSRR Model
The QSSR model was derived from peptides obtained from the digestion of 8 model proteins. The amino acid sequences of these peptides were proved by MS/MS analysis and identified by Sequest software (Bioworks 3.0 package Thermo Fisher Scientific Inc., Waltham, MA, USA). Only peptides with the highest scores were taken into account in the model set of peptides used to derive the QSRR model. Peptides were assumed and considered as true positives according to their cross-correlation score to a tryptic peptide Xcorr values with over 2.0 for 1+ and 2+ and over 4.5 for 3+ charged peptides. Peptides with lower values of Xcorr were excluded from the model set of peptides, due to higher possibility of their false positive identification. Hence, the peptides included in the study were divided into five groups: one set of model peptides (Table 1) and four testing sets of peptides (Tables 2–5). 50 model peptides used to derive QSRR model and collected in Table 1 originated from 8 model proteins. The 21 peptides reported in Table 2 were used to check the general validity of the proposed QSRR model. In view of the main objective of this work, three other sets of testing peptides originating from B. subtilis proteome digestion were used. One set includes 54 peptides belonging to proteins identified on the basis of more than one peptide with Xcorr above 1.5 (Table 3). A second set comprises 41 peptides belonging to proteins identified again with Xcorr above 1.5, but on the basis of just one peptide (Table 4). And the third set comprises 40 peptides belonging to proteins identified on the basis of just one peptide, but with Xcorr below 1.5 (Table 5).
| Peptide sequence | Protein | m/z | Missed cleavages | Charge | Xcorr | log Sum(k+1)AA | clog P | tR exp |
|---|---|---|---|---|---|---|---|---|
| ALKALPMHIR | 1 | 575.73 | 1 | 2 | 3.06 | 1.3542 | −1.74 | 25.12 |
| LFTFHADICTLPDTEKQIK | 3 | 1111.28 | 1 | 2 | 3.21 | 1.6674 | −4.6 | 32.60 |
| KIKVYLPR | 4 | 509.15 | 2 | 2 | 2.27 | 1.3005 | −0.95 | 24.06 |
| LVNEVTEFAK | 6 | 575.65 | 0 | 2 | 3.42 | 1.3540 | −2.44 | 24.53 |
| YTRKVPQVSTPTLVEVSR | 1 | 1031.19 | 2 | 2 | 3.01 | 1.4758 | −5.67 | 25.80 |
| ALHVTNIK | 8 | 896.07 | 0 | 1 | 2.48 | 1.2148 | −3.14 | 19.69 |
| DTHKSEIAHR | 3 | 597.64 | 1 | 2 | 2.8 | 1.1246 | −7.77 | 13.15 |
| AAFTECCQAADK | 6 | 687.7 | 0 | 2 | 3.07 | 1.2657 | −5.69 | 19.31 |
| ALPGEQQPLHALTR | 8 | 766.37 | 0 | 2 | 3.37 | 1.4145 | −5.22 | 24.36 |
| VKEAMAPK | 5 | 874.08 | 1 | 1 | 2.13 | 1.0165 | −2.19 | 14.40 |
| QHMDSSTSAASSSNYCNQMMK | 7 | 789.84 | 0 | 3 | 4.75 | 1.4546 | −11.8 | 20.46 |
| AFDEKLFTFHADICTLPDTEK | 3 | 814.91 | 1 | 3 | 5.11 | 1.7127 | −4.51 | 34.45 |
| HIIVACEGNPYVPVHFDASV | 7 | 1113.73 | 0 | 2 | 5.15 | 1.6128 | −3.51 | 32.15 |
| VHTECCHGDLLECADDRADLAK | 6 | 1295.34 | 1 | 2 | 4.47 | 1.5447 | −9.95 | 24.98 |
| AFDEKLFTFHADICTLPDTEKQIK | 3 | 1406.60 | 2 | 2 | 4.42 | 1.7629 | −4.95 | 34.77 |
| DLGEENFK | 6 | 952 | 0 | 1 | 2.09 | 1.2654 | −4.48 | 19.46 |
| CCAAADPHECYAK | 6 | 778.79 | 0 | 2 | 3.46 | 1.2195 | −5.77 | 16.98 |
| IPGSPEIR | 8 | 869.00 | 0 | 1 | 2.18 | 1.1657 | −2.43 | 19.40 |
| LKPDPNTLCDEFKADEKKFWGKYLYEIAR | 1 | 1174.00 | 5 | 3 | 4.49 | 1.8691 | −5.45 | 39.20 |
| ALHVTNIKK | 8 | 1024.24 | 1 | 1 | 2.01 | 1.2405 | −4.64 | 18.27 |
| VLPVPQKAVPYPQR | 5 | 796.96 | 1 | 2 | 3.32 | 1.3948 | −3.24 | 24.20 |
| AEFAEVSK | 6 | 880.97 | 0 | 1 | 2.10 | 1.1909 | −2.94 | 18.06 |
