Characterization of porcine endogenous retrovirus particles released by the CRISPR/Cas9 inactivated cell line PK15 clone 15
Funding Information
This study was supported by grant SFB TRR CRC 127: “Biology of xenogeneic cell and organ transplantation-from bench to bedside” from the Deutsche Forschungsgemeinschaft, DFG, Bonn, Germany.
Abstract
The infection of human transplant recipients by porcine endogenous retrovirus (PERV) is a safety issue for xenotransplantation (XTx). CRISPR/Cas9 technology has enabled the generation of pigs free of functional PERVs, and the susceptibility of these animals to reinfection by PERVs remains unclear. To assess virological safety, we characterized a cell line in which PERVs have been inactivated by CRISPR/Cas9 (PK15 clone 15) for its susceptibility to infectious PERV. First, basal expression of PERV pol, the porcine PERV-A receptor (POPAR), and reverse transcriptase (RT) activity of PERV were determined. PK15 clone 15 cells were inoculated with PERV and monitored post infection for virus expression and RT activity. Particles were visualized by electron microscopy. Our data show that PK15 clone 15 cells still produce viral proteins that assemble to produce impaired viral particles. These virions have an irregular morphology that diverges from that of mature wild type. The particles are no longer infectious when tested in a downstream infection assay using supernatants of PK15 clone 15 cells to infect susceptible swine testis-IOWA (ST-IOWA) cells. The expression of POPAR was quantified to exclude the possibility that lack of susceptibility to reinfection, for PERV-A, is caused by absence of viral host receptor(s). PK15 and PK15 clone 15 cells do, in fact, express POPAR equally. PERV RT inactivation mediated by CRISPR/Cas9 does not compromise virus assembly but affects virion structure and proviral integration. The constitutive virion production seems to maintain cellular resistance to superinfection and possibly indicates a protective side effect of this specific CRISPR/Cas9 mediated RT inactivation.
1 INTRODUCTION
Genome-wide inactivation of PERV using CRISPR/Cas9 is potentially a great breakthrough in XTx.1, 2 All 62 copies of the PERV pol gene, targeting the highly conserved catalytic center of the RT coding region, were disrupted in a porcine kidney epithelial cell line (PK15) demonstrating a >1000-fold reduction in PERV transmission to human cells from the engineered cells.1 Subsequently, PERV-inactivated pigs were generated by somatic cell nuclear transfer (SCNT)2 using primary porcine cells where 25 copies of functional PERVs were inactivated by targeting the PERV pol gene using CRISPR/Cas9.
This strategy is an important step in overcoming existing organ shortage for allotransplantation, provided that PERV inactivation and gene editing on donor animals have no detrimental, systemic off-target effects.2-5 To generate these animals, two issues must be considered: (a) the likelihood of recombination events that could theoretically restore the mutated PERV pol sites resulting in functional retroviral reverse transcriptase (RT) activity; and (b) the prevention of new and potentially unapparent reinfection (superinfection, SI), with exogenous PERV.6, 7
Host range interference studies show polytropism for PERV-A and PERV-B and their capacities to infect human cells in vitro.8, 9 PERV-C is an ecotropic virus that is restricted to pig cells. The recombination of ecotropic PERV-C with human-tropic PERV-A results in PERV-A/C that replicates to higher levels and can infect selected human cell lines.10, 11 Although in vivo infection of human xenotransplant recipients with PERV has not yet been demonstrated, it cannot be excluded since PERV is expressed in pig organs.12-14 Given the potential consequences, the transplantation of xenogeneic tissue containing functional PERV into an immunosuppressed patient should be avoided.
It is not fully understood why the PERV mechanism of target cell entry varies in efficacy. Both binding efficacy and host range are mediated by the viral surface envelope protein (Env) and its receptor binding domain (RBD), which has been well characterized for PERV-A and PERV-C Env.11, 15-18
In humans, two receptors (HuPAR1 and HuPAR2) have been described for PERV-A. The porcine homologue is the Sus scrofa solute carrier family 52 member 2 (SLC52A2 or POPAR).19 Other receptors or putative cofactors for PERV-B, PERV-C, and PERV-A/C have not been described yet.10, 19
To address the safety aspects of PERV inactivation for XTx, we tested the susceptibility of a PERV RT-inactivated porcine kidney cell line, PK15 clone 15, to different classes of exogenous PERV and investigated the influence of CRISPR/Cas9-mediated indels on virus particle formation and residual infectivity. As such the basal expression of PERV pol and POPAR as well as viral RT activity was determined. Subsequently, PK15 clone 15 cells have been inoculated with different classes of PERV to check re-infectivity. Parameters such as RT activity, virus expression, and virus release were monitored weekly for 55 days comparing the mutant cell line, PK15 clone 15, with PK15 wild-type (wt). In addition, cells and supernatants were analyzed by Western blot (WB) for presence of viral capsid protein p30. Furthermore, the supernatant of PK15 clone 15 cells was tested in a downstream infection assay using ST-IOWA cells as target for RT-inactivated PERV.
