A novel approach for rapid detection of bacterially contaminated platelet concentrates via sensitive measurement of microbial DNA polymerase activity
Abstract
Background
Transfusion of bacterially contaminated platelet concentrates (PCs) can result in serious health consequences for the affected patient. Before being released from blood banking facilities, PCs are routinely screened for bacterial contamination by culture-based tests. However, culture-based PC screening methods require extended holding and incubation periods and are prone to false-negative results due to sampling error. Screening PCs closer to the time of transfusion using rapid point-of-issue tests represents an alternative approach; however, FDA-approved assays generally suffer from a lack of sensitivity.
Study Design and Methods
Presented herein is the feasibility of a novel approach toward rapid, sensitive, and universal detection of bacterially contaminated PCs via selective measurement of microbial DNA polymerase activity. This approach is achieved using a differential cell lysis procedure in combination with enzymatic template generation and amplification (termed ETGA-PC assay).
Results
Serial dilution spiking experiments revealed an approximate sensitivity of 30 to 200 colony-forming units (CFUs)/mL (mean, 85 CFUs/mL) for Staphylococcus epidermidis, Staphylococcus aureus, Escherichia coli, and Klebsiella pneumoniae. An additional 22 clinically relevant strains of bacteria were also detected below 200 CFUs/mL after spiking into PC aliquots. Furthermore, the ETGA-PC assay was able to accurately monitor the presence and growth of seven clinically relevant bacterial species that were spiked into PC units.
Conclusion
Together, the data presented here demonstrate that the ETGA-PC assay is a feasible approach for rapid and sensitive detection of bacterially contaminated PCs. Experiments, aimed at simplification and/or automation of the assay procedure, are under way.
Abbreviations
-
- ALR
-
- apheresis leukoreduced
-
- ANLR
-
- apheresis nonleukoreduced
-
- DPE
-
- DNA polymerase extension
-
- ETGA
-
- enzymatic template generation and amplification
-
- LOD
-
- limit of detection
-
- NLC(s)
-
- nonlysed control(s)
-
- NSC
-
- no spike control
-
- PC(s)
-
- platelet concentrate(s)
-
- POI
-
- point of issue
-
- qPCR
-
- quantitative polymerase chain reaction
-
- WBNLR
-
- whole blood–derived nonleukoreduced
Transfusion of blood components such as red blood cells and platelet concentrates (PCs) is an important part of patient care around the world. Due to numerous factors, blood components are at risk of containing a variety of infectious agents, including viruses and bacteria.1 Despite significant reductions to the risk of transfusion-related viral infections,2, 3 bacterial contamination remains a persistent problem for blood banking facilities and hospitals.4-8 In particular, PCs represent the highest risk among all blood components, with bacterial contamination rates as high as 1 in 1000 to 3000 units.6-10 Since millions of PC units are transfused annually, many patients are therefore at risk of acquiring life-threatening transfusion-related illnesses, such as sepsis.6
In a continued effort to reduce these risks, the AABB adopted a new standard in 2004 recommending the implementation of measures to detect and limit bacterial contamination of PC.11 To meet this mandate, blood banking facilities have implemented procedures such as improved skin disinfection, diversion of the initial blood draw, and bacterial detection tests to screen for contaminated PC.12 Currently, broth-based blood culture is the most commonly used test for screening PC before release from blood banking facilities,13, 14 and its implementation has been associated with a reduced risk of transfusion-related fatalities.15 However, culture-based testing does not always detect PC units containing low levels of bacteria, resulting in release of contaminated units (i.e., false negatives).16 Since PCs are stored at 22 to 24°C before transfusion, bacteria are able to proliferate to dangerous levels within contaminated units missed by culture-based tests. Unfortunately, such scenarios have resulted in transfusion of contaminated PCs and have been associated with fatal septic reactions.15, 17 In addition to these safety risks, prerelease testing by culture-based methods presents logistic challenges to blood banking facilities. For example, the testing procedures associated with these methods can require up to 48 hours of holding and incubation before releasing PCs for transfusion.9, 18 Since PCs have a shelf life of only 5 days from the time of collection, such delays limit their availability for transfusion. Together, the aforementioned limitations associated with prerelease testing have sparked the development of rapid point-of-issue (POI) assays for detection of bacterially contaminated PCs just before transfusion.9, 19
We recently reported the characterization of a novel enzymatic template generation and amplification (ETGA) method that enables rapid, highly sensitive, and universal detection of viable microbes via measurement of endogenous DNA polymerase extension (DPE) activity.20 In a subsequent study, we demonstrated that a differential cell lysis procedure, followed by ETGA analysis, enabled reduced time-to-detection of simulated bloodstream infections when compared to a continuous monitoring blood culture instrument.21 Herein, we set out to determine the feasibility of using ETGA to detect bacterially spiked PCs. Since anucleated cells such as platelets (PLTs) are known to contain mitochondrial DNA polymerase,22 early experiments were aimed at evaluating and controlling PC-derived ETGA background signals via a modified differential cell lysis procedure. The goal of subsequent experiments was to assess the ability of a modified differential cell lysis-ETGA procedure (referred to hereafter as the ETGA-PC assay) to sensitively detect bacterially spiked PCs.