| ALHVTNIKK | 8 | 512.62 | 1 | 2 | 2.70 | 1.2405 | −4.64 | 18.24 |
| TCVADESAENCDK | 6 | 751.23 | 0 | 2 | 4.05 | 1.1488 | −7.23 | 15.32 |
| NECFLQHK | 6 | 1077.16 | 0 | 1 | 2.33 | 1.2654 | −3.74 | 19.67 |
| TCVADESHAGCEK | 3 | 675.73 | 0 | 2 | 3.47 | 1.1488 | −7.76 | 14.99 |
| CASIQKFGER | 3 | 570.16 | 1 | 2 | 2.78 | 1.2958 | −4.66 | 19.81 |
| VHTECCHGDLLECADDR | 6 | 1046.05 | 0 | 2 | 6.23 | 1.4161 | −8.42 | 22.86 |
| LFTFHADICTLPDTEK | 3 | 926.55 | 0 | 2 | 4.79 | 1.6039 | −3.25 | 32.90 |
| RIPGSPEIR | 8 | 513.09 | 1 | 2 | 2.63 | 1.1944 | −3.5 | 20.00 |
| HLVDEPQNLIK | 3 | 653.75 | 0 | 2 | 3.48 | 1.3690 | −4.68 | 24.56 |
| TCVADESHAGCEKSLHTLFGDELCK | 3 | 1348.00 | 1 | 2 | 3.77 | 1.6483 | −7.37 | 31.14 |
| YPNCAYK | 7 | 916.99 | 0 | 1 | 2.58 | 1.1508 | −2.34 | 16.78 |
| LRCASIQKFGER | 1 | 704.83 | 2 | 2 | 3.82 | 1.4107 | −4.96 | 22.79 |
| WKEPCRIELYR | 8 | 747.38 | 2 | 2 | 2.70 | 1.4991 | −3.52 | 26.67 |
| TPEVDDEALEKFDK | 1 | 818.87 | 1 | 2 | 5.09 | 1.4067 | −6.12 | 24.94 |
| LDELRDEGK | 6 | 538.08 | 1 | 2 | 2.52 | 1.2299 | −5.34 | 16.62 |
| YICDNQDTISSKLK | 1 | 814.91 | 1 | 2 | 3.80 | 1.4504 | −6.1 | 22.75 |
| YICDNQDTISSKLK | 3 | 814.91 | 1 | 2 | 4.03 | 1.4504 | −6.1 | 22.75 |
| QTALVELLKHKPK | 3 | 753.42 | 2 | 2 | 2.89 | 1.4159 | −3.08 | 27.89 |
| VKEAMAPKHK | 5 | 570.20 | 2 | 2 | 2.90 | 1.0930 | −3.41 | 13.48 |
| ELINSWVESQTNGIIR | 4 | 930.53 | 0 | 2 | 3.93 | 1.6097 | −5.86 | 31.95 |
| ISQAVHAAHAEINEAGR | 4 | 887.96 | 0 | 2 | 3.19 | 1.3932 | −7.56 | 19.51 |
| YICDNQDTISSK | 3 | 694.25 | 0 | 2 | 3.8 | 1.3468 | −6.61 | 18.79 |
| SHCIAEVEKDAIPENLPPLTADFAEDKDVCK | 1 | 1133.93 | 2 | 3 | 5.65 | 1.7342 | −3.53 | 33.13 |
| GGLEPINFQTAADQAR | 4 | 844.91 | 0 | 2 | 4.19 | 1.4735 | −6.71 | 27.41 |
| EAMAPKHKEMPFPK | 5 | 821.49 | 2 | 2 | 3.87 | 1.3625 | −3.31 | 21.52 |
| FYLPNCNKNGFYHSR | 8 | 931.04 | 0 | 2 | 2.76 | 1.5912 | −5.83 | 26.57 |
| YICENQDSISSK | 6 | 723.25 | 0 | 2 | 4.04 | 1.3468 | −6.77 | 18.06 |
| SLHTLFGDELCK | 3 | 682.28 | 0 | 2 | 3.55 | 1.4829 | −3.32 | 30.69 |
| Peptide sequence | Protein | m/z | Missed cleavages | Charge | Xcorr | log Sum(k+1)AA | clog P | tR exp | tR pred | Dt R |
|---|---|---|---|---|---|---|---|---|---|---|
| WKEPCR | 8 | 818.97 | 1 | 1 | 1.50 | 1.1950 | −2.52 | 17.32 | 19.00 | 1.69 |
| VVESLAK | 8 | 745.89 | 0 | 1 | 1.52 | 1.1192 | −2.11 | 16.42 | 16.88 | 0.46 |
| VLPVPQK | 5 | 780.98 | 0 | 1 | 1.57 | 1.1192 | −1.30 | 19.19 | 17.49 | 1.70 |
| IELYR | 8 | 693.81 | 0 | 1 | 1.57 | 1.2011 | −0.67 | 20.90 | 20.61 | 0.29 |
| LDELR | 6 | 645.73 | 0 | 1 | 1.62 | 1.1133 | −2.48 | 17.06 | 16.40 | 0.66 |
| RIPGSPEIR | 8 | 1025.19 | 1 | 1 | 1.68 | 1.1944 | −3.50 | 19.97 | 18.24 | 1.74 |
| NGFYHSR | 8 | 880.93 | 0 | 1 | 1.72 | 1.2308 | −3.98 | 16.06 | 19.04 | 2.99 |
| SLGKVGTR | 3 | 817.96 | 1 | 1 | 1.79 | 1.1164 | −4.29 | 15.54 | 15.13 | 0.41 |
| AQETSGEEISK | 8 | 1179.22 | 0 | 1 | 1.83 | 1.1560 | −7.53 | 13.32 | 13.94 | 0.62 |
| NVACK | 7 | 592.65 | 0 | 1 | 1.90 | 0.7843 | −3.04 | 8.38 | 5.39 | 2.99 |
| ETCFAEEGKK | 6 | 600.63 | 1 | 2 | 2.00 | 1.2158 | −5.18 | 16.58 | 17.65 | 1.07 |
| CCAADDKEACFAVEGPKLVVSTQTALA | 1 | 1371.57 | 2 | 2 | 2.01 | 1.6500 | −6.31 | 32.44 | 30.76 | 1.68 |
| FYLPNCNK | 8 | 500.08 | 0 | 2 | 2.02 | 1.3425 | −2.34 | 24.31 | 23.88 | 0.43 |
| HLKTEAEMK | 2 | 544.14 | 1 | 2 | 2.12 | 1.1551 | −4.21 | 16.09 | 16.43 | 0.34 |
| HKEMPFPK | 5 | 507.61 | 1 | 2 | 2.25 | 1.1970 | −0.82 | 20.27 | 20.36 | 0.09 |