2 MATERIALS AND METHODS
2.1 Cell lines and viruses
A mixture of PERV-A and PERV-B was harvested from porcine kidney epithelial cell line PK15, purchased by American Type Culture Collection (ATCC, CCL-33™). PERV-B was harvested from a PERV-B transfected human embryonal kidney cell line (HEK 293FT/B(33)/ATG).21 HEK 293/5°, provided by Robert Koch-Institut (RKI), Berlin, Germany,11, 22 was used as producer cell line for recombinant PERV-A/C. Cells were routinely sequenced for presence of PERV-A/C. PERV-C was harvested from the producer cell lines ST-IOWA carrying either molecular clone PERV-C(1312), (accession number AM229312.220 or PERV-C(5683), (accession number KY352351.1; Paul-Ehrlich-Institut, Germany). PERV-inactivated PK15 clone 15 was provided by Harvard Medical School, Boston, MA, USA.1 PERV-susceptible, RT-negative ST-IOWA cells,23 and PERV-negative HEK 293T (ATCC, CRL-3216™) were used as controls. Cells were cultured as described previously,1, 24 or as recommended by ATCC. Cells and RT activity of virus-containing supernatants used for infection experiments are listed in Table 1.
2.2 In vitro infection assay
For in vitro infection, 2 × 105, cells/well were seeded in triplicate in 6-well cell culture plates (Sarstedt, Germany). 24 hours later the cells were incubated with 1 mL cell-free supernatant per well containing replication-competent PERV in different amounts according to RT activity values (Tables 1 and 2). The supernatant was harvested from producer cell lines, stored at −80°C, and tested for RT activity before use. To maintain virus integrity and infectivity, the inoculum was not further processed. Incubation with inoculum was performed for 24 hours. Cells were then washed three times with phosphate-buffered saline (PBS) to remove input virus and were cultivated for a further 8 weeks and were passaged every 3 to 4 days in equal split ratios. The expression of PERV was measured weekly by monitoring the cell-free supernatant for PERV RT activity and viral RNA (vRNA). The infectivity of the virus inoculum was tested on susceptible ST-IOWA and HEK 293T cells, respectively. All samples tested were pooled from triplicates. Cell-free supernatant aliquots were stored at −80°C until testing.
| Cell line | Reference | Inoculation with |
|---|---|---|
| PK15 clone 15 | Provided by G. Church, Harvard Medical School, Boston, MA | PERV-B(33), PERV-A/C, PERV-A and PERV-B, PERV-C(1312), PERV-C(5683) |
| ST-IOWA | 23 | PERV-C(1312), PERV-C(5683) |
| HEK 293T | ATCC, CRL-3216™ | PERV-B(33), PERV-A/C, PERV-A and PERV-B |
Note
- Cell lines are tested routinely for absence of RT activity.
2.3 Expression analysis and quantification of PERV
For expression analysis, total cellular RNA was purified by RNeasy Plus Mini Kit (Qiagen) from 106 cells according to the manufacturer's instructions. Isolation of vRNA from cell-free supernatant was performed using the QIAamp Viral RNA Mini Kit (Qiagen), according to manufacturer's instructions. For detection and quantification of vRNA, a one-step qRT-PCR was performed on PERV pol, env, and POPAR in a LightCycler® 480 (Roche Life Science) using the QuantiTect SYBR Green RT-PCR Kit (Qiagen) as recommended by the manufacturer (Table 3). The cycling conditions were as follows: RT-reaction (50°C, 30 minutes); denaturation (95°C, 15 minutes); 40 cycles of amplification (94°C, 15 seconds, denaturation; °C primer specific, 30 seconds, annealing; and 72°C, 30 seconds, elongation); melting curve analysis (95°C, 10 seconds, 65°C, 10 seconds, slowly heating up to 97°C with continuous measuring of the fluorescence); and cooling (40°C, 30 seconds). Annealing temperatures were chosen as follows: pol 58°C; envA and envB 56°C; envC 62°C; and POPAR 61°C. RNA copy number was determined using in vitro transcribed external homologue standards with pGEM®-T Easy (Promega) as backbone. Samples were measured in triplicate, and the mean value and standard deviation was determined. Homologous in vitro transcribed RNA standards were generated according to manufacturer's instructions utilizing dilutions from 109 to 103 copies/µL.
