Origin of endemic species in a moderately isolated ancient lake: The case of a snakehead in Inle Lake, Myanmar
Abstract
Inle Lake is an ancient lake in Myanmar, which is an important area with unique and diverse fauna. Its ichthyofauna is believed to have formed non-radiatively, but the historical processes are poorly understood. To elucidate the mechanisms that shape species diversity in this moderately isolated biogeographical ‘island’, this study focused on a typical endemic fish of Inle Lake, Channa harcourtbutleri (Channidae, Anabantiformes), with its widely distributed sister species, C. limbata, and estimated the historical distribution and diversification processes of the endemic fish based on genome-wide polymorphism (MIG-seq) and mitochondrial DNA data. Channa harcourtbutleri contained two genetically and morphologically distinct groups inhabiting Inle Lake and the surrounding rivers respectively. These two groups were genetically the closest to each other; however, the riverine group showed some similarity to the closely related species, C. limbata from Southeast Asia. The mtDNA haplotypes of the endemic species were not monophyletic; most of the riverine group had haplotypes identical or close to those of C. limbata from the upper Irrawaddy and Salween rivers. The time tree suggested that C. harcourtbutleri diverged from C. limbata in the early Pleistocene and then experienced secondary contact with C. limbata in the late Pleistocene. Genetic and morphological differentiation within C. harcourtbutleri suggests that local adaptation to different environments has played an important role for the coexistence of its two forms with some reproductive isolation. Further, the results highlight the importance of multiple colonization and allopatric speciation in shaping biodiversity in the long-term, moderately isolated environments.
1 INTRODUCTION
Understanding how local faunas form and transition is one of the major topics in biogeography. In the study of community formation, island-like habitats with distinct boundaries, such as islands, lakes and mountain tops, have long been a focus of attention (Brooks, 1950; Losos & Ricklefs, 2009; MacArthur & Wilson, 1963, 1967; Rahbek et al., 2019). Island biogeographical theory has attempted to explain species composition in an ‘island’ using the dynamics of colonization from the species pool ‘continent’ and speciation and extinction within the island (Whittaker et al., 2008, 2017). When the distance between an island and the continent (source area) increases, the contribution to species composition shifts from colonization to speciation (Emerson & Gillespie, 2008). In strongly isolated oceanic islands and ancient lakes, in situ diversification often accounts for the species diversity in various taxa (Gillespie et al., 2020). However, in more general communities with moderate isolation, the continent (source) and timing of colonization should have a more important role in the community assembly (Hauffe et al., 2020; Mittelbach & Schemske, 2015; Rosindell & Phillimore, 2011). Colonization can occur multiple times and from multiple source regions (Johnson et al., 2019; Kimura et al., 2022; Šlechtová et al., 2021); therefore, detailed phylogeographical studies using molecular markers based on regionally exhaustive sampling are needed for identifying the processes of community formation with colonization (Cox et al., 2020).
Ancient lakes, often defined as those that have existed since 130,000 BP (Hampton et al., 2018), have unique biota because they maintain a relatively closed, stable aquatic environment for long periods. They have attracted the attention of many biologists as ‘natural experiments’ for understanding the processes and mechanisms that lead to adaptation and speciation (Cristescu et al., 2010). Adaptive radiation is the prominent process underlying the biodiversity in ancient lakes; it is a factor for the explosive in situ speciation (Henning & Meyer, 2014; Schluter, 2000). Non-radiative processes, such as colonization and long-term isolation of multiple ancestors, also greatly contribute to the formation of unique fauna (Hauffe et al., 2020; Tabata et al., 2016). However, this has received little attention compared to radiation (Seehausen, 2006). Ancient lakes where explosive radiation did not occur are good systems to examine the combined effects of isolation and colonization from surrounding water systems on the formation of the lake fauna.
Inle Lake is the only ancient lake in the continental part of Southeast Asia; it is located in Shan Plateau of Myanmar. The age of Inle Lake is estimated at about 1.5 million years (Hampton et al., 2018), although the geological evidence remains unclear. Inle Lake is a small lake with an area of 116 km2 and a depth of 2 m (Toke et al., 2013); however, it has rich biodiversity and several endemic species (Abell et al., 2008; Annandale, 1918; Fuke et al., 2021). Fifteen endemic fish species have been reported from the lake, and there might still be several undescribed species (Kano et al., 2016, 2022). The native fish fauna of Inle Lake is composed of a wide range of taxa (28 genera from 13 families; Kano et al., 2016, 2022), which are believed to be formed through non-radiative processes (Fuke et al., 2022). Annandale (1918) suggested that the unique fish fauna of Inle Lake includes widely distributed species that colonized the lake from surrounding areas and some endemic species derived through geographical isolation. In addition, several species treated as endemic species of Inle Lake also occur in the surrounding areas, and some of them are known to show genetic differentiation among regions (Fuke et al., 2022; Kano et al., 2022). The fishes of the Inle region, defined here as the area including Inle Lake and surrounding regions in Shan Plateau, will provide good materials for exploring the processes by which the interactions of in situ diversification and colonization shape local biodiversity. However, detailed historical biogeographical studies are lacking in this region.