| ALKAWSVAR | 3 | 501.60 | 1 | 2 | 2.28 | 1.3755 | −2.65 | 23.31 | 24.71 | 1.40 |
| LFTFHADICTLPDTEKQIKK | 1 | 783.91 | 2 | 3 | 2.57 | 1.6767 | −4.86 | 31.65 | 32.72 | 1.07 |
| TPEVDDEALEKFDKALK | 1 | 650.38 | 2 | 3 | 2.58 | 1.5119 | −4.06 | 29.46 | 28.03 | 1.43 |
| LYAEERYPILPEYLQCVKELYR | 4 | 930.74 | 2 | 3 | 2.59 | 1.7534 | −3.66 | 39.18 | 36.10 | 3.08 |
| LFTFHADICTLPDTEKQIKKQTALVELLK | 1 | 1115.98 | 3 | 3 | 3.22 | 1.8423 | −5.65 | 37.13 | 37.45 | 0.32 |
| LKECCDKPLLEK | 1 | 710.37 | 2 | 2 | 4.32 | 1.3797 | −3.12 | 21.94 | 24.49 | 2.55 |
| Peptide sequence | m/z | Missed cleavages | Charge | Xcorr | log Sum(k+1)AA | clog P | tR exp | tR pred | Dt R |
|---|---|---|---|---|---|---|---|---|---|
| ALDMLEASPVQGFDAK | 846.96 | 0 | 2 | 4.90 | 1.52 | −4.71 | 32.56 | 27.66 | 4.90 |
| ITGTSNYEDTAGSDIVVITAGIAR | 1213.32 | 0 | 2 | 4.87 | 1.63 | −6.38 | 34.61 | 30.19 | 4.42 |
| RHDDYDSKK | 582.61 | 1 | 2 | 2.57 | 1.10 | −7.90 | 12.26 | 11.84 | 0.42 |
| KPHHHCDDYK | 640.70 | 1 | 2 | 3.17 | 1.13 | −6.12 | 14.34 | 14.26 | 0.08 |
| DYLYQEPHGK | 625.67 | 0 | 2 | 2.60 | 1.33 | −4.72 | 24.86 | 21.52 | 3.34 |
| EGLKDYLYQEPHGK | 839.42 | 1 | 2 | 2.81 | 1.46 | −5.99 | 30.89 | 25.03 | 5.86 |
| KEGLKDYLYQEPHGK | 903.51 | 2 | 2 | 3.71 | 1.48 | −4.79 | 32.18 | 26.41 | 5.77 |
| YYKKPHHHCDDYK | 867.96 | 2 | 2 | 1.94 | 1.38 | −4.29 | 33.93 | 23.46 | 10.47 |
| GTAMAYDQIDGAPEER | 862.92 | 0 | 2 | 3.63 | 1.38 | −8.06 | 23.43 | 20.86 | 2.57 |
| TVGSGVVSTITE | 1150.26 | 0 | 1 | 1.42 | 1.27 | −5.07 | 24.54 | 19.44 | 5.10 |
| GITISTAHVEYETETR | 904.47 | 0 | 2 | 4.86 | 1.44 | −7.39 | 24.43 | 23.06 | 1.37 |
| GQVLAKPGTITPHSK | 767.89 | 1 | 2 | 2.77 | 1.37 | −6.82 | 27.57 | 21.33 | 6.24 |
| VGDEVEIIGLQEENKK | 900.99 | 1 | 2 | 4.59 | 1.46 | −6.08 | 29.49 | 24.80 | 4.69 |
| HYAHVDCPGHADYVK | 856.94 | 0 | 2 | 4.62 | 1.39 | −4.95 | 30.91 | 23.30 | 7.61 |
| DLLSEYDFPGDDVPVVK | 955.04 | 0 | 2 | 4.92 | 1.58 | −4.46 | 35.03 | 30.00 | 5.03 |
| LLDYAEAGDNIGALLR | 852.95 | 0 | 2 | 3.63 | 1.59 | −4.49 | 36.17 | 30.20 | 5.97 |
| NMITGAAQMDGAILVVSAADGPMPQTR | 1359.07 | 0 | 2 | 5.04 | 1.64 | −7.20 | 37.02 | 29.78 | 7.24 |
| SHANIGTIGHVDHGKTTLTAAITTVLHKKSGK | 1099.25 | 3 | 3 | 3.27 | 1.72 | −11.83 | 39.57 | 28.71 | 10.86 |
| NVGVPYIVVFLNKCDMVDDEELLELVEMEVR | 1806.60 | 1 | 2 | 4.26 | 1.85 | −4.97 | 53.42 | 38.15 | 15.27 |
| ALAPEIVGEEHYAVAR | 863.46 | 0 | 2 | 2.54 | 1.46 | −4.87 | 30.36 | 25.85 | 4.51 |
| EGNDLFYEMSDSGVINK | 960.03 | 0 | 2 | 4.61 | 1.56 | −6.57 | 31.77 | 27.82 | 3.95 |
| GMEAVDTGAPISVPVGDVTLGR | 1071.71 | 0 | 2 | 5.50 | 1.54 | −5.68 | 32.56 | 27.81 | 4.75 |
| VFNVLGENIDLNEPVPADAK | 1078.20 | 0 | 2 | 5.91 | 1.61 | −5.15 | 34.41 | 30.44 | 3.97 |
| KLTEMGIYPAVDPLASTSR | 1025.68 | 1 | 2 | 3.85 | 1.56 | −4.80 | 34.63 | 29.00 | 5.63 |
| VQPGQQHLKR | 596.18 | 1 | 2 | 2.37 | 1.18 | −6.77 | 15.36 | 15.23 | 0.13 |
| IVSINPADKEEVVGR | 813.92 | 0 | 2 | 3.32 | 1.40 | −5.93 | 27.21 | 22.88 | 4.33 |
| AGGPDYLALHMQAK | 736.85 | 0 | 2 | 3.68 | 1.43 | −4.63 | 29.65 | 24.93 | 4.72 |
| VSDFDEALEVANNTEYGLTGAVITNNRK | 1014.75 | 1 | 3 | 4.37 | 1.72 | −9.43 | 36.85 | 30.66 | 6.19 |
| GYFIKPTIFADLDPK | 863.50 | 1 | 2 | 3.55 | 1.62 | −1.12 | 38.36 | 33.88 | 4.48 |
| LMQEEIFGPVVAFCK | 856.54 | 0 | 2 | 3.25 | 1.59 | −0.08 | 40.34 | 33.48 | 6.86 |
| QQNQSAEQNKQQNS | 816.82 | 1 | 2 | 2.81 | 1.15 | −13.83 | 9.13 | 8.82 | 0.31 |