| Primer | Target Gene | Sequence 5′-3′ | Accession No. | Nucleotide pos. | Amplicon |
|---|---|---|---|---|---|
| F | PERV pol | TCTCCCCAAGTAAAGCCTGAT | AJ133816.1 | 3119-3139 | |
| R | PERV pol | ACTAGGATGCCCTGTTGGATTA | AJ133816.1 | c3345-3324 | 227 nt |
| PERV-A_for | PERV envA | GTCTCAATGACCAGGCCACACC | AJ133817.1 | 6439-6460 | |
| PERV-A2_rev | PERV envA | GCTCCATTGCTTGCAAAAGA | AJ133817.1 | c6563-6544 | 125 nt |
| EnvB-F_AWG | PERV envB | AAACTCCCCAGGTGTTCCTGTTAA | AJ133816.1 | 7052-7075 | |
| EnvB-R_AWG | PERV envB | GCTCTTTGGTCACATTGAATTTTCC | AJ133816.1 | c7233-7209 | 182 nt |
| EnvC for2 | PERV envC | GTGCTCTCCTTCAGACCTAGATTAC | AM229312.2 | 9383-9407 | |
| EnvC-R3_AWG | PERV envC | AGCCATTGGAGGCTCCAGCTGG | AM229312.2 | c9566-9545 | 184 nt |
| poPAR_Intron_F | POPAR | AGTGCTCTGACCAACGGCGT | NM_001004033.1 | 862-881 | |
| poPAR_Intron_R | POPAR | CCAGACACAGAGCCCACAA | NM_001004033.1 | c1153-1135 | 292 nt |
2.4 RT activity assay
RT activity in cell-free supernatants of infected cells and producer cells was detected using the C-Type RT activity Kit (Cavidi) according to the manufacturer's instructions, protocol B. Samples were measured in triplicate (10 µL each), and the mean and standard deviation was determined. The standard dilution series covered RT activity values from 407.1 mU/mL to 1.39 mU/mL. RT activity values below this were considered to be RT negative and defined the limit of detection.
2.5 Immunoblot
PERV isolated from supernatants of positive cells and cell lysates were analyzed for the presence of structural proteins by WB as described.25 Briefly, samples were subjected to SDS-PAGE and transferred onto Immobilon-P PVDF membranes (Merck Millipore). Membranes were blocked with 0.1% TBS-T containing 5% (w/v) skimmed milk (AppliChem) for 1 hour at room temperature. Proteins were detected with polyclonal rabbit anti-PERV capsid directed antibody p3025, 26 diluted 1:2,000 in TBS-T and goat anti-rabbit peroxidase-conjugated secondary antibody (Dianova; 1:10,000). Rabbit pre-immune serum (1:2,000) and anti-beta-actin (Abcam; 1:10,000) were used as controls. For detection, an Amersham Hyperfilm™ and ECL Western Blotting Detection Reagents (GE Healthcare) were used.
2.6 Indirect immunofluorescence microscopy
Immunofluorescent detection of PERV particles in PERV producer cells was performed with polyclonal rabbit anti-PERV nucleocapsid antibody (p10, 1:200 in PBS) as described previously.25, 26 Brief semi-confluent to confluent cells, plated on glass coverslips, were washed with PBS without Ca and Mg, fixed with freshly prepared 2% paraformaldehyde (Sigma-Aldrich) in PBS for 45 minutes, washed again, and stored at 4-8°C. For indirect immunofluorescence, cells were first permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 10 minutes, washed three times with PBS, blocked with 1% BSA in PBS for 10 minutes, and incubated either with p10 pre-antiserum or with p10 antiserum for 30 minutes. Cy2-conjugated polyclonal anti-rabbit IgG (Dianova) was used for detection according to the manufacturer's instructions. All samples were embedded in Fluoroshild™ mounting medium (Sigma-Aldrich), and images were acquired using the AxioVision imaging system (Zeiss).