In this study, we aimed (1) to estimate the historical patterns of colonization from the surrounding water systems to Inle Lake and (2) to characterize the in situ diversification of endemic fish species within the region. We focused on Channa harcourtbutleri (Annandale, 1918) (Channidae, Anabantiformes), an endemic snakehead species occurring in and around Inle Lake (Rüber et al., 2020). Channa harcourtbutleri is one of the local populations derived from Channa limbata (Cuvier in Cuvier & Valenciennes, 1831), which is distributed widely from Southeast Asia to southern China except the Inle region (Courtenay & Williams, 2004; Rüber et al., 2020). Channa limbata is genetically differentiated by region (Conte-Grand et al., 2017; Rüber et al., 2020); therefore, it would be possible to estimate the origin of C. harcourtbutleri, to understand from which region and when the ancestral population of this endemic species colonized the Inle region. We first estimated the phylogeographical patterns of C. harcourtbutleri and C. limbata based on genome-wide SNPs and mitochondrial DNA (mtDNA) to infer the origin of the populations in the Inle region. Second, we examined the genetic population structure and morphological characters of C. harcourtbutleri to determine whether population diversification occurred within the region. These results assess the formation history of the non-radiative biodiversity in this region and provide insights into the dynamic relationships between regional biodiversity and connectivity with and isolation from the surrounding areas.
2 MATERIALS AND METHODS
2.1 Taxonomic background and identification
In this study, we focused on two species of snakeheads, Channa harcourtbutleri, an endemic species in Inle Lake and its surroundings, and C. limbata, widely distributed in Southeast Asia. These species belong to Channidae, which includes two genera with 56 species distributed in Africa and South to East Asia (Adamson et al., 2010; Fricke et al., 2022). The species of this family have a suprabranchial organ for air-breathing and are adapted to a variety of environments, including streams, lakes and wetlands (Musikasinthorn, 2003). Among them, C. harcourtbutleri and C. limbata are small snakehead species with a maximum body size of about 20 cm, inhabiting hill streams, lakes and ponds. Channa limbata had been regarded as a junior synonym of Channa gachua (Hamilton, 1822) (Roberts, 1993); however, considering the distinct phylogenetic divergence, the western populations are referred to as the C. gachua group and the eastern populations, as the C. limbata group, bound by the Indo-Burman ranges (Conte-Grand et al., 2017). Channa harcourtbutleri is a regional clade nested within the C. limbata group (Rüber et al., 2020).
Ng et al. (1999) documented that C. harcourtbutleri can be morphologically distinguished from C. gachua (including C. limbata) by body colour and head shape. However, they did not provide detailed quantitative comparisons that account for allometry and geographical variation. Channa harcourtbutleri occurs not only in Inle Lake but also in stream habitats surrounding Inle Lake (Kano et al., 2016, 2022). Although intraspecific differentiation in morphological and genetic features is often observed among freshwater fishes inhabiting different environments (e.g. lakes and streams; Ravinet et al., 2013; Theis et al., 2014), no detailed comparisons have been made for the Channa species. Therefore, we tentatively treated the specimens of the C. limbata group based on their locality as follows: specimens from the Inle region as C. harcourtbutleri following Annandale (1918) and those from Indochina, China and Myanmar except the Inle region as C. limbata following Conte-Grand et al. (2017). Recently, Laskar et al. (2023) treated specimens from the Andaman Islands and mainland India as C. harcourtbutleri based on mtDNA barcoding only. However, this cannot be validated considering the mtDNA diversity of the C. limbata group (Conte-Grand et al., 2017; Rüber et al., 2020; see also the results of this study), as well as the nomenclatural priority.
2.2 Fish sampling
We used a total of 152 specimens of C. harcourtbutleri collected from various habitats or purchased from local markets in and around Inle Lake by Kano et al. (2016) and Kano et al. (2022) from 2014 to 2016, and 2020 (Tables S1 and S2). Among these, 137 individuals were used for genetic analysis, of which 53 were used for morphological analysis. Specimens obtained from local markets could potentially have been collected far from the market areas, but based on local interviews and the style of selling, we judged that they were not transported long distances and used these samples (Table S1). We grouped the sampling sites into the following 10 areas (Figure 1): Inle Lake, collected at four sites on the lake and purchased at two local markets; streams north of the lake (North, seven sites); streams west of the lake (West, two sites); Inn Dein Stream (Inndein, three sites and one local market), a stream flowing into the lake from the southwest; Min Ywar Village (South, one site), about 15 km downstream from the lake; Moe Byel Dam Lake (Dam, one local market), 60 km downstream from Inle Lake; Loikaw (two sites and one local market), 20 km downstream from the dam; Heho (three sites), about 20 km northwest of Inle Lake separated by highlands; Aungpan (one site), about 10 km further west of Heho, separated from Inle Lake by highlands; and Hopong (two sites), about 30 km northeast of Inle Lake, separated from the lake through the river by about 400 km (Kano et al., 2022).
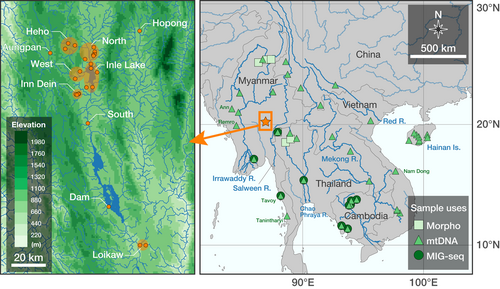
A total 33 specimens of C. limbata were collected from Myanmar, Thailand and Cambodia for genetic analysis. In addition, we used 22 C. limbata specimens from Myanmar and Thailand for morphological analysis, which were not used for genetic analysis (Figure 1).