Materials and Methods
Source of PLTs
Three types of PCs were used in this study. Single-donor whole blood–derived nonleukoreduced (WBNLR) PCs were purchased from Innovative Research (Novi, MI). Apheresis nonleukoreduced (ANLR) PCs were purchased from Biological Specialty (Colmar, PA). Innovative Research and Biological Specialty received informed consent from all donors before collecting PCs. Apheresis leukoreduced (ALR) PCs were collected at Miller Keystone Blood Center (Bethlehem, PA) for use at St Luke's University Hospital (Bethlehem, PA) and transported to our laboratory after their 5-day expiration period. St Luke's University Hospital and Health Network Internal Review Board (Bethlehem, PA) approved this study protocol.
ETGA-PC assay procedure
The ETGA-PC assay consists of a differential cell lysis procedure followed by sensitive measurement of DPE activity. We previously described a hematopoietic cell lysis procedure that enabled reduction of the DNA polymerase signal associated with 0.75-mL aliquots of whole blood–inoculated blood culture bottles.21 For this study, we modified this procedure for PC as follows. Briefly, 0.5-mL PC aliquots were added to microfuge tubes containing 0.5 mL of a detergent mix (1% Triton X-100/Tween 20/saponin). In an effort to begin lysing PLT, the tubes were then capped, inverted four times to mix, and incubated at room temperature for 5 minutes. Samples were then centrifuged at 8000 × g for 3 minutes. The supernatant was poured off into a waste container, and the opening of each tube was tapped onto a plastic-backed wipe to remove any residual liquid. The pellets were then resuspended in 5 mmol/L sodium hydroxide by pipetting up and down 10 times in an effort to complete PLT lysis and inactivate extracellular DNA polymerase activity. Samples were incubated at room temperature for 5 minutes, and centrifuged at 8000 × g for 3 minutes. The supernatant was poured off into a waste container, and the opening of each tube was tapped onto a plastic-backed wipe to remove any residual liquid. Next, 0.6 mL of a Tris-based wash buffer was added to each tube, pipetted up and down five times, simultaneously transferred to a beadmill lysis tube, and centrifuged at 8000 × g for 3 minutes to concentrate bacteria. After spinning, the supernatant was removed and DNA polymerase activity was measured by ETGA (see below).
Measurement of DNA polymerase activity via ETGA
ETGA is a highly sensitive method that links microbial DNA polymerase activity to quantitative polymerase chain reaction (qPCR) amplification. Complete details regarding measurement of DNA polymerase activity via ETGA (including oligonucleotide sequences, buffer formulations, and qPCR conditions) have been previously reported.20 Briefly, 50 μL of a DPE reaction mix is added to a beadmill tube containing preconcentrated microbes. The core components of the DPE mix are a synthetic DNA substrate, a dNTP mix (dATP, dCTP, dTTP, dGTP), Tris buffer, and magnesium sulfate. The beadmill tube containing the DPE mix is mixed vigorously at 2800 rpm for 6 minutes using a 2-mL disrupter attachment (Scientific Industries, Inc., Bohemia, NY) to lyse the microbes and release endogenous DNA polymerase. After disruption, the beadmill tubes are transferred to a 37°C heat block and are incubated for 20 minutes, during which the synthetic DNA substrate is modified by microbial-derived DNA polymerase. Samples are then transferred to 95°C for 5 minutes to inactivate bacterial DNA polymerase and subsequently centrifuged at 12,000 × g for 30 seconds. Four microliters from each beadmill tube is then transferred to the associated qPCR procedure, and positivity is determined automatically by the thermocycler's software.