2.7 Electron microscopy
Electron microscopy of PK15 clone 15 and PK15 cells was performed according to standard procedures as described previously.20 Briefly, for electron microscopy cells were fixed with 2.5% glutaraldehyde (Sigma-Aldrich) in culture medium for 45 minutes at room temperature. After washing with PBS, cells were scraped off the culture flask and gently mixed with 2% warm liquid agarose. After cooling and gelling, small agarose blocks containing the cells were cut. These blocks were post-fixed with 2% osmium tetroxide (Sigma-Aldrich) in PBS and treated with 1% tannic acid (Carl Roth).27 Cells were then gradually dehydrated in a series of ethanol baths and finally embedded in Epoxy resin (Sigma-Aldrich). After polymerization for 48 hours at 60°C, ultrathin sections were cut and stained with 2% uranyl acetate (Merck) for 15 minutes followed by 2% lead citrate (Serva) for 5 minutes. Sections were examined in a Zeiss CEM902 electron microscope using electron filtering (ESI) mode.
3 RESULTS
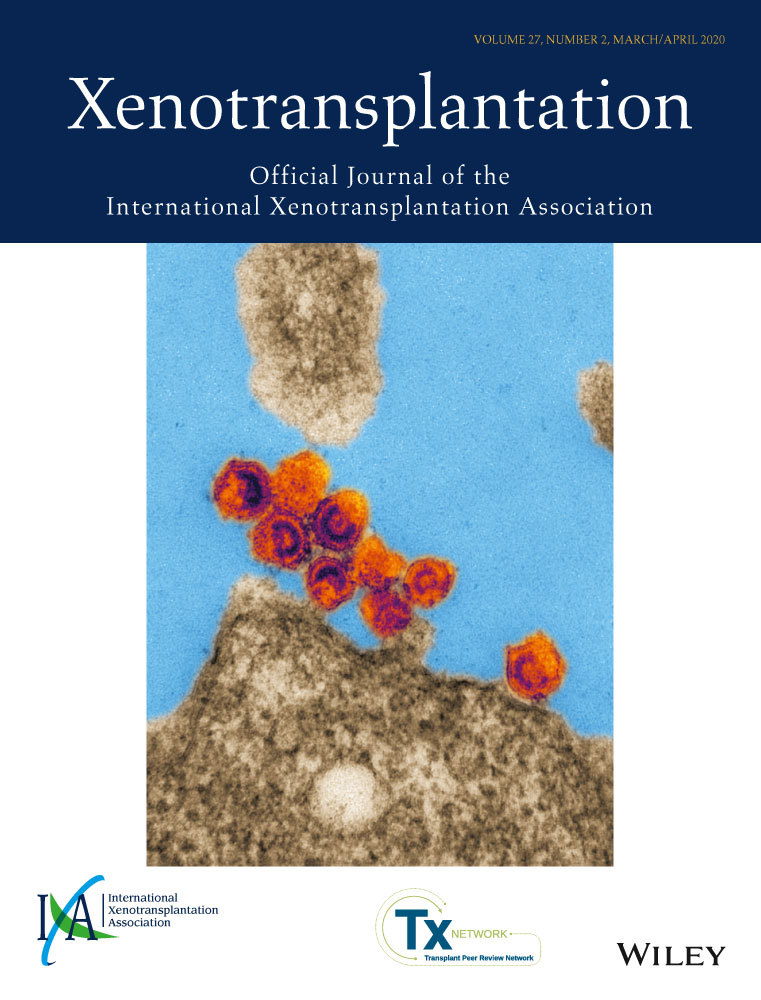
3.1 Quantification of PERV expression and RT activity
The efficacy of CRISPR/Cas9 gene editing on PERV was initially tested by quantification of pol comparing vRNA from cell supernatant from the positive control PK15 with PK15 clone 15 cells. Porcine ST-IOWA and human 293T cells were used as negative controls. In addition, the RT activity and cellular expression of POPAR were determined in these cell lines (Figure 1). Despite indel mutagenesis, both the PK15 and the PK15 clone 15 cell lines release comparable amounts of PERV pol RNA, approximately 106 to 107 copies/µL. Porcine ST-IOWA shows a basal expression of less than 104 copies/µL, which is consistent with what has been previously reported for this cell line.28 As expected, human 293T cells show no PERV expression. All porcine cell lines tested express comparable amounts of the PERV-A receptor POPAR, approximately 104 copies/100 ng total RNA. As expected, human 293T cells do not express POPAR (Figure 1B). Despite PERV pol expression, no RT activity was found in PK15 clone 15. Only PK15 which is a known producer cell line for PERV-A and PERV-B showed RT activity of approximately 150 mU/mL (Figure 1C), while neither ST-IOWA nor 293T showed any RT activity.