We used the DNA samples shared with Kano et al. (2016, 2022). DNA from some samples was newly extracted using the Wizard Genomic DNA Purification Kit (Promega).
The mtDNA cytochrome b (cytb) sequences of 67 individuals of C. limbata for which locality information was available were obtained from the International Nucleotide Sequence Database (Figure 1; Table S1). We additionally included eight Channa species and 30 other anabantiform species as outgroups from the database (see Table S1).
2.3 DNA sequencing
To assess the genome-wide polymorphism, we used the multiplexed ISSR genotyping by sequencing method (MIG-seq; Suyama & Matsuki, 2015; Suyama et al., 2022). The MIG-seq library was prepared according to Watanabe et al. (2020) and Onuki and Fuke (2022) for 136 individuals of C. harcourtbutleri and 31 of C. limbata. The library was sequenced on MiSeq (Illumina) with MiSeq Reagent Kit v3 (150 cycle) or outsourced to Novogene and sequenced on NovaSeq 6000 (Illumina).
The mtDNA cytb region was amplified using PCR with the following primers: L-Glu (5′- CTA ACC AGG ACT AAT GGC TTG AA-3′) and H-Thr (5′-CGG CTT ACA AGA CCG GCG CTC TGA-3′) (Takahashi et al., 2016). The PCR conditions were as follows: initial denaturation at 94°C for 120 s; 30 cycles of denaturation at 94°C for 15 s, annealing at 52°C for 15 s, extension at 72°C for 30 s and final extension at 72°C for 420 s. Sequencing was performed on an ABI 3130xl Genetic Analyzer (Applied Biosystems) following the method of Fuke et al. (2022) or outsourced to Macrogen Japan and sequenced on ABI 3730xl DNA analyser. The sequences were checked for quality and edited using the mapping function of Unipro UGENE 41.0 (Okonechnikov et al., 2012); multiple alignments were performed using MAFFT (Katoh & Standley, 2013). Finally, the cytb sequences were obtained for a total of 137 and 98 specimens for C. harcourtbutleri and C. limbata respectively.
2.4 MIG-seq data treatment and SNP detection
The raw data sequenced by NovaSeq 6000 were demultiplexed using the process_shortread program in Stacks 2.49 (Rochette et al., 2019). Quality control and trimming of the obtained raw reads were performed using fastp program (Chen et al., 2018). All reads were trimmed to 80 bases: the first 14 bases of Read 2 were trimmed for Miseq data; the first 35 and last 29 bases of Read 1 and 17 and 52 bases of Read 2 were trimmed for NovaSeq data. In addition, the row quality bases (<Q30) and primer sequences were removed.
Detection of single nucleotide polymorphisms (SNPs) was performed using the denovo_map.pl pipeline of Stacks with paired-end mode with the setting: maximum distance allowed between stacks (M) = 5; a minimum depth of coverage required to create a stack (m) = 5. Outfiles for the following analyses were generated using populations program in Stacks with the following setting: minimum percentage of variant sites shared by all individuals (R) = 0.8; only the first SNP per locus (--write-single-snp). We excluded sites with heterozygosity greater than 75% (--max-obs-het = 0.75) and less than two minor alleles (--min-mac = 2). All other parameters were set as the default setting.
2.5 Population structure and phylogenetic analyses based on SNPs
To estimate the population structure of C. harcourtbutleri and C. limbata, we conducted principal component analysis (PCA) using PLINK (Purcell et al., 2007). To estimate individual admixture proportions, we performed unsupervised clustering using the likelihood model-based program ADMIXTURE 1.3.0 (Alexander et al., 2009) at each number of genetic populations (K) from 1 to 10 with a convergence criterion set to 10−4 (C = 0.0001). These analyses were repeated 100 times with different seed values, and the optimal K value was estimated based on the lowest mean cross-validation error (CV-error) value. The outputs of the runs with the lowest CV-error for each K-value were plotted. In the following analysis, individuals with a major ancestry proportion below 90% in the best K were treated as putative hybrids. Summarization of CV-error values and visualization of the output data were performed in R 4.1.1 (R Core Team, 2021).
To estimate the relationship among clades, the network tree inference was performed using SplitsTree 4.14.7 (Huson & Bryant, 2006) based on the Neighbour-net method with the uncorrected p-distance.
2.6 Haplotype network and phylogenetic analyses based on mtDNA
To estimate the population structure of C. harcourtbutleri and C. limbata based on mtDNA cytb sequences, we detected the haplotypes and conducted a haplotype network analysis using POPART 1.7 (Leigh & Bryant, 2015) based on the TCS algorithm (Clement et al., 2000). The networks were generated based on two datasets, one with C. harcourtbutleri only and the other with both species. Channa limbata used in the network analysis were limited to samples from the Salween and Irrawaddy River systems, which formed a monophyletic group including C. harcourtbutleri.