| KQNQQSAAGQGQFGTEFASETNAQQVR | 1456.52 | 1 | 2 | 5.75 | 1.61 | −11.78 | 25.84 | 25.38 | 0.46 |
| YDDYDKK | 946.98 | 1 | 1 | 1.90 | 1.15 | −2.97 | 13.79 | 17.24 | 3.45 |
| DYDCDYDKK | 583.11 | 1 | 2 | 2.76 | 1.21 | −5.81 | 16.52 | 16.93 | 0.41 |
| DYDYVVEYK | 597.64 | 0 | 2 | 3.30 | 1.34 | −3.31 | 25.78 | 23.08 | 2.70 |
| DYDYVVEYKK | 661.72 | 1 | 2 | 2.57 | 1.36 | −3.32 | 26.07 | 23.70 | 2.37 |
| VGNDGVITIEESK | 681.24 | 0 | 2 | 2.66 | 1.34 | −6.20 | 23.92 | 20.83 | 3.09 |
| FGSPLITNDGVTIAK | 767.38 | 0 | 2 | 2.73 | 1.52 | −4.16 | 30.33 | 28.23 | 2.10 |
| EIELEDAFENMGAK | 798.87 | 0 | 2 | 3.31 | 1.46 | −5.96 | 32.86 | 25.01 | 7.85 |
| ESIAQVAAISAADEEVGSLIAEAMER | 1331.46 | 0 | 2 | 6.41 | 1.64 | −8.79 | 46.55 | 28.48 | 18.07 |
| WNTNAGDDYVSNGPFK | 893.43 | 0 | 2 | 2.63 | 1.57 | −5.83 | 27.55 | 28.40 | 0.85 |
| GVIMPGTGEVYFR | 713.83 | 0 | 2 | 2.42 | 1.47 | −1.26 | 32.23 | 28.95 | 3.28 |
| ADYTGPDKQK | 562.10 | 1 | 2 | 2.85 | 1.13 | −5.89 | 14.13 | 14.44 | 0.31 |
| MLTEIGEVENAEPYIR | 933.05 | 0 | 2 | 4.38 | 1.51 | −4.61 | 31.8 | 27.65 | 4.15 |
| EDYGIAENFLYTLNGEEPSPIEVEAFNK | 1595.71 | 0 | 2 | 3.18 | 1.80 | −7.22 | 40.25 | 35.03 | 5.22 |
| MSGWLAHILEQYDNNRLIRPR | 861.99 | 2 | 3 | 3.54 | 1.73 | −7.62 | 43.05 | 32.45 | 10.60 |
| VLQQPNCLEVTISPNGNK | 978.11 | 0 | 2 | 4.43 | 1.50 | −5.93 | 28.75 | 26.15 | 2.60 |
| YRDNNYLDDEHEVIAK | 998.05 | 1 | 2 | 4.61 | 1.50 | −6.85 | 29.35 | 25.49 | 3.86 |
| IVVQAEREFLAEVVGETK | 1009.65 | 1 | 2 | 2.99 | 1.56 | −4.48 | 38.2 | 29.17 | 9.03 |
| IVNPLGQPVDGLGPILTSK | 960.13 | 0 | 2 | 5.18 | 1.60 | −3.36 | 35.86 | 31.39 | 4.47 |
| KGRNPQTGEEIEIPASKVPAFKPGK | 894.02 | 4 | 3 | 4.53 | 1.59 | −7.21 | 33.22 | 28.24 | 4.98 |
| MNKTELINAVAEASELSK | 975.11 | 1 | 2 | 5.11 | 1.50 | −6.38 | 35.32 | 25.91 | 9.41 |
| MNKTELINAVAEASELSKK | 1039.19 | 2 | 2 | 5.25 | 1.51 | −6.58 | 35.64 | 26.19 | 9.45 |
| AVDSVFDTILDALKNGDKIQLIGFGNFEVR | 1099.57 | 2 | 3 | 5.81 | 1.87 | −5.30 | 50.01 | 38.68 | 11.33 |
| Peptide sequence | m/z | Missed cleavages | Charge | Xcorr | log Sum(k+1)AA | clog P | tR exp | tR pred | Dt R |
|---|---|---|---|---|---|---|---|---|---|
| NIAEMVK | 754.79 | 0 | 1 | 1.62 | 1.1042 | −2.43 | 14.25 | 16.15 | 1.90 |
| INIMSAR | 1463.68 | 0 | 1 | 1.87 | 1.1746 | −2.86 | 14.23 | 18.09 | 3.86 |
| NLLFAAR | 707.86 | 0 | 1 | 1.81 | 1.3307 | −1.12 | 14.27 | 24.44 | 10.17 |
| LNSLDSR | 896.11 | 0 | 1 | 1.64 | 1.1755 | −4.79 | 14.29 | 16.65 | 2.36 |
| DIMSPSR | 1380.53 | 0 | 1 | 1.53 | 1.0654 | −4.27 | 14.30 | 13.50 | 0.80 |
| LALDLESKK | 922.17 | 1 | 1 | 1.62 | 1.3216 | −2.73 | 18.01 | 22.92 | 4.91 |
| IDIALESKK | 1020.08 | 1 | 1 | 1.54 | 1.2931 | −2.77 | 18.04 | 21.97 | 3.93 |
| SHTGKAAVLNR | 524.07 | 1 | 1 | 1.52 | 1.2061 | −6.85 | 24.54 | 16.07 | 8.47 |
| GHNPGQPEPLSGSK | 718.86 | 0 | 2 | 3.54 | 1.2550 | −8.65 | 17.05 | 16.27 | 0.78 |
| VVVSVNTDQDQAQAQSQDGED | 868.09 | 0 | 2 | 4.73 | 1.4038 | −14.74 | 19.37 | 16.42 | 2.95 |
| GNQVSENLQQAAR | 694.78 | 0 | 2 | 2.03 | 1.2571 | −9.4 | 20.52 | 15.76 | 4.76 |
| LIDKHKKYVYHRINK | 920.72 | 4 | 2 | 2.60 | 1.5299 | −4.29 | 29.00 | 28.43 | 0.57 |
| EAEELIPNVTTAAVK | 1025.52 | 0 | 2 | 2.43 | 1.3889 | −6.37 | 29.18 | 22.31 | 6.87 |
| ELQEKFLIPAVEQKK | 1044.81 | 2 | 2 | 2.24 | 1.5292 | −3.62 | 29.27 | 28.92 | 0.35 |
| QDIPIEARMNEIVHSLK | 1098.25 | 1 | 2 | 2.15 | 1.5231 | −5.67 | 29.33 | 27.16 | 2.17 |
| AAEMAVARQNEQKVKK | 617.20 | 3 | 2 | 2.20 | 1.2894 | −7.16 | 29.44 | 18.51 | 10.93 |
