Kuri: A Simulator of Ecological Genetics for Tree Populations
Abstract
This paper presents Kuri, a software package developed to simulate the temporal and spatial dynamics of genetic variability in populations and multispecies communities of trees, as well as their interactions with environmental factors. A conceptual model using agents inspired on Echo models is used to define the environment, the hierarchical structures, and the low-level rules of the system. At the individual agent (tree) level a genetic algorithm is used to model the genotypic structure and the genetic processes, from a small set of simple rules, complex higher-order population, and environmental interactions emerge. The program was written in Delphi for the Windows environment, and was designed to be used for educational and research purposes.
1. Introduction
Computational simulations have been widely used to represent and simulate genetic processes. Some examples that fall within the scope of this work include simulations that were mainly developed for educational purposes, such as Populus [1], WinPop [2], Sigex [3], and Genup [4]. Others were developed for practical applications and are used in, for example, programs for forestry management [5, 6]. Simulations are also used to understand complex adaptive systems from a “first-principles’’ approach. Conceptual models such as Holland’s Echo model are widely used [3, 7]. In this paper we discuss the software Kuri, a simulator of ecological genetics for tree populations. The program allows investigation of genetic and microevolutionary phenomena of tree populations or entire forest communities. Kuri can be used to study the dynamics of neutral genetic markers under certain biological factors and environmental constraints, such as dispersion mechanisms and geographical barriers, among others. Either real field data or artificial genetic and environmental parameters can be used for a given simulation. The latter allows creation and testing of hypothetical situations for theoretical and/or educational purposes.
Along the same lines used in the Sigex simulator [3], Kuri mechanistically implements low level elementary biological rules, for example, Mendelian segregation and mating, which interact to produce patterns that are analogous to those observed in natural populations, such as Hardy-Weinberg equilibrium. Thus, population data generated in Kuri is not obtained from sampling from a distribution, but is instead, a quantifiable element at the population level which emerges from the low level mechanistic interactions at the genetic level.
2. Software Kuri
Kuri was developed using the Delphi programming language, an object oriented derivative of Pascal. It uses a modular construct which allows easy implementation of new functions and applications and also enables seamless integration with the other modules. The program needs limited computational resources and will run on a 1.2 GHz processor with 512 M RAM and 2 GB free space on the hard disk. The operating system can be Windows XP or above. The current version of Kuri consists of three main modules: the graphical user interface (GUI), a dispersion module, and a genetic operators (KGOP) module.
In Kuri, environmental factors that affect germination/viability of seeds are combined to create a heatmap in which the colors represent different germination probabilities (Figure 1 shows a screenshot of Kuri with a probability heatmap based on satellite images). The GUI allows the user to import images, such as satellite photographs or schematic pictures to represent features of interest in a given area. Up to five images at a time can be used to represent different environmental parameters in a given simulation. Each image could represent, for example: (1) inhospitable areas where seeds cannot germinate, (2) areas of human intervention, (3) soil depth, (4) soil quality, and (5) hydrology. Note that each environmental parameter can be altered by the user. For instance, the map of soil depth can be replaced by a topographic map of the region, if it is more relevant for a particular research topic. Currently Kuri works with bitmap image files which are easy to generate or to convert from other file formats with available imaging software.
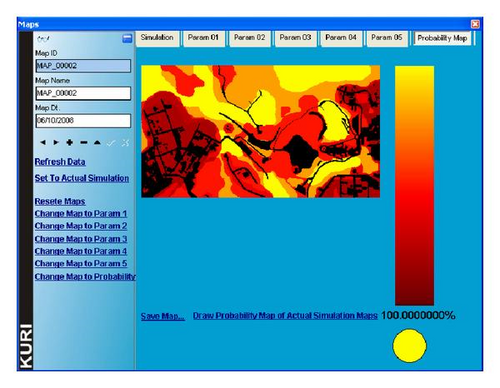
For each of these (up to 5) environmental parameters, probabilities of germination success on its respective map can be assigned to either discrete features or interval ranges for continuous features. Probabilities are color coded on the map and resolved at the pixel level. This means that each pixel can be assigned its own independent probability, irrespective of neighboring probabilities, allowing for a discontinuous probability landscape. The color scheme of probabilities is user defined which makes it easy to identify features. For example, areas where the germination of seeds is impossible such as buildings, streets, water masses, or rocky terrain are by default represented in black (Figure 1). Since colors and probabilities are linked, it is simply a matter of changing the probability associated with a specific color to update all points in the map to a new probability.