To estimate phylogenetic relationships among mtDNA cytb haplotypes detected in C. harcourtbutleri and C. limbata, a maximum likelihood (ML) tree inference was performed using IQ-TREE 2.1.3 (Minh et al., 2020) with 38 outgroup sequences. The cytb sequences were partitioned by codon position, and a substitution model was selected for each position using ModelFinder (Kalyaanamoorthy et al., 2017). The estimated trees were visualized using FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
2.7 Divergence time estimation
To estimate the divergence time of C. harcourtbutleri from C. limbata, a time-calibrated tree was inferred based on mtDNA using BEAST 2.6.7 (Bouckaert et al., 2019). All haplotypes of C. harcourtbutleri and C. limbata were used in the analysis. The cytb sequences were partitioned by codon position, and the following setting was applied for all codon positions; the substitution and site models were set to the same as in the IQ-TREE2 analysis, the clock model to the Relaxed Clock Log Normal model, and the tree prior to the Birth-death model. The analysis was carried out with three calibration points according to Rüber et al. (2020) and Wu et al. (2019): concestor of Parachanna stem (log-normal, offset = 33.0, log mean = 0.1, log stdev = 0.8), concestor of Osphronemus stem (log-normal, offset = 28.5, log mean = 0.23, log stdev = 1.0), concestor of Anabantidae stem (uniform, lower = 24.3 Ma, upper = 163.5 Ma). All other parameters were set at the default setting. MCMC was performed for 300,000,000 generations with sampling every 10,000 generations. We confirmed that all parameters had ESS values above 200 using Tracer 1.7.1 (Rambaut et al., 2018). TreeAnnotator excluded the first 10% of trees as burn-in and combined all trees into a single tree.
2.8 Morphological analyses
Three phenotypic datasets were obtained to identify morphological characteristics of C. harcourtbutleri: body shape based on geometric morphometrics, and six counting and 11 metric traits. Specimens were fixed in 10% formalin and then placed in 70% ethanol. For C. harcourtbutleri, only specimens grouped by genetic analysis were used for morphological analysis. For C. limbata, all specimens were pooled and treated as one group, regardless of whether they were genetically analysed or not. Specimens smaller than 100 mm in total length were not included in the morphological analysis, with referring to the mature body size of C. gachua, the closely related species to the two species (Silva, 1991).
The number of fin-rays and lateral line scales (LLS) were counted, as counting traits. The length of dorsal fin base (LDFB), anal fin base (LAFB), pectoral fin (PFL), ventral fin (VFL), caudal fin (CFL), 1st ray of dorsal fin (DFL1), and nasal tube (NTL), mouth width (MW), head width (HW), body width (BW) and inter orbital width (IOW) were measured as metric traits and calculated as a percentage of standard length (SL) (Hubbs & Lagler, 1958; Musikasinthorn, 1998). The final dataset included 53 C. harcourtbutleri and 22 C. limbata specimens.
Sample images for geometric morphological analysis were taken using a digital camera under the same condition. These images are deposited in ffish.asia (https://ffish.asia), a biodiversity database of freshwater fish and freshwater organisms in Asia (Kano et al., 2013; Watanabe et al., 2010). Landmarks were plotted on tpsDIG2 2.3 (https://www.sbmorphometrics.org/soft-dataacq.html) for the ventral and left images of the specimens. The ventral side and left side of the body were plotted with eight and nine landmarks respectively. In order to alleviate the potential effects of body axis curvature, the position of the landmark of the ventral side was limited to the front half of the body, which was hardly affected by the curvature. For the lateral side analysis, specimens with obviously distorted body axes were not included. The final datasets consisted of n = 72 (51 C. harcourtbutleri and 21 C. limbata) for the ventral side and n = 51 (37 C. harcourtbutleri and 14 C. limbata) for the lateral side. The differences in position, direction and size of the samples were corrected using Procrustes analysis; the principal component analysis (PCA) was carried out using MorphoJ 1.06 d (Klingenberg, 2011).
The results were compared among the genetic groups based on the SNP analysis. The Kruskal–Wallis test was performed for each PC score, counting and metric trait to identify the differences among groups. We performed multiple comparisons using the Steel–Dwass test to examine which groups differed. The significance level was set at 1% in all statistical tests. These calculations and graphical visualization were performed using R 4.1.1.
3 RESULTS
3.1 Population structure and phylogenetic relationships based on SNPs data
PCA results based on 231 SNPs for 167 individuals of Channa harcourtbutleri (n = 136) and C. limbata (n = 31) showed that C. harcourtbutleri formed a single cluster, while C. limbata clustered by location, with the lower Irrawaddy River, middle Salween River and Indochina clusters in PC1 in this order (Figure 2a). For the C. harcourtbutleri only data, PCA based on 236 SNPs for 136 individuals showed two dense clusters and some intermediate individuals in PC1. Some of the intermediate individuals from Hopong separated from the others in PC2. One cluster consisted mainly of individuals from Inle Lake-to-Loikaw (outflow river); another consisted of individuals from the surrounding rivers. Hereafter, these two clusters of the endemic species, which were also supported by ADMIXTURE analysis (see below), are referred to as SNP Cluster 1 (Inle Lake and its outflow) and 2 (surrounding rivers) respectively. Some sites, such as North, Inndein and Hopong, had individuals from both clusters. The intermediate individuals in PC1 consisted of some specimens from North and Hopong.