| EGTVIKELIGAGQLDEK | 817.41 | 1 | 2 | 2.40 | 1.5147 | −5.74 | 29.49 | 26.84 | 2.65 |
| EVMIEGVLSVLEGQAPK | 731.84 | 0 | 2 | 2.38 | 1.5167 | −4.34 | 29.51 | 27.97 | 1.54 |
| DRVFIAPVGGGPR | 580.16 | 1 | 2 | 2.68 | 1.3967 | −3.64 | 29.82 | 24.64 | 5.18 |
| SGETEDSTIADIAVATNAGQIK | 865.45 | 0 | 2 | 3.33 | 1.5192 | −9.39 | 30.05 | 24.21 | 5.84 |
| IDNLSYYIEQEYK | 952.72 | 0 | 2 | 2.13 | 1.5361 | −4.63 | 30.89 | 28.37 | 2.52 |
| SGSIESIDVSLTDLR | 613.73 | 0 | 2 | 2.53 | 1.4873 | −6.49 | 33.05 | 25.39 | 7.66 |
| LEIASEFGVNLGADTTSR | 1481.93 | 0 | 2 | 4.30 | 1.5661 | −5.87 | 33.12 | 28.39 | 4.73 |
| HSSDEEPFSALAFK | 531.67 | 0 | 2 | 2.95 | 1.4894 | −5.34 | 33.16 | 26.33 | 6.83 |
| AVLSPLFPTATEGGENMDSNLK | 1146.78 | 0 | 2 | 4.62 | 1.6314 | −6.36 | 34.13 | 30.12 | 4.01 |
| VCELQKVAVLNINDLANAVK | 1078.27 | 1 | 2 | 2.00 | 1.5981 | −4.6 | 34.35 | 30.39 | 3.96 |
| TEWRQERLNPLQRLTGR | 1077.71 | 3 | 2 | 2.48 | 1.5870 | −8.41 | 34.47 | 27.13 | 7.34 |
| GVSNNIIELINASGEPVIWK | 1077.73 | 0 | 2 | 2.25 | 1.6913 | −4.42 | 34.49 | 33.52 | 0.97 |
| LSLKSIIIGGRIPNYHK | 955.65 | 2 | 2 | 2.06 | 1.6217 | −5.35 | 35.03 | 30.58 | 4.45 |
| ANVPLDQIAVLSIGTGEAPTR | 1062.20 | 0 | 2 | 4.11 | 1.5774 | −5.59 | 35.54 | 28.97 | 6.57 |
| DQDISGEKATADQLLKDVK | 1038.13 | 2 | 2 | 2.09 | 1.4968 | −8.56 | 35.61 | 24.12 | 11.49 |
| LIDIVNPTPQTVDALMR | 949.11 | 0 | 2 | 4.64 | 1.5453 | −4.51 | 36.18 | 28.76 | 7.42 |
| AEELGAIIVDPSKTDDVVAEIAER | 1271.40 | 1 | 2 | 2.49 | 1.6150 | −6.83 | 36.41 | 29.24 | 7.17 |
| GGGFLIEDVTYDQMYTPEDFTDEHK | 1455.04 | 0 | 2 | 2.46 | 1.7382 | −7.29 | 36.58 | 32.85 | 3.73 |
| AIDSAVEELTFIAGQKPVVTR | 1123.28 | 1 | 2 | 2.89 | 1.6162 | −5.29 | 37.19 | 30.45 | 6.74 |
| TYNLSLDNGGDFIQIGSDGGLLPR | 1262.37 | 0 | 2 | 3.54 | 1.7529 | −7.58 | 37.31 | 33.10 | 4.21 |
| TIPLNITPYYASLMDPDNPR | 1146.80 | 0 | 2 | 2.01 | 1.6343 | −5.13 | 37.49 | 31.15 | 6.34 |
| IVPISEIPSDLEAIDIGTK | 1006.15 | 0 | 2 | 2.95 | 1.6095 | −3.93 | 37.73 | 31.27 | 6.46 |
| IQNGDPIAGLFDEFTQTVQR | 1125.73 | 0 | 2 | 2.68 | 1.6493 | −5.7 | 42.90 | 31.20 | 11.70 |
| KVKTINRQIKISIRAEDQAFYR | 893.71 | 5 | 3 | 2.54 | 1.6664 | −7.19 | 33.22 | 30.62 | 2.60 |
| SLEEGQEVSFEIVEGNRGPQASNVVKL | 973.06 | 2 | 3 | 2.52 | 1.6909 | −8.47 | 34.44 | 30.43 | 4.01 |
| Peptide sequence | m/z | Missed cleavages | Charge | Xcorr | log Sum(k+1)AA | clog P | tR exp | tR pred | Dt R |
|---|---|---|---|---|---|---|---|---|---|
| RADGSINQHPQER | 754.79 | 1 | 2 | 1.4014 | 1.2128 | −10.28 | 14.94 | 13.67 | 1.27 |
| KGTDWNLYFWTAASYNAVIFVFVLV | 1463.68 | 1 | 2 | 1.0082 | 1.9367 | −5.79 | 42.39 | 40.38 | 2.01 |
| ALECFKEMTTKI | 707.86 | 2 | 2 | 1.0212 | 1.4322 | −5.63 | 26.63 | 24.27 | 2.36 |
| VKVIKPDP | 896.11 | 2 | 1 | 0.9391 | 1.1301 | −1.63 | 15.64 | 17.59 | 1.95 |
| AQLSEKKGADGYL | 1380.53 | 2 | 1 | 1.1544 | 1.3902 | −5.26 | 26.46 | 23.20 | 3.26 |
| TRLMGLILAVVAVGMIGAG | 922.17 | 1 | 2 | 1.0633 | 1.6382 | −6.61 | 33.61 | 30.15 | 3.46 |
| SDNNIDKTL | 1020.08 | 1 | 1 | 1.2258 | 1.2125 | −8.54 | 18.87 | 14.98 | 3.89 |
| EEKENWVL | 524.07 | 1 | 2 | 0.9181 | 1.3569 | −5.1 | 26.26 | 22.25 | 4.01 |
| SWIGLPAPIFAGIAAIFAIQP | 718.86 | 0 | 3 | 1.2819 | 1.8072 | −3.13 | 33.64 | 38.24 | 4.60 |
| LLGILTGFFMIGAKRP | 868.09 | 2 | 2 | 0.9776 | 1.6883 | −3.32 | 39.42 | 34.27 | 5.15 |
| ELSASMG | 694.78 | 0 | 1 | 1.1685 | 1.0896 | −5.98 | 18.33 | 12.98 | 5.35 |
| KHGVHIVAGSVAVRKNSDVYNTMYI | 920.72 | 3 | 3 | 1.2386 | 1.6583 | −11.81 | 33.61 | 26.84 | 6.77 |