Assessment of PC-derived background signal
To assess the level and DNA polymerase dependence of PC-derived background signal, 0.5 mL of PC (n = 6 aliquots) was pelleted at 8000 × g, resuspended in 0.5 mL of sterile saline (150 mmol/L), and simultaneously transferred to beadmill tubes. Beadmill tubes were centrifuged at 8000 × g for 3 minutes, and the supernatant was carefully removed. The DNA polymerase activity was measured via standard ETGA method in three of the six beadmill tubes. Since dideoxyCTP has been previously demonstrated to block DPE of the ETGA assay substrate,20, 21 a DPE reaction mix (containing dATP, dGTP, dTTP, and dideoxyCTP) was added to the remaining three bead mill tubes in an effort to demonstrate that any observed assay background signal is specific for DNA polymerase and not other enzymatic activities associated with PC lysates. Complete methodologic details regarding assessment of DNA polymerase dependence of ETGA signal via dCTP versus ddCTP have been previously published.20, 21
Reduction of PC-derived background signal by a selective cell lysis procedure
For background reduction experiments, 0.5-mL aliquots of WBNLR, ANLR, and ALR PCs were subjected to the ETGA-PC assay procedure as described above. For each unit, an additional 0.5-mL aliquot was washed once with saline as described above and was included as a nonlysed control (NLC). The NLC was included for each assayed PC unit in an effort to demonstrate the potential ETGA background signal for each PC aliquot when not subjected to the differential cell lysis portion of the of the ETGA-PC assay. For each incoming PC unit, sterility was verified by inoculating 8 mL of PC into a blood culture bottle and also by plating 100 μL of PC onto blood agar. Each PC-inoculated blood culture bottle was continuously monitored by a blood culture incubator for 5 days before designating as negative.
Spiking of serially diluted bacteria into PC aliquots
Staphylococcus epidermidis (ATCC 12228), Staphylococcus aureus (ATCC 25923), Escherichia coli (ATCC 25922), and Klebsiella pneumoniae (ATCC 700721) were used for serial dilution spiking studies due to the established prevalence of these species among contaminated PC units associated with transfusion-related fatalities.9, 23 From a fresh plate of each bacterial species, colonies were suspended in 3 mL of saline until an OD625 = 0.100 ± 0.010 was achieved. This OD reading yielded bacterial stocks at approximately 1 × 108 colony-forming units (CFUs)/mL. Dilutions were then made in saline to obtain stock concentrations of 1 × 106, 1 × 105, 2 × 104, 1 × 104, and 5 × 103 CFUs/mL. Next, 0.99 mL of PC was added to six sterile microfuge tubes. Ten microliters of each bacterial stock was added to five of these tubes, resulting in spiked PC stocks targeted at 1 × 104, 1 × 103, 200, 100, and 50 CFU/mL. Ten microliters of sterile saline was added to the final tube of PC, designated as the no spike control (NSC). Next, 0.5 mL of each spike level, and the NSC, was subjected to the ETGA-PC assay procedure as described above. One-hundred microliters of each PC stock was also plated to determine the approximate CFU levels within spiked samples and to confirm the sterility of the NSC. For each of the four bacterial species, serial dilution spikes were performed as described above using three different ANLR PC units and three different ALR PC units, totaling six independent experimental replicates.
Stocks of 22 additional bacterial species (see Table 3 for microbial IDs) were prepared in saline and spiked into PC aliquots at three target-input levels around the 200 CFUs/mL level. All spikes were compared to a donor-matched NSC. One-hundred microliters of PC stocks (spiked and nonspiked) was also plated in parallel to determine the approximate CFU-input levels and to confirm the sterility of nonspiked PC.