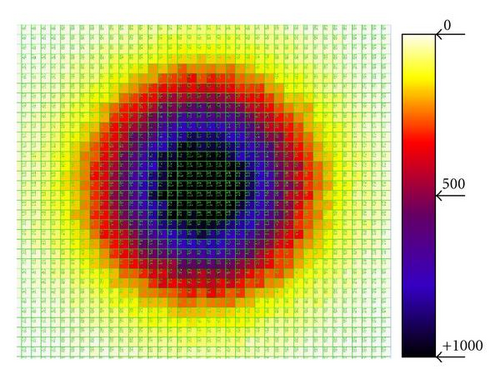
To simulate the dispersion of pollen and seeds, the total simulation area is divided into cells of user defined granularity, with height and width in pixels defined by the user. For each grain of pollen and for each seed in a particular cell, the probability of dispersing to another cell depends on the wind. This is achieved through a simple probabilistic function, where an integer ranging between 0 and n (n is a user defined parameter between zero and the total number of grid cells) is randomly sampled from a uniform distribution and multiplied by the probability of the wind direction (Figure 2). The value of n effectively sets the dispersion boundaries. Wind direction is also a user defined parameter consisting of a set of probabilities for each cardinal point and a decay rate from the center of dispersion.
The KGOP module is essentially a relational database that holds information on the biological community, the various species and their respective biological features, the genetic features of the species, and the genetic composition (essentially all allelic frequencies across all genes) of the population of each species, including the chromosome sets for each species with the number of loci in each chromosome, the linkage map between loci, and the number of alleles in each locus.
For each species the following biological parameters can be stored: the individual occupation range (species boundaries), the dispersion of pollen and seeds, the maximum and minimum ages of reproduction and images for each age group of the specimens. For this last parameter, Kuri’s image collection can be used, or the user can import and add his/her own images. All parameters relate directly back to their original biological meaning and can be used quite intuitively.
For each new species added to the database, the user should specify the number of chromosomes that will be used in the simulation and the number of loci per chromosome. Up to 26 allele slots are available for each locus. The chromosomes and genes that will effectively be used in a simulation can be selected prior to a run. Recombination frequencies between genes should also be specified by the user. Mutation rates are the same for all genes/alleles, but can be changed across runs. Note that mutation in Kuri does not generate new allelic variants; it simply swaps an allele for another one from the database with a uniform probability. Initial populations are by default generated in Hardy-Weinberg equilibrium based on the given allelic frequencies (allelic and genotypic frequencies and chi-squared values for Hardy-Weinberg equilibrium tests are given in Kuri), but different initial population structures can be defined.
Computationally, the genetic mechanisms of the species are simulated using a Genetic algorithm (GA) [8]. In previous work we have [3] detailed how to implement these genetic processes and shown that they conform to theoretical predictions of population genetics. But briefly, GAs are the class of Evolutionary Computation algorithms which most closely mimic evolutionary processes at the genetic level. GA organisms are represented as linear strings which are referred to as chromosomes. The value in each position of the string is an allele and the position itself is a gene or locus. The combination of values (alleles) in the string (chromosome) can be mapped to a phenotypic expression (note that in Kuri all alleles are neutral). Thus GAs operate at two structural levels: a genotypic and a phenotypic one. Crossover swaps chromosome parts between selected parents to form the offspring while mutation changes the value of alleles at randomly selected loci.
The practical limits for the software (i.e., number of individuals, size of geographic area, number of generations, etc.) relate to the limits of the MySQL database. The effective size of the tables for the database is normally restricted by the operating system′s filesystem. The total number of loci are limited to 128.
3. A Simulation Example: Dispersion Effects
In this section we discuss a simple simulation of seed dispersion effects to illustrate the use of Kuri in population genetics. We created a single species population in a homogeneous environment with a single locus and two segregating alleles of interest. Initially all plants were heterozygous. We ran the simulation under two scenarios with different wind intensities (strong and mild winds). Wind intensities affect the dispersion process and, consequently, the distribution of genetic variability.
For each scenario, five simulation runs of 25 generations each were performed. In Figure 3(a) the distribution pattern of the plants across generations is depicted under strong winds for the first replicate. Note that the distribution pattern remains homogeneous over the generations, meaning that dispersion occurs with a high level of panmixia, that is, random matting. Figure 3(b) shows the mild wind scenario over generations for the first replicate. Note the formation of endogamic groups, that is, most matings occur within subpopulations, which are to be expected in an environment that does not favor dispersion.