| DGWKVCLGKVGSMDAHKVVAAIETASKKSG | 1025.52 | 5 | 3 | 1.2037 | 1.7212 | −13.46 | 37.72 | 27.61 | 10.11 |
| EYLDLLEKNVPYPAPSDLIFWSNEDY | 1044.81 | 1 | 3 | 1.1465 | 1.8645 | −4.77 | 48.33 | 38.83 | 9.50 |
| KAEDLLRKVGLFEKRNDY | 1098.25 | 5 | 2 | 1.0209 | 1.6135 | −5.98 | 39.52 | 29.83 | 9.69 |
| LLFKPNEERS | 617.20 | 2 | 2 | 1.1289 | 1.3877 | −3.86 | 10.37 | 24.18 | 13.81 |
| EVTPEIEAAAGKGFTI | 817.41 | 1 | 2 | 1.0159 | 1.4795 | −9.51 | 9.03 | 22.84 | 13.81 |
| NRVEYVKAEIQI | 731.84 | 2 | 2 | 1.0301 | 1.3873 | −5.22 | 40.09 | 23.13 | 16.96 |
| LEEFKKDLH | 580.16 | 2 | 2 | 0.8629 | 1.3695 | −3.24 | 6.94 | 24.07 | 17.13 |
| AGQHERLKEMNVTDT | 865.45 | 2 | 2 | 1.0166 | 1.3300 | −8.47 | 37.37 | 18.82 | 18.55 |
| TGALIVYTSADSVLQIAAHEEVVPLEE | 952.72 | 0 | 3 | 1.1387 | 1.7286 | −5.69 | 52.12 | 33.76 | 18.36 |
| KIDKSIFPGIQGGPLMH | 613.73 | 2 | 3 | 1.0526 | 1.5877 | −4.11 | 12.03 | 30.43 | 18.40 |
| QMLRMMMMQMGMKPSQKKINQMMK | 1481.93 | 4 | 2 | 1.3223 | 1.6338 | −9.93 | 49.34 | 27.48 | 21.86 |
| RILLSLFLS | 531.67 | 1 | 2 | 0.9681 | 1.5406 | −3.02 | 8.19 | 29.74 | 21.55 |
| LTELQVRHII | 1222.46 | 1 | 1 | 1.3716 | 1.4101 | −5.64 | 48.06 | 23.55 | 24.51 |
| EPIQSFFQID | 1224.34 | 0 | 1 | 1.1369 | 1.4701 | −4.17 | 51.92 | 26.60 | 25.32 |
| NRAVGFISFVI | 1223.45 | 1 | 1 | 1.1616 | 1.5144 | −4.62 | 58.59 | 27.68 | 30.91 |
| IHTLEHLLAFTI | 1408.67 | 0 | 1 | 1.0082 | 1.5688 | −5.13 | 81.04 | 29.04 | 52.00 |
| GQEQLIPPLIL | 1221.47 | 0 | 1 | 1.3977 | 1.4715 | −4.79 | 81.34 | 26.17 | 55.17 |
| PIITVAKEAWPTL | 1439.72 | 1 | 1 | 0.9968 | 1.5364 | −6.34 | 83.17 | 27.08 | 56.09 |
| IIGYLDQME | 541.63 | 0 | 2 | 1.0583 | 1.3894 | −2.12 | 83.84 | 25.56 | 58.28 |
| IGLLIFLP | 886.16 | 0 | 1 | 1.2041 | 1.5192 | 1.17 | 93.97 | 32.24 | 61.73 |
| IVLKY | 635.82 | 1 | 1 | 0.9641 | 1.2298 | 0.91 | 86.20 | 22.73 | 63.47 |
| GIIAAYG | 664.77 | 0 | 1 | 1.0865 | 1.2361 | −0.43 | 89.29 | 21.92 | 67.37 |
| PKCPV | 543.70 | 1 | 1 | 0.934 | 0.7843 | −2.24 | 77.72 | 6.00 | 71.72 |
| PQTPVP | 638.74 | 0 | 1 | 1.1985 | 0.8503 | −2.83 | 80.37 | 7.68 | 72.69 |
| LAAGISTI | 745.89 | 0 | 1 | 1.1611 | 1.2704 | −5.24 | 92.62 | 19.36 | 73.26 |
| IDFPTNITMD | 1167.31 | 0 | 1 | 1.3316 | 1.3871 | −5.3 | 96.80 | 23.07 | 73.73 |
| DGITDVL | 732.80 | 0 | 1 | 1.0216 | 1.1875 | −6.2 | 93.5 | 15.96 | 77.54 |
| HGGSLSAPAIH | 1047.15 | 0 | 1 | 1.2372 | 1.2628 | −6.4 | 97.03 | 18.23 | 78.80 |
The model set consisting of 50 peptides with the highest values of Xcorr was used to create a model to predict further retention times of the peptides from proteome of Bacillus subtilis cells. Among this group differences between experimental and predicted retention times ranged from 0.01 to 2.81 min. 42% (21 peptides) of the results were characterized by differences between experimental and predicted retention times lower than 1 min, and for the remaining 58% (29 peptides), these values ranged from 1 to 3 min (Table 2). Taking into account retention times and the values of descriptors for those 50 model peptides, the following specific equation was derived:
The description of tR by (2) was good as documented by the following criteria of statistical quality. All the regression coefficients were highly statistically significant as was the whole equation. Multiple correlation coefficient, R, standard error of estimate, s, and the value of the F-test of significance, F, all were also satisfactory.