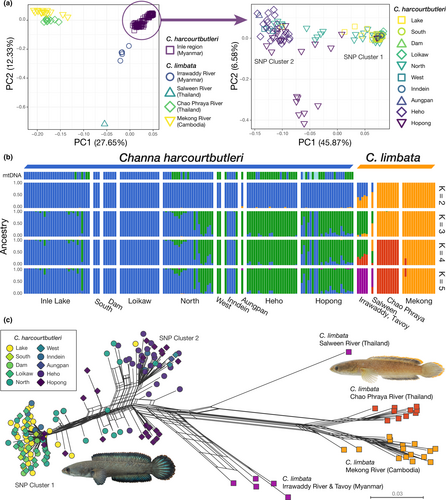
ADMIXTURE analysis, including the two species, showed the optimal number of clusters was K = 4 or 5 where the mean or minimum CV-error value was minimized respectively (Figure 2b). The two clusters of C. harcourtbutleri shown in K = 3–5 corresponded to the two clusters in the PCA results (i.e. SNP Clusters 1 and 2); individuals consisting of both elements corresponded to the intermediate individuals of the two clusters in the PCA results. Individuals from the two clusters were observed sympatrically in Inle Lake, Inndein, North and Hopong. Among them, individuals having both cluster elements, as putative hybrids between the two clusters, were found in Inle Lake, North, Heho and Hopong. North and Hopong were particularly notable for the sympatry and admixture of the two clusters. These results were also supported in the dataset for endemic species only (Figure S3). In the case of K = 5, C. limbata was composed of three clusters that were divided by region: Myanmar, Thailand and Cambodia. Channa limbata from the Irrawaddy and Salween rivers partially contained a genetic component of SNP Cluster 2 of C. harcourtbutleri in K = 2–4.
The Neighbour-net network showed two genetically differentiated groups in C. harcourtbutleri corresponding to SNP Clusters 1 and 2 with some putative hybrids between them (Figure 2c). SNP Cluster 2 and C. limbata were closer in the network than SNP Cluster 1 and C. limbata. Four populations of C. limbata were detected, in which the population most closely related to C. harcourtbutleri was from the Irrawaddy River, followed by that from the Salween River.
3.2 Phylogenetic relationships based on mtDNA
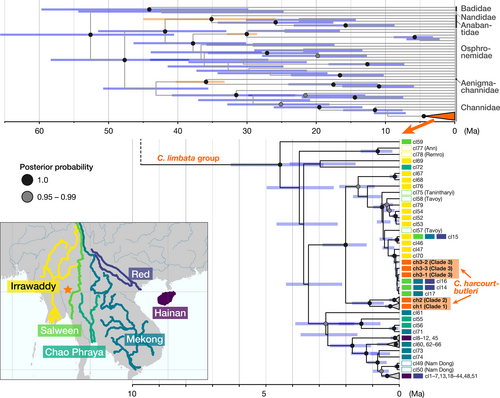
Haplotype analysis based on the cytb sequences (1134 bp) for 137 individuals of C. harcourtbutleri and 98 of C. limbata detected a total of 27 and 84 haplotypes respectively. Haplotypes of C. harcourtbutleri were included in three clades (Clades 1, 2 and 3); 21 haplotypes in Clade 1, 3 in Clade 2 and 3 in Clade 3 (Figure S2b,c). All haplotypes in Clade 2 were found in Hopong specimens. Haplotypes in Clades 1 and 3 were found at several sites, the former mainly from Inle Lake-to-Loikaw and the latter mainly from the surrounding rivers, such as Heho and Hopong. In Clade 3, the most major haplotype ch3-1 was identical to the haplotype widely found in C. limbata in the upper Salween, upper Mekong and upper Red rivers (cl16; Hap 16 in Wang et al., 2021). The haplotype ch3-2 had one substitution with the haplotype of C. limbata from the upper Irrawaddy River (cl70). The uncorrected p-distances between Clade 1 and 2 haplotypes were 1.9%–2.5%, and those between Clades 1 + 2 and 3 were 3.1%–3.6%.
The ML tree based on the 27 mtDNA cytb haplotypes of C. harcourtbutleri and 84 haplotypes of C. limbata revealed polyphyly of mtDNA in C. harcourtbutleri. The tree supported that Clades 1 and 2 were monophyletic, and Clade 3 was involved in a clade of C. limbata from the upper regions of the Salween, Irrawaddy, Mekong and Red river systems. This mtDNA polyphyly of C. harcourtbutleri contrasted with the results of the SNP-based analysis, which showed a clear segregation of C. harcourtbutleri and C. limbata. Specimens having Clades 1 or 2 haplotypes were generally included in SNP Cluster 1 (62/68, 3/3, respectively, calculated excluding putative hybrids), and those having Clade 3 mtDNA were included in SNP Cluster 2 (30/34). The correspondence between mtDNA clades and SNP clusters was unclear in the North and Hopong populations (Figure 2b).
Channa limbata included several deeply divergent clades that showed geographical cohesion. The basal clades were found from the middle Salween River and the western coast of Myanmar (i.e. cl59, 77, 78; Figure S2a). There were several haplotypes (e.g. cl14, 16 and 17) found from different water systems; they were all distributed in the upper reaches of these rivers (Figure 3; Figure S2a).
3.3 Divergence time estimation
Using all 27 haplotypes of C. harcourtbutleri and 84 haplotypes of C. limbata, the divergence time was estimated based on the partitioned cytb region (Figure 3). The sister species to C. limbata group that included C. harcourtbutleri was estimated as C. barca; their divergence time was estimated at 9.69 Ma [95% highest posterior density (HPD), 5.87–12.87]. The most recent common ancestor (tMRCA) of the C. limbata group was estimated at 4.47 Ma (95% HPD, 2.84–6.31). The divergence time of Clade 1 and Clade 2 of C. harcourtbutleri was estimated at 1.11 Ma (95% HPD, 0.58–1.77), and Clade 1 + 2 and C. limbata including Clade 3 was estimated at 2.01 Ma (95% HPD, 1.22–2.88). The tMRCA of Clade 3 was estimated at 0.13 Ma (95% HPD, 0.03–0.18).