Analysis of bacterial growth within PC units
Immediately before spiking bacteria, a 0.5-mL aliquot of PC was subjected to the ETGA-PC assay to determine the baseline background level for each respective unit. For analysis of bacterial growth in PC units, the panel of four microorganisms (listed above) was expanded to include Pseudomonas aeruginosa (ATCC 27853), Serratia marcescens (ATCC 29021), and Bacillus cereus (ATCC 11778). Except for B. cereus, stock solutions of each bacterial species were prepared as described above. For B. cereus, two colonies from a freshly grown plate were inoculated into brain heart infusion broth and incubated at 30°C with shaking until reaching an OD600 of 1.00 ± 0.10. Using stock solutions of each bacterial species (n = 7 total), a target of 100 CFUs was spiked into each PC unit. The actual amount of input CFU was determined by plating analysis of the stock bacterial suspensions. Once spiked, the PC unit was placed into a PC incubator (Model PC900, Helmer, Noblesville, IN) and incubated at 22°C with shaking. During incubation, aliquots of PC were removed and immediately analyzed by the ETGA-PC assay after 24, 48, 72, 144, and 168 hours of incubation. In an effort to monitor the CFU levels, parallel plating was performed on serial dilutions of each PC aliquot taken during the time course incubation.
Results
Characterization and reduction of PC-derived ETGA assay background signal
In this study we set out to determine the feasibility of using ETGA to detect bacterially contaminated PCs. A basic schematic diagram of the proposed approach is presented in Fig. 1. Since PLTs are a potential source of DNA polymerase activity,22 we first set out to verify that 0.5-mL aliquots from a sterile PC unit were detectable by the ETGA assay. As shown in Fig. 2, triplicate analysis of nonlysed PC aliquots contributed significant amounts of ETGA assay background signal. In contrast, ETGA assay signal was undetectable from triplicate parallel preparations of nonlysed PC aliquots that were incubated with a dNTP mix containing dideoxyCTP instead of dCTP (Fig. 2).
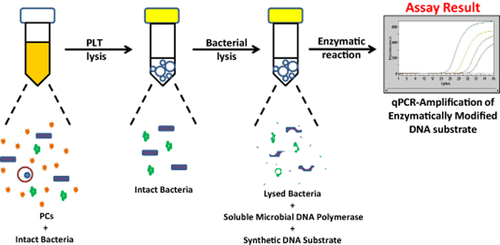
Schematic of a novel approach for detecting contaminated PCs. A schematic overview of the ETGA-PC assay is depicted. First, PLTs are lysed via a differential cell lysis procedure, leaving intact bacteria. Bacterial DNA polymerase is then exposed to a synthetic DNA substrate during a beadmill-induced microbial lysis. During a 37°C incubation, bacterial DNA polymerase then modifies the synthetic DNA substrate. The modified DNA substrate is detected by qPCR and indicates the presence of bacterial contamination.
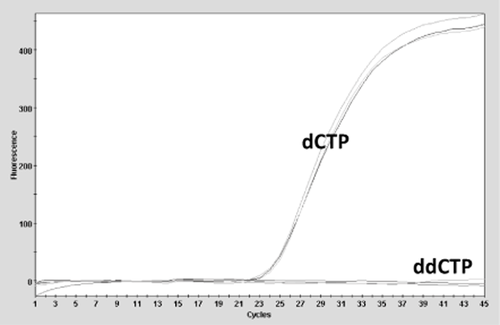
Characterization of PC-derived background signal. ETGA assay signals from nonlysed PC aliquots were compared using DPE reactions containing standard nucleotides (dATP, dGTP, dATP, and dCTP) versus a nucleotide mix containing dATP, dGTP, dATP, and dideoxyCTP.
Next, we assessed the ability of a differential cell lysis procedure to reduce the amount of background signal associated with 0.5-mL aliquots taken from WBNLR, ANLR, and ALR PC units. As shown in Figs. 3A through 3C, the differential cell lysis procedure effectively reduced the ETGA assay signals associated with all three types of PC units, relative to the respective NLCs. In addition to the PC units presented in Figs. 3A to 3C, n = 6 × 0.5-mL aliquots from WBNLR (n = 3 units), ANLR (n = 7 units), and ALR (n = 34 units) were subjected to the ETGA-PC assay. In Fig. 3D, a compilation of the background signals (obtained from all PC units analyzed) is presented, relative to a representative NLC signal. An experimental positivity threshold was established using the assembled background data and is presented in Fig. 3D. This positivity threshold was used as an objective assay result criteria for subsequent bacterial spiking experiments. Parallel blood culture analysis of an 8-mL inoculation from each PC unit was also performed and yielded negative results after a 5-day incubation period.