Equation (2) provides the predictive model based on experimentally obtained descriptor (log Sum (k+1)AA) and improved by the implementation of molecular-modeling-based descriptor (clog P). Experimentally obtained descriptor (log Sum (k+1)AA) appeared to possess significant contributions into peptides’ retention. However, the log Sum (k+1)AAhas little in common with n-octanol/water partition coefficient—neither for individual amino acids nor for the peptide. The considered analytes were highly ionizable and only minute fraction of molecules can exist in nonionized form in solution. Only for that fraction log P (clog P) properly reflects the ability to partition between aqueous and hydrophobic phase. Therefore, the log Sum (k+1)AA parameter was not considered to mimic clog P; actually it reflects differences in peptides polarities. Instead, clog P was an auxiliary peptide structure descriptor: a correction for log Sum (k+1)AA.
In order to check the correctness of the model, the set of 21 peptides (Table 2), derived from 8 model proteins, was used as the validation set. The predicted retention times, calculated from (2), were then compared to the experimental retention times and the differences between these two retention times were calculated. Differences varied from 0.09 to 3.08 minutes in retention time (mean value 1.29 min, Table 2). For 9 peptides the range of differences between experimental and predicted retention times (42.86%) was from 0.09 to 0.46 min; for 11 peptides (52.38%) the range was 1.07–2.99 min; for 1 peptide (4.76%) this value was over 3 min. Correlation (R = 0.979) between experimental and predicted retention times confirmed additionally the validity of the model (Figure 1), proving that similar values of predicted and experimental retention times of analyzed peptides correlate also with higher probability of identification correctness using Sequest algorithm (Figure 5).
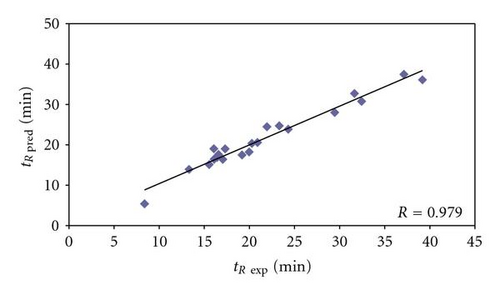
3.2. QSRR-Based Analysis of Peptides from Bacillus subtilis Proteome
Using (1), the predicted retention times for peptides identified for proteome of Bacillus subtilis cells were further calculated (Tables 3–5). The experimental retention times for these peptides were obtained in LC-MS/MS analysis and compared to the calculated ones. Here, the special attention on peptides with low Xcorr (around 1.5) was taken into account to check the applicability of the proposed model and to indicate the potential false positives. In this case, the most important were the attempts to provide the QSRR-based tool to confirm true and false positively identified peptides.
The derived accurate model, as confirmed in Figure 1, was applied to calculate also the retention times of peptides from the real proteome sample of Bacillus subtilis cells. Its correctness was proved first by calculating the predicted retention times of peptides belonging to proteins identified on the basis of more than one peptide with Xcorr above 1.5, that is, those ones that are assumed to be the most confident true positives. It is clearly seen on correlation plot depicted in Figure 2 that the predicted retention times and experimental retention times do not vary significantly, and so it can be concluded that those peptides, and the proteins, to which they are assigned, are correctly identified and really present in the analyzed sample. The detailed accuracy of the peptide identification can be further examined in Table 3. In the set of 54 peptides obtained from digestion of Bacillus subtilis proteome and belonging to proteins identified on the basis of more than one peptide with Xcorr above 1.5, the differences between experimental and predicted retention times varied from 0.08 to 18.07 min (mean value 5.13 min). For 8 peptides, being 14.82% of the set, the difference between experimental and predicted retention times was lower than 1 min. There were 6 peptides (11.11%), which retention times differences ranged between 1 and 3 min. In most cases, differences between experimental and predicted retention times were from 3 to 5 min and then from 5 to 10 min, for 18 (33.33%) and 16 (29.63%) peptides, respectively. 4 peptides (7.41%) were characterized by difference in experimental and predicted retention times ranging from 10 to 15 min. There were even also 2 cases, for which these values varied between 15 and 20 min. The correlation between experimental and predicted retention times can be considered good with correlation coefficient equaled 0.936 (Figure 2). However, some peptides in this set could be considered probably as false positives (e.g., ESIAQVAAISAADEEVGSLIAEAMER, or MSGWLAHILEQYDNNRLIRPR). Generally, at that moment, it was proved that it is again possible to predict the retention times of unknown peptides of Bacillus subtilis proteome, based on retention data obtained experimentally only for the limited number of known model peptides originating from 8 known model proteins.
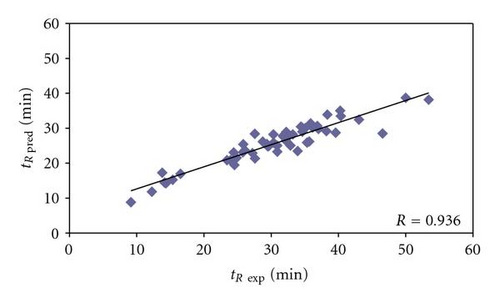
Among 41 Bacillus subtilis peptides belonging to proteins identified on the basis of only just one peptide with Xcorr above 1.5 (Table 4), the difference between experimental and predicted retention times varied from 0.35 to 11.7 min and the mean value was 4.92 min. The predicted retention times of 5 peptides varied from the experimental ones less than 1 min, which refers to 12.20% of the investigated set. For other 8 peptides (19.51%) the difference between experimental and predicted retention times was higher than 1 min, but lower than 3 min. The range from 3 to 5 min in retention time difference was characteristic for 11 peptides, constituting 26.83% of the studied set. The highest numbers of peptides (13) were characterized by 5 to 10 min difference in retention times (31.76%). On the other hand, the highest values, over 10 min, of the difference between predicted and experimental retention times were characteristic for 4 peptides (9.76%) and the largest difference was 11.7 min (Table 4). The correlation between experimental and predicted retention times is still reasonably with correlation coefficient equaled 0.8405 (Figure 3). Some peptides in this set seem to be also false positives (e.g., DQDISGEKATADQLLKDVK or IQNGDPIAGLFDEFTQTVQR), even though they fulfill the established level of Xcorr criterion for proper peptide identification. The differences between predicted and experimental retention times (here 11.49 and 11.70 minutes, resp.) suggest that these peptides, and proteins, from which they originate, may not be really present in the analyzed sample.