3.4 Morphological analyses
The ranges of standard length (SL) of each genetically classified group (C. harcourtbutleri SNP Clusters 1 and 2, putative hybrid and C. limbata) largely overlapped (Figure S4; Table S3) and showed no significant differences except for one group pair (Steel–Dwass test, p = .007 for SNP Cluster 2 vs. hybrid, p > .07 for other pairs). Among the six counting and 11 metric traits, two counting traits (dorsal fin rays and LLS) were more abundant in the endemic species than in C. limbata (Figure 4c; Figure S5; Table S3). SNP Cluster 2 showed intermediate values between SNP Cluster 1 and C. limbata in three traits (anal fin rays, LDFB/SL and MW/SL), and more similar values to C. limbata than to SNP Cluster 1 in five traits (LAFB/SL, CFL/SL, IOW, HW/SL and BW/SL). Ventral fin rays showed differences between SNP Cluster 2 and C. limbata (Steel–Dwass test, p = .004); however, the ranges were consistent among all groups (5–6). PFL/SL showed differences among the groups (Kruskal–Wallis rank sum test, p = .006) but did not reach significance levels (1%) in any multiple comparison (Steel–Dwass test, the smallest p = .02 between SNP Clusters 1 and 2). No significant difference among the groups was found in the other five traits (Kruskal–Wallis rank sum test, p > .013).
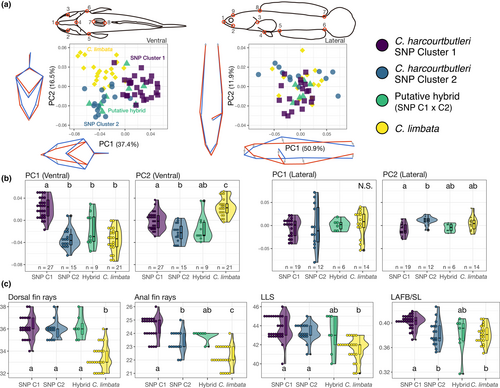
Morphometric analysis based on the eight landmarks on the ventral side of Channa species showed significant differences among the genetically classified groups in PC1 (36.0% contribution) and PC2 (16.2%) (Kruskal–Wallis rank sum test, p < .001). Negative correlations were found between body size and PC1 in SNP Cluster 1 and C. limbata (Spearman's rank correlation test, ρ = −.58, −.67, p = .002, .001 respectively); however, no correlations were detected in other groups. In PC1, SNP Cluster 1 had a longer and narrower head, while SNP Cluster 2, hybrid and C. limbata had a shorter and wider head (Steel–Dwass test, p < .001; Figure 4). In PC2, SNP Cluster 2 had a narrower mouth width and more anterior isthmus position, whereas C. limbata had a wider mouth width and more posterior isthmus position; the SNP Cluster 1 was found to be intermediate between them (Steel–Dwass test, p < .01)
The analysis for the nine landmarks on the left lateral side of the Channa species showed significant differences among groups in PC2 (11.9% contribution; Kruskal–Wallis rank sum test, p < .001). No significant correlations between body size and PC scores were found in any of the four groups (Spearman's rank correlation test, p > 0.03). The variation in PC1 (50.9% contribution) probably reflected the curvature of the specimens during fixation (Figure 4a). The PC2 results indicated that the distance between the snout and the dorsal fin origin was shorter in SNP Cluster 1 and longer in SNP Cluster 2 (Steel–Dwass test, p = 0.002).
4 DISCUSSION
Species diversity on an ‘island’ is hypothesized to be formed through the dynamics of species colonization through dispersal from ‘continent(s)’ and extinction and speciation within the island (Heaney, 2000; Whittaker et al., 2008). Especially on moderately isolated ‘islands’, where immigration could occur multiple times through multiple routes (Hirao et al., 2015; Spironello & Brooks, 2003), detangling the complex patterns of isolation, colonization and in situ speciation over the islands' history is necessary to understand the mechanism underlying the formation of the regional fauna (Cavender-Bares et al., 2009; Ricklefs, 1987). In this study, to understand the mechanisms underlying the formation of the unique fauna in and around Inle Lake, a moderately isolated biogeographical island, we examined the phylogeography, population structure and morphological divergence of the endemic fish Channa harcourtbutleri. The results provide important insights into the evolutionary history including colonization, divergence and secondary contact events experienced by the species in this region (Figure 5).