3.2 Infection of PK15 clone 15 with PERV
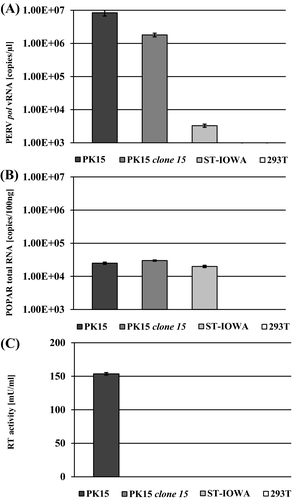
PK15 clone 15 cells were infected with PERV of all three classes (PERV-A and PERV-B in combination, PERV-B(33), PERV-A/C, PERV-C(1312), or PERV-C(5683); Tables 1 and 2). PERV-susceptible ST-IOWA cells were infected with either PERV-C(1312) or PERV-C(5683), and PERV-A- and PERV-B-susceptible human 293T cells were infected with either PERV-B(33), PERV-A/C, or PERV-A, PERV-B to confirm infectivity of the virus inoculum used. As indicators for positive infection the RT activity (Figure 2A-C) and viral pol RNA isolated from culture supernatant were quantified (Figure 3A-C). At day 55 post infection, the number of transcripts of envA, envB, and envC RNA from supernatant of infected cells was determined (Figure 4A-C).
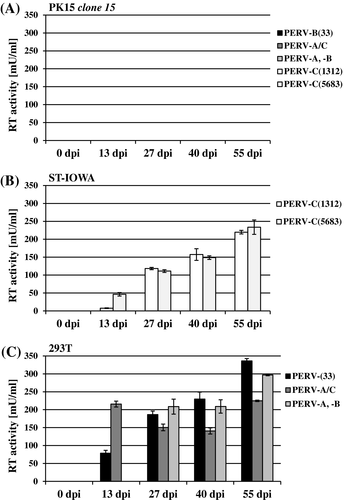
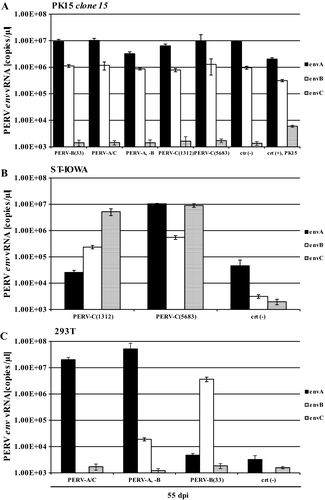
PK15 clone 15 cells remained RT-negative compared with the controls (Figure 2A), independent of the virus inoculum used for infection. ST-IOWA (Figure 2B) and 293T cells (Figure 2C) became productively infected within 13 dpi, reaching RT activities greater than 200 mU/mL at the end of the experiment, which represents approximately 5 × 106 vRNA copies/µL. The controls demonstrate that the virus inoculum that was used did in fact efficiently infect the cells. Similar results were obtained for expression of PERV pol (Figure 3A-C). PK clone 15 cells expressed similar levels of pol expression as the non-infected control (crt (-); Figure 3A). The positive controls ST-IOWA and 293T cells infected with PERV showed increasing pol copy numbers within 13 dpi, which corresponds to the increase in RT activity seen in Figure 2, and reached a plateau of 106 vRNA copies/µL by 27 dpi (Figure 3B,C). The specific quantification of env at 55 dpi confirmed that both PK15 clone 15 and PK15 express comparable amounts of envA and envB RNA (Figure 4A). PERV infection does not appear to increase the basal expression of env. PK15 clone 15 cells express envA and envB at levels of 107 copies/µL envA and 106 copies/µL envB, respectively, independent of the viral inoculum used (Figure 4A). Expression of envC was generally very low in these cells. ST-IOWA cells that were infected with PERV-C(1312) or with PERV-C(5683) showed elevated envC expression up to 107 copies/µL, as well as increases of envA and envB expression compared with the uninfected control (crt (-); Figure 4B). This was expected since ST-IOWA are known to harbor non-functional proviral PERV-A and PERV-B sequences that are affected upon PERV-C infection29 (N. Fischer, unpublished data). Infection of 293T cells resulted in elevated envA and envB levels (Figure 4C). As expected, the 293T cells remained negative for expression of envC.