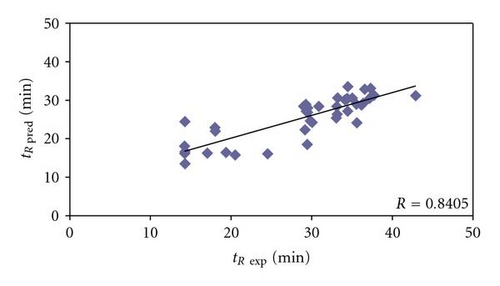
Finally, in the group of 40 Bacillus subtilis peptides, belonging to proteins identified again on the basis of just one peptide, but with Xcorr below 1.5 (Table 5), the differences between experimental and predicted retention times range from 1.27 to 78.80 min (mean value equaled 29.41 min). There were only 4 peptides (10%) with predicted and experimental retention times varied less than 3 min. In next 5 cases this difference was over 3 but lower than 5 min, which makes 12.5%. There were 3 peptides (7.5%) in the range between 10 and 15 min of difference in predicted and experimental retention times. For other 5 peptides, the difference in predicted and experimental retention times was from 15 to 20 min (12.5%). Next 4 (10%) peptides in the group belonging to proteins identified on the basis of one peptide with Xcorr below 1.5 were characterized by 20 to 30 min difference between predicted and experimental retention times. There was 1 case (2.5%), where this difference in retention times ranged between 30 and 50 min. For last 13 peptides (32.5%) in this set the experimental and predicted retention times varied even over 50 min: there were 4 cases (10%), where these values differed between 50 and 60 min; 3 peptides (7.5%) in the 60 to 70 range of retention time difference and 6 (15%) varying more than 70 min (Table 5). It must be stated that for peptides belonging to proteins identified on the basis of one peptide with Xcorr below 1.5, correlation between experimental and predicted retention times cannot be observed (Figure 4). Therefore it may be concluded that a large number of peptides in this set should be classified as false positives, especially those ones with extremely high difference between experimental and predicted retention times (e.g., HGGSLSAPAIH, DGITDVL, IDFPTNITMD, or LAAGISTI, where these differences are 78.80, 77.54, 73.73, and 73.26 minutes, resp.).
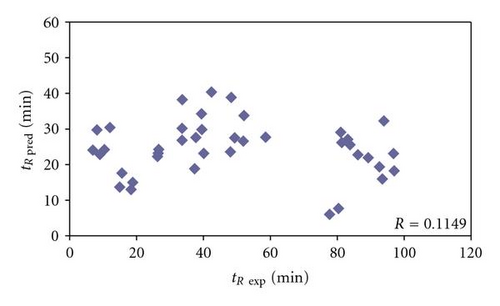
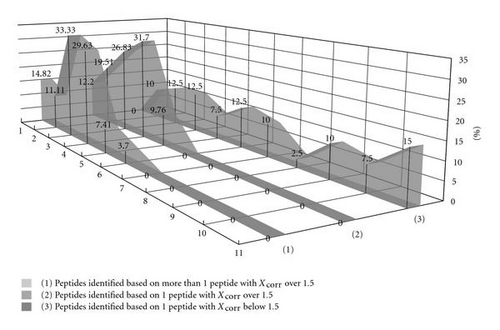
Generally, it can be noticed that lower values of Xcorr correlate with the higher percentage of peptides are characterized by larger difference between experimental and predicted retention times (Figure 5). In particular, it is observed, when comparing the percentage of cases, where differences between predicted and experimental retention times are higher than 15 min, that in each group of Bacillus subtilis peptides belonging to proteins and identified on the basis of the following: one peptide with Xcorr below 1.5 (Table 5), one peptide with Xcorr over 1.5 (Table 4), and more than one peptide with Xcorr over 1.5 (Table 3). The percentages of peptides characterized by higher than 15 min difference in experimental and predicted retention times in these groups are 57.5%, 0%, and 3.7%, respectively. On the other hand, in model and testing sets of peptides obtained from model proteins all differences between predicted and experimental retention times were lower than 15 min (Tables 1 and 2). It is noticeable that high percent of peptides with low values of Xcorr was characterized by differences between predicted and experimental retention times larger than 15 min, what can provide an additional indication that they could be considered as potential false positives and in fact were not identified in the analyzed sample. Therefore, QSRR equation to predict peptides retention times might be useful tool to increase throughput of the protein identification in LC-MS/MS.
4. Conclusions
Quantitative structure-retention relationships (QSRRs) model derived with the use of set of peptides identified with the highest scores and originated from 8 known proteins was tested with regards to its predictive capability of the retention time prediction. Bacillus subtilis proteome digest was used to check the predictive ability of the novel QSRR model proposed in the study. It was found that the QSRR approach can be applied as the additional constraint in proteomic research verifying results of MS/MS ion search and confirming the correctness of the peptides identifications along with the indication of the potential false positives. The results suggested that due to the QSRR used for the prediction of peptide retention, liquid chromatography separation stage of proteomic research could be useful in the final identification of peptides, especially considering the most uncertain protein identifications based on findings for just one peptide.
Acknowledgments
The work was supported by the Polish State Committee for Scientific Research Projects N N405 1040 33 and by Polish-Italy bilateral scientific and technological cooperation project 2007–2009.