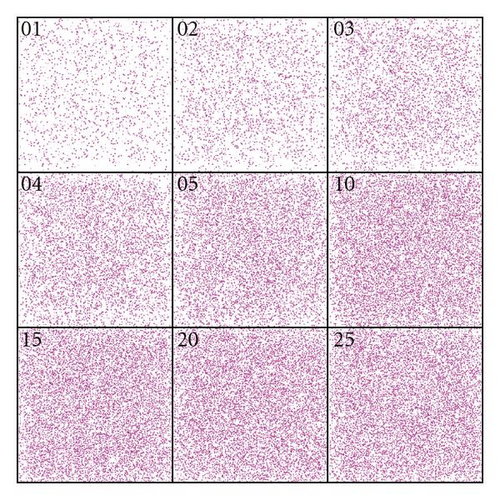
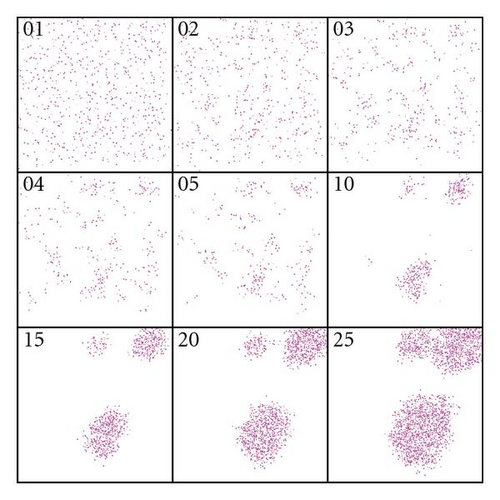
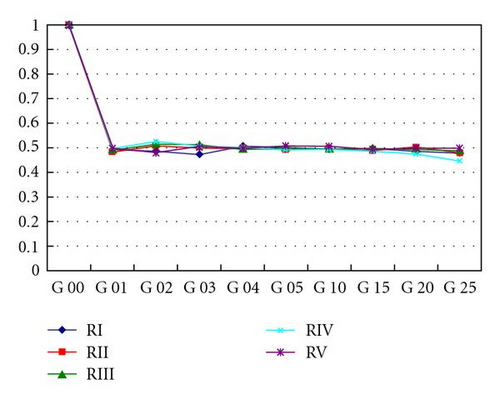
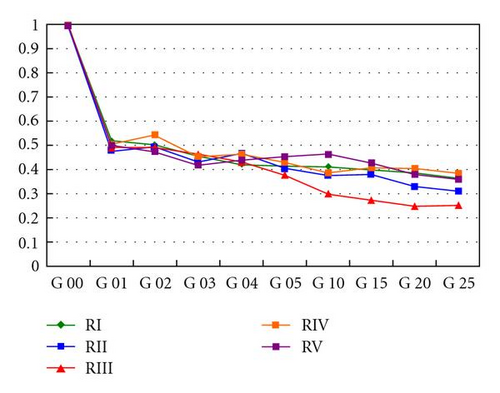
The dynamics over time of the frequencies of heterozygotes for the five repeats are shown in Figure 4(a) (strong wind) and Figure 4(b) (mild wind). In the former, the frequencies of the heterozygotes reach equilibrium after the first generation, oscillating around 0.5. In the second case, a decrease in heterozygosity is noticeable since the subdivision creates a new population structure—an example of a genetic phenomenon known as Wahlund effect. In all strong wind repeats, equilibrium was reached and maintained across generations whilst with mild winds the number of homozygotes increases over time.
Even this simple scenario can provide insights about natural populations. Jump and Penuelas [9] showed that habitat fragmentation caused by human activity led to high levels of inbreeding due to a Wahlund effect. This was the first study showing that even widespread wind-pollinated trees are negatively affected by habitat fragmentation. Argumentatively, Kuri could be used to estimate genetic effects under different scenarios. For example, a satellite image of a forested area can be artificially fragmented in different patterns and these used to estimate the genetic effects of deforestation. This has implications for urbanization decisions and can assist in finding a solution that minimizes human impact. Clearly, for realistic results, there has to be reliable data and detailed knowledge of the ecology of the species.
For population studies the simulated data can be treated and analyzed as if it were real data, with the advantage of having full knowledge of the population structure and a handle on the mechanisms that yielded the dataset. For example, data from only the last generation could be used to make inferences about the evolutionary processes that were acting on the population. The degree of deviation from HW equilibrium can be calculated and used to estimate parameters such as FST [10]. These results can then be compared to the original experimental model to provide insights about the dynamics of the system.
Kuri was designed to simulate microevolutionary phenomena which can be detected through molecular markers which are usually selectively neutral. Neutral markers have the advantage that since they are not being selected for or against, any observed fluctuations in allelic frequencies are only due to population structure and environmental effects.
4. Concluding Remarks
Kuri can be used to simulate a wide range of biological scenarios. It allows manipulation of the genomes, alleles, and genotypes of different plant species and the interactions of these populations with the ecosystem. Kuri’s database can be used to store different genetic models of species, being these based on real data of species or virtual organisms tailored for educational purposes. Alongside the biological parameters, the user can manipulate and/or create environmental parameters based on field data (such as satellite imagery) to study how these affect the genetic composition and size of populations. The software meets theoretical expectations, but it still has to be tested under realistic scenarios for which real data is available and results can be compared. Due to the lack of real data testing it is still unclear how detailed field data and knowledge of the ecology of the species has to be able to make valid inferences. Future work and user feedback may assist in answering these questions.
The software is modular. It was designed so that it can be modified and expanded to simulate other phenomena. For example, in the current version all genes/alleles are neutral, but it is straightforward to implement environmental constrains associated to the genotypes in order to simulate natural selection, or even simulate molecular evolution by adding another module that allows handling each allele as a DNA base pair. Kuri is open source and freely available from the web address: http://www.allesys.com.br/kuri/.