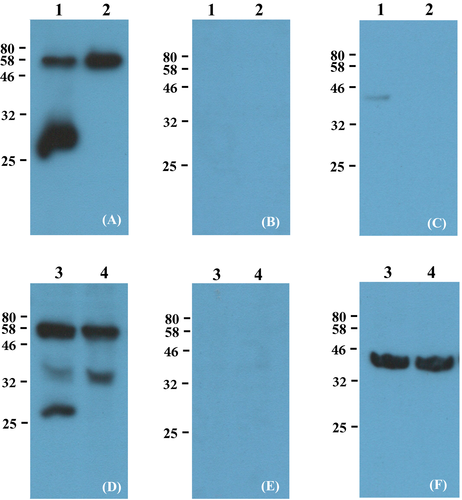
3.3 Western blot analysis of viral capsid protein
Supernatants and cell lysates of PK15 and PK15 clone 15 were analyzed for the presence of PERV viral capsid (CA) protein (PERV p30) using a PERV CA-directed antibody p30 (Eurogentec)25, 26 (Figure 5A,D). The Gag precursor polyprotein was expected at 60 kDa, an intermediate consisting of CA (p30) and NC (p10) at 40 kDa and the mature CA at 30 kDa. Results show bands at 80 kDa representing the unprocessed Gag precursor and a second at 30 kDa representing the mature CA protein (Figure 5A, lane 1 and Figure 5D, lane 3). Virus isolates from PK15 clone 15 revealed only one signal at 80 kDa, and mature processed p30 was not detected (Figure 5A, lane 2). Similar results were observed in cellular lysates (Figure 5D, lane 4). PK15 clone 15 does not express mature p30 but instead had a signal at 40 kDa that was stronger than for the PK15 cell lysate, which corresponds to the intermediate, and showed that the mature CA protein was not produced. The pre-immune serum controls performed for all samples tested revealed no non-specific binding (Figure 5B,E). Comparability of protein concentrations for cell lysates was shown using anti-beta-actin directed antibodies as a control (Figure 5F). Although the loading control for the virus isolates is very weak depending on divergent co-packaging (Figure 5C), we assume that even a weak signal for p30 of PK15 clone 15 (Figure 5A) would have been detected since the Gag precursor protein (Figure 5A, 80 kDa band) was even stronger than for PK15. The loading control of the cell lysates shows equal amounts of total protein (Figure 5F).
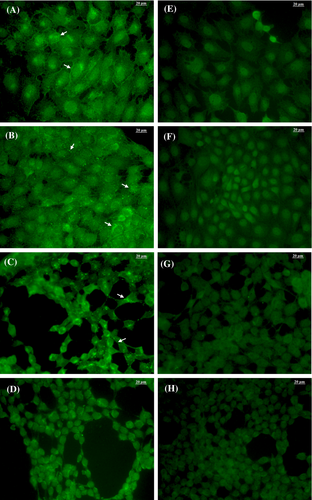
3.4 Detection of PERV expression in different cell lines by immunofluorescence
PK15 and the mutant PK15 clone 15 cells produce and release PERV particles at comparable amounts (Figure 6A,B) as revealed by immunofluorescence staining using a polyclonal p10-directed antibody (Eurogentec)26 and p10 pre-immune serum as a control (Figure 6E-H). The comparison with PERV-B positive 293T/B(33)ATG cells (Figure 6C) and PERV-negative 293T cells (Figure 6D) clearly shows that PK15 and PK15 clone 15 cells produce viral protein. Both cell lines release PERV particles that are visible at the cell membranes as green shading and dots.