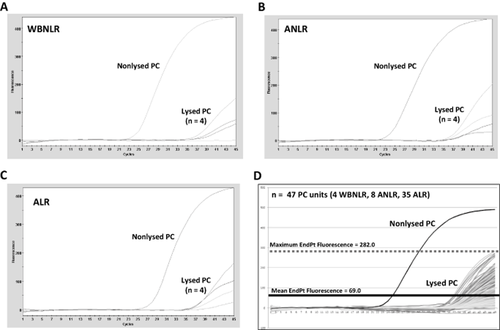
Assessment and reduction of ETGA assay background derived from different types of PC units. (A-C) ETGA-PC assay signal was determined for WBNLR, ANLR, and ALR PC units. A nonlysed PC control was included to determine the potential background signal associated with each respective unit. (D) ETGA-PC assay signals derived from 47 PC units (multiple replicates per unit) were combined into one graph. A positivity threshold was subsequently generated by taking the highest endpoint fluorescence value obtained from the background curves.
Detection of clinically relevant bacteria from spiked PC aliquots
After demonstrating that the differential cell lysis procedure was effective at reducing PC-derived background signal, we set out to verify that it was also compatible with detection of DNA polymerase activity from clinically relevant bacterial species via ETGA. Two Gram-positive (S. aureus and S. epidermidis) and two Gram-negative (E. coli and K. pneumoniae) bacterial species were initially chosen for spiking experiments due to their established prevalence among contaminated PC units.9, 23 The ETGA-PC assay was performed on bacterially spiked PC aliquots from three independent ANLR and ALR units as described under Materials and Methods, and a representative set of ETGA-qPCR curves (along with the positivity threshold) is presented for each of the four bacterial species in Fig. 4. Complete data sets from ETGA-PC assay analysis of serially diluted bacterial spikes into ANLR and ALR PC units is presented in Tables 1 and 2, respectively. Together, the serial dilution spiking experiments revealed an approximate sensitivity of 30 to 200 CFUs/mL (mean, 85 CFU/mL) for S. epidermidis, S. aureus, E. coli, and K. pneumoniae.
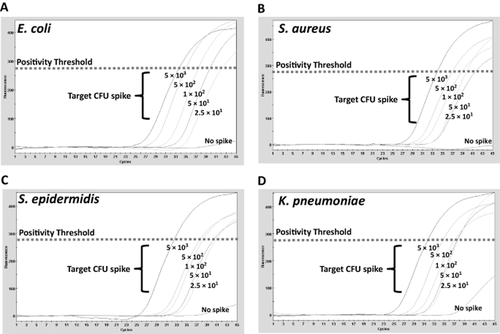
Examples of ETGA-PC assay curves from bacterial spikes. (A-D) Four different bacterial species were serially diluted and spiked into PC aliquots. Examples of the ETGA-PC assay results are presented for each bacterial species versus nonspiked PC aliquots. In each graph, the positivity threshold is included as a visual reference.