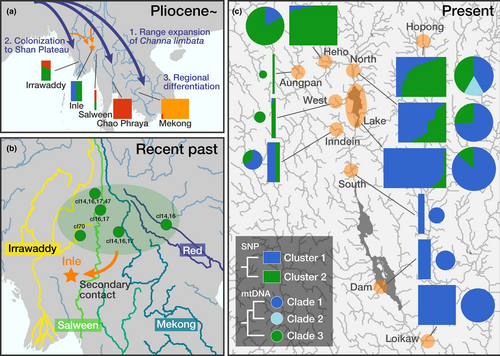
4.1 The origin and secondary contact with closely related species
Genome-wide SNP and mtDNA sequence data successfully reconstructed the complex history of C. harcourtbutleri, which was derived from and experienced secondary contact with the widely distributed species C. limbata in a geological time framework. Channa limbata exhibited clear geographical differentiation, with its population in the Irrawaddy or Salween River system, which were geographically near the Inle region, being most closely related to C. harcourtbutleri (Figures 2 and 5a). SNP-based results suggest that the colonization originated from the Irrawaddy River system, but it is not conclusive due to insufficient samples from the Salween River. The mtDNA monophyletic group (Clade 1 + 2) endemic to the Inle region was estimated to have diverged from their sister lineage at 2.0 Ma (Early Pleistocene), suggesting that the ancestral population of C. harcourtbutleri colonized the Inle region from the Irrawaddy or Salween River system around this time. This timing was slightly older than that estimated by Rüber et al. (2020) (1.4 Ma), which was obtained based on the phylogeny of mtDNA cytb and ribosomal RNA regions' and RAG1 sequences calibrated using the fossil records common to this study. However, both overlapped at credible intervals and supported their differentiation in the Early Pleistocene. The tMRCA for the C. limbata group including C. harcourtbutleri was estimated at ca. 4.5 Ma (Early Pliocene), also similar to (or slightly older than) the estimation (3.7 Ma) by Rüber et al. (2020), even though our samples included the basal populations of C. limbata from the Salween River system not used in the previously analysis. The C. limbata group is inferred to have originated in the Eastern Himalayan region (see Figure 3a) in Rüber et al., 2020. Following the range expansion of C. limbata towards Sundaland and East Asia probably from the Early Pliocene (tMRCA of the species group), the ancestor of C. harcourtbutleri could have been isolated on the Shan Plateau since at 2.0 Ma (Pleistocene). After the Indo–Eurasian collision in ca. 35 Ma (Aitchison et al., 2007), the Shan Plateau continued regional uplifting during the Oligocene and the Pliocene (Morley, 2009). Such geological formation processes of the plateau might have impacted the isolation of the ancestral population of C. harcourtbutleri.
In addition to Clades 1 and 2 haplotypes, a part of C. harcourtbutleri specimens had highly differentiated haplotypes that composed the third clade (Clade 3) together with several haplotypes of C. limbata (Figure 5b). The occurrence of mtDNA Clade 3 in C. harcourtbutleri suggests that C. limbata re-immigrated to the Inle region after speciation of the former (i.e. secondary contact). This event would have occurred in the recent past, probably in the late Pleistocene, because the Clade 3 haplotypes from C. harcourtbutleri were identical or closely related (1–2 nucleotide differences, <0.2% in uncorrected p-distance) to those of C. limbata from the upper Irrawaddy, Salween, Mekong and Red rivers (Figure 3). This similarity is in contrast with their greater differentiation from the haplotypes of Clades 1 and 2 (3.1%–3.6% in uncorrected p-distance, roughly corresponding to 2.0 Ma). Two clusters revealed by genome-wide SNP data would reflect these multiple invasions of the C. limbata group; SNP Cluster 2, which mainly occurs outside Inle Lake and the outflow river, is inferred to have originated from the second colonizers. The morphological similarity between the riverine populations and C. limbata supported this scenario. However, the genetic cohesion in the nuclear DNA of C. harcourtbutleri (Figure 2) suggested the partial genetic admixture in the Inle region after the second colonization. In this scenario, mtDNA Clade 2, the rare group that is a sister group of Clade 1, occurring only in Hopong together with Clades 1 and 3 (Figure 5), could be a trace of past geographical isolation.
The gene flow observed in the C. limbata group across river systems in the Indo-Burma region could be explained by the historical water system connections via river captures. The common haplotype found in the Inle region, upper Salween, upper Mekong and upper Red rivers (ch3-1 = cl16; this study; Wang et al., 2021) provides evidence of river captures in the upper regions of these three adjacent rivers (Clark et al., 2004) and dispersal of C. limbata among the water systems. There is no direct geological evidence of a river capture between the Irrawaddy and Salween rivers (Clark et al., 2004; Robinson et al., 2014); however, the shared haplotype in C. limbata in those areas (cl15; Wang et al., 2021) suggests that these water systems were connected in the past. The complex history of connections between the major rivers surrounding the Shan Plateau could have contributed to the exchange and differentiation of species in the Indo-Burma region (Glaubrecht & Köhler, 2004; He & Chen, 2006; Ratmuangkhwang et al., 2014; Rüber et al., 2004).
4.2 Genetic and morphological differentiation within the Inle region
This study revealed the intraspecific differentiation of C. harcourtbutleri, that is, the genetic and morphological differences between the lake–outflow populations and the surrounding river populations. The two groups were inferred to have been formed via secondary invasion of the closely related species rather than through in situ differentiation. However, it should be noted that the second colonizers have rarely intruded into Inle Lake and its outflowing rivers. The association among genetic cohesion, morphological similarity and habitats in the two groups suggests that the intraspecific differentiation has been maintained by genetic isolation under divergent selection for adaptation to different environments (Heaney, 2000; Nosil, 2012; Schluter, 2000).