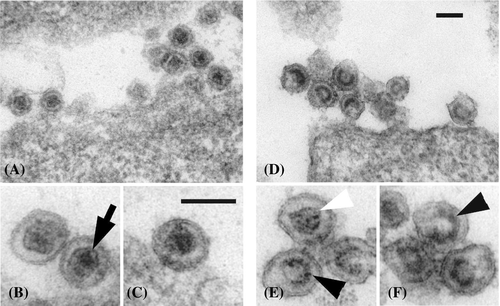
3.5 Detection of impaired PERV virions in PK15 clone 15 by electron microscopy
Electron microscopy showed that both PK15 and PK15 clone 15 cells produce and release PERV viruses at comparable levels (Figure 7). Visualizing particle structure at higher magnification revealed a possible side effect of CRISPR/Cas9 mediated indel mutagenesis that results in insufficient protein maturation. PERV particles carrying variant mutations in the catalytic center of the RT coding region have less dense structures and have a particular horseshoe-like capsid conformation. The structure differs from the common PERV morphology where the capsid is collapsed, dense, and round (Figure 7A-C). With the exception of a few particles, most of the envelopes of viruses from PK15 clone 15 cells were irregularly shaped and the viruses appeared immature (Figure 7D-F).
3.6 Infection assay with impaired PERV released by PK15 clone 15
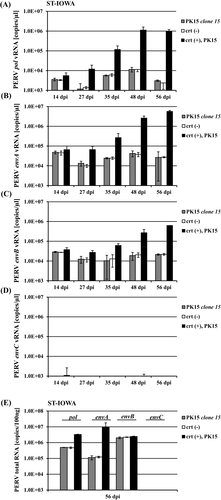
The residual infectivity of virus particles released from PK15 clone 15 cells was tested in a downstream infection assay using PERV-susceptible ST-IOWA cells as recipients (Figure 8. Virus isolated from PK15 (wt) was used as positive control. Monitoring the changes in the PERV pol and PERV-A, PERV-B, and PERV-C env expression over 8 weeks revealed that infection PERV wt particles resulted in increased envA and envB expression. However, when virus particles released by PK15 clone 15 were used, envA and envB expression remained at baseline levels, comparable to the negative control (Figure 8A-D). As expected, expression of PERV envC remained negative because PK15 wt does not harbor PERV-C. The final quantification of cellular PERV-A, PERV-B, and PERV-C env at 56 dpi showed productive infection for cells infected with PK15 wt virus (Figure 8E), while ST-IOWA cells inoculated with supernatant from PK15 clone 15 cells only showed basal PERV expression comparable to the uninfected control (crt (-)).
4 DISCUSSION
Like HERV in the human, genome PERV is an integral part of the pig genome that evolved over approximately 7.6 × 106 years, while being vertically transmitted.30 Multiple proviral copies encoding functional and non-functional PERV are spread throughout the pig genome. Previous studies have shown that PERV is expressed in pig tissues14 and is capable of exogenous replication in vitro.31 The safety of pig-derived xenotransplants requires the generation of donor animals and porcine cells that are free of functional PERV to avoid infection of immune-suppressed transplant recipients. Many attempts have been made to minimize the viral burden, including treatment with antiretroviral drugs, vaccination, neutralizing antibodies, RNA interference, and genome editing, first by using zinc finger nucleases, and more recently with CRISPR/Cas9 technology.31, 32 Despite these efforts, PERV still remains a potential safety risk for xenotransplantation that should not be ignored.33-35
Our results show that despite CRISPR/Cas9 mediated indel mutagenesis which inactivated the viral RT, PK15 clone 15 cells do constitutively express and release high levels of vRNA (Figure 1A) but remain RT negative (Figure 1C). These cells are protected from fresh PERV infection.
The porcine PERV-A receptor POPAR was examined to address the question of whether PERV inactivation, and therefore reduction of the viral load, might stimulate or even enhance its expression. Experiments revealed that POPAR RNA levels are not affected and remain constant at 104 RNA copies/100ng total RNA. We did not investigate the surface expression of the SLC transporter in this work since we focused on superinfection. Nonetheless, since virus infection and Env-mediated host receptor regulation are mutually interacting processes36 it would be important to correlate receptor presence and virus expression/release by binding studies using recombinant Env (rEnv) or PERV. After inoculation of the PK15 clone 15 cells with four different PERV classes, no productive infection was observed (Figure 2A and Figure 3A) whereas ST-IOWA and 293T cells could easily be infected using the same virus stocks. The control cells became PERV positive within 8 weeks (Figure 2B,C and Figure 3B,C). Even though ST-IOWA cells contain non-functional PERV-A and PERV-B sequences and show a basal PERV expression without producing infectious particles (N. Fischer,unpublished data), they are not protected against exogenous virus infection. This is an important difference to PK15 clone 15 cells that are descendants of PK15 cells, which carry and release functional viruses.37 This suggests that release of virus particles, or possibly the release of mature Env protein, covers the cellular receptors required for adherence and virus uptake as have been demonstrated for other retroviruses.36 Since Env-mediated superinfection interference has been described for retroviruses like HIV, MuLV, and FeLV, a similar mechanism for PERV may be reasonably assumed here.
Furthermore, the infection of ST-IOWA cells with PERV-C seems to influence the expression of proviral PERV (Figure 4B). PERV-A and PERV-B env were increased in PERV-C-infected cells, a finding that is often observed in ST-IOWA cells (N. Fischer, unpublished data). It is unclear whether the infection process affects the LTR regions of endogenous PERV by using similar integration sites or if this is caused by recombination events, such as those leading to the generation of PERV-A/C, or perhaps both. These questions are addressed separately by sequence analysis of virus released.
Since PK15 clone 15 cells expressing PERV as shown by one-step qRT-PCR for pol and env (Figures 3A and 4A), virus from cell supernatants and cell lysates expressed an unprocessed precursor of the viral capsid protein p30 (Figure 5A). The mature p30 was not detected in culture supernatants nor in cell lysates of PK15 clone 15 cells. The introduction of indels thus seems to adversely affect protein maturation. Although the Gag region that codes for the matrix (MA), capsid (CA), and nucleocapsid (NC) proteins is upstream of the Pol-coding region that harbors the viral enzymes protease (pro), polymerase (pol), integrase (int), and RT, the p30 protein is not efficiently cleaved from its precursor. Further protein analyses of conformational changes in the viral proteins would be required to determine whether the effect described here is mediated by indels.
PERV expression in cells was detected by immunofluorescence using the nucleocapsid-directed antibody p10 (Figure 6). Virus morphology was determined by electron microscopy (Figure 7). The inactivation of the viral RT by mutagenesis does not seem to adversely affect the formation of virus particles. PERV assembly and release still occurs at the cell membrane. However, in contrast to functional PK15 the mutant viral particles are defective, incapable of productive infection. Since CRISPR/Cas9 site-directed mutagenesis of PERV in PK15 clone 15 cells introduced indels that differ in length,1 the observed effect is not homogeneous and viral phenotypes appear different. Env is not affected since it is encoded by one reading frame that is spliced from the viral mRNA (Figure 4A).
Using a second approach, the infective potential of virus particles released from PK15 clone 15 was tested in a downstream infection assay using PERV-susceptible ST-IOWA cells, but these particles did not establish a productive infection (Figure 8). Our data do not exclude that the impaired virions released by PK15 clone 15 may adhere to or even enter cells, since we only tested for productive infection. In any case, the particles were no longer present a few days after inoculation since the vRNA is not reverse transcribed and does not integrate into the host genome due to the defective RT. Were this not the case, an increase in viral expression of PERV pol and env from positive control experiments with viruses from PK15 cells would have been observed, but the expression of PERV pol and env remained at the same levels as the uninfected control.
Based on these results and the assumption that viral Env or virus particles protect against superinfection, it could be tested if (a) stable transduction of viral env into the host genome, which is constitutively expressed and released, would lead to retroviral resistance. Alternatively, (b) the inactivation via CRISPR/Cas9 of functional PERV released by the cells should maintain resistance to superinfection without being infective. Both approaches need to be focused and should confirm the data obtained in this study that PERV producing cells, which are RT-inactivated remain protected against superinfection.
ACKNOWLEDGMENTS
We thank our colleagues from the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich Transregio Collaborative Research Centre 127 (DFG SFB TRR CRC 127) for discussion as well as Regina Eberle (Paul-Ehrlich-Institut) for her technical assistance conducting electron microscopy sample preparation. In addition, we thank Bevan Sawatsky (Paul-Ehrlich-Institut) for his help with editing of the manuscript.
CONFLICT OF INTEREST
The authors declare that they have no competing interests. The activities of George M. Church (GC), including Technology Transfer, Advisory Roles, and Funding Sources, are documented at http://arep.med.harvard.edu/gmc/tech.html. GC is the co-founder and scientific advisor of eGenesis.
AUTHOR CONTRIBUTIONS
All authors contributed substantially to research design, acquisition, analysis, and interpretation of data. All authors also contributed to the drafting of the manuscript as well as critical revision and approval of the final submitted version.