The specimens composing SNP Cluster 1, mainly distributed in the lake–outflow, have more elongated head and anal fin base and larger number of anal fin rays than those in specimens composing SNP Cluster 2, mainly occurring in the surrounding rivers, as well as C. limbata. The latter two resembled each other in multiple morphological traits representing body and head shape; the results will be helpful in future taxonomic studies. The differences in these traits could reflect in some of the adaptations to flow velocity, space for swimming and food resources in different habitats (Friedman et al., 2020; Langerhans, 2008; Walker, 1997). Adaptation to contrasting environments often leads to reproductive isolation via restriction of dispersal and maladaptation in another environment (Nosil et al., 2005; Waters et al., 2020). Indeed, genetic admixture between the two SNP clusters was only partially observed at the sites where individuals of both clusters co-occurred (e.g. Hopong, North); overall, these SNP clusters were distributed exclusively to each other in the Inle region. This occurrence pattern suggests the presence of some degree of reproductive isolation between them. The pre-establishment of partial reproductive isolation between C. harcourtbutleri and C. limbata before secondary contact cannot be ruled out; however, the contrasting habitats of the two groups (lake vs. stream) associated with morphological differences imply that local adaptation has contributed to their isolation. On the other hand, SNP Clusters 1 and 2 showed similarities in some traits (dorsal fin rays and LLS), or the latter showed intermediate values between the former and C. limbata (anal fin rays and MW/SL). These could reflect local adaptation to unique environments common to the lake and rivers in this region and/or partial gene flow; however, the relationships between their morphology and adaptation could not be determined in this study.
The presence of mtDNA Clade 2 in Hopong provided insight into the formation of regional faunas in the Inle region and the diversity of distribution history among members of the faunas. Clade 2 showed differentiation corresponding to 1.11 (0.58–1.77) Ma from Clade 1, widely distributed in the Inle region; this range includes the formation of Inle Lake (1.5 Ma; Hampton et al., 2018). Similar to that in C. harcourtbutleri, regional genetic differentiation was reported in several endemic fish species in the Inle region (Fuke et al., 2022; Kano et al., 2022). Three of the five species examined showed genetic differentiation between Inle Lake and Hopong populations (Kano et al., 2022). Among these, the red dwarf rasbora, Microrasbora rubescens (Cyprinidae), endemic to the Inle region, showed a contrasting pattern to C. harcourtbutleri. In the latter, two genetic groups co-occur with morphological divergence, whereas the rasbora exhibited a large genetic differentiation (roughly corresponding to 2.7 Ma) among regions, without any morphological differentiation (Fuke et al., 2022). This morphological invariability could be explained through niche conservatism. The rasbora was adapted to a stagnant water environment; therefore, dispersal through rivers has been strongly limited. On the other hand, snakeheads, including C. harcourtbutleri, have airbreathing organs and most of them have some degree of terrestrial migration ability (Musikasinthorn, 2003). These examples suggest the presence of several different mechanisms that could have generated the genetic, morphological and species diversity in the Inle region. Further studies on more species are necessary to better understand the biodiversity formation in this region. In addition, the adaptive significance of morphological and ecological characteristics and their contribution to reproductive isolation should be studied to determine the maintenance mechanisms of differentiated populations.
5 CONCLUSION
The phylogeographical study of Channa harcourtbutleri, endemic to the Inle region, with closely related species specifically characterized Annandale's (1918) insights that the ichthyofauna of this region was formed via colonization and isolation from adjacent areas. The colonization events could have occurred at least twice; first as an immigration of the ancestral population of C. harcourtbutleri from the Irrawaddy or Salween River system to the Inle region (ca. 2 Ma), followed by the secondary contact of C. limbata in the relatively recent past. The coexistence of the old and new lineages could have been maintained through divergent adaptation to different environments. These multiple colonization patterns suggest that the Inle region is not strictly isolated from the adjacent large river systems and that historical connectivity of the upper Salween or Irrawaddy River system has influenced the formation of its ichthyofauna. However, the mountainous topography and the presence of several endemic species and genera in the Inle region imply that the degree of isolation was high enough to exclude frequent colonization. The long-lived, possibly stable environment and moderate isolation of the Inle region might have contributed to its unique ichthyofauna, where endemic and widely distributed non-endemic species coexist. Future studies on the origins of other endemic species will allow for an interspecific comparative approach and contribute to further understanding of the community assembly and evolutionary history of fish diversity in this region.
ACKNOWLEDGEMENTS
We deeply appreciate the kind assistance from Nyi Nyi Kyaw (Forest Department, Ministry of Natural Resources and Environmental Conservation, Myanmar [FD-MOECAF]), Win Naing Thaw (Nature and Wildlife Conservation Division, FD-MOECAF), the late Than Htay (Inle Lake Wildlife Sanctuary, FD-MOECAF), the late Ohn and Than Nwai (Forest Resource Environment Development and Conservation Association, Myanmar [FREDA]), Nyi Nyi Lwin (Green Leaf, Myanmar) and Akihisa Iwata (Kyoto University) in the fieldwork in and around Inle Lake. We are grateful to members of the Inland Fisheries Research and Development Institute (IFReDI, Cambodia), Yoshinori Kumazawa (Nagoya City University, Japan) and members of Research Laboratory of Ichthyology, Faculty of Fisheries, Kasetsart University (RLIKU, Thailand) for supporting the fieldwork in Southeast Asia. We also thank members of the Animal Ecology Laboratory, Kyoto University for supporting the molecular work and data analyses. This study was supported by the Research Foundation of Kurita Water and Environment Foundation (17B028, 18 K013, 19 K008), JSPS KAKENHI Grant (26304007, 19 J23130) and the Sumitomo Foundation (the Grant for Environmental Research Project, 193271).