| Unit A | Unit B | Unit C | |||||
|---|---|---|---|---|---|---|---|
| Strain | CFUs targeted per 0.5 mL of PC* | CFUs assayed | Result | CFUs assayed | Result | CFUs assayed | Result |
| E. coli | 5,000 | 2,800 | Positive | 5,000 | Positive | 4,500 | Positive |
| E. coli | 500 | 280 | Positive | 500 | Positive | 450 | Positive |
| E. coli | 100 | 56 | Positive | 100 | Positive | 90 | Positive |
| E. coli | 50 | 28 | Positive | 50 | Positive | 45 | Positive |
| E. coli | 25 | 14 | Positive | 25 | Positive | 23 | Positive |
| K. pneumoniae | 5,000 | 4,500 | Positive | 2,800 | Positive | 3,450 | Positive |
| K. pneumoniae | 500 | 450 | Positive | 280 | Positive | 345 | Positive |
| K. pneumoniae | 100 | 90 | Positive | 56 | Positive | 69 | Positive |
| K. pneumoniae | 50 | 45 | Positive | 28 | Positive | 34 | Negative |
| K. pneumoniae | 25 | 23 | Negative | 14 | Positive | 17 | Negative |
| S. aureus | 5,000 | 4,500 | Positive | 11,000 | Positive | 3,330 | Positive |
| S. aureus | 500 | 450 | Positive | 1,100 | Positive | 333 | Positive |
| S. aureus | 100 | 90 | Positive | 220 | Positive | 67 | Positive |
| S. aureus | 50 | 45 | Positive | 110 | Positive | 34 | Positive |
| S. aureus | 25 | 23 | Positive | 55 | Positive | 17 | Negative |
| S. epidermidis | 5,000 | 3,000 | Positive | 4,000 | Positive | 3,500 | Positive |
| S. epidermidis | 500 | 300 | Positive | 400 | Positive | 350 | Positive |
| S. epidermidis | 100 | 60 | Positive | 80 | Positive | 70 | Positive |
| S. epidermidis | 50 | 30 | Positive | 40 | Negative | 35 | Positive |
| S. epidermidis | 25 | 15 | Positive | 20 | Positive | 18 | Positive |
- * Multiply by 2 to obtain CFUs/mL.
| Unit A | Unit B | Unit C | |||||
|---|---|---|---|---|---|---|---|
| Strain | CFUs targeted per 0.5 mL of PC* | CFUs assayed | Result | CFUs assayed | Result | CFUs assayed | Result |
| E. coli | 5000 | 4150 | Positive | 4000 | Positive | 3650 | Positive |
| E. coli | 500 | 415 | Positive | 400 | Positive | 365 | Positive |
| E. coli | 100 | 83 | Positive | 80 | Positive | 73 | Positive |
| E. coli | 50 | 42 | Negative | 40 | Positive | 37 | Positive |
| E. coli | 25 | 21 | Positive | 20 | Positive | 19 | Negative |
| K. pneumoniae | 5000 | 5000 | Positive | 3500 | Positive | 4700 | Positive |
| K. pneumoniae | 500 | 500 | Positive | 350 | Positive | 470 | Positive |
| K. pneumoniae | 100 | 100 | Positive | 70 | Positive | 94 | Positive |
| K. pneumoniae | 50 | 50 | Positive | 35 | Positive | 47 | Positive |
| K. pneumoniae | 25 | 25 | Negative | 18 | Positive | 24 | Positive |
| S. aureus | 5000 | 8750 | Positive | 4250 | Positive | 5850 | Positive |
| S. aureus | 500 | 875 | Positive | 425 | Positive | 585 | Positive |
| S. aureus | 100 | 175 | Positive | 85 | Positive | 117 | Positive |
| S. aureus | 50 | 88 | Positive | 43 | Negative | 59 | Positive |
| S. aureus | 25 | 44 | Positive | 22 | Negative | 30 | Positive |
| S. epidermidis | 5000 | 3600 | Positive | 5000 | Positive | 1800 | Positive |
| S. epidermidis | 500 | 360 | Positive | 500 | Positive | 180 | Positive |
| S. epidermidis | 100 | 72 | Positive | 100 | Positive | 36 | Positive |
| S. epidermidis | 50 | 36 | Negative | 50 | Positive | 18 | Negative |
| S. epidermidis | 25 | 18 | Positive | 25 | Negative | 9 | Negative |
- * Multiply by 2 to obtain CFUs/mL.
| Bacterial species | ATCC number | Lowest level of CFUs detected per 0.5 mL of PC |
|---|---|---|
| S. marcescens | 29021 | 28 |
| P. aeruginosa | 27853 | 34 |
| Y. enterocolitica | 9610 | 60 |
| K. oxytoca | 700324 | 30 |
| B. cereus | 11778 | 100 |
| B. subtilis | 6633 | 60 |
| P. acnes | 6919 | 95 |
| C. perfringens | 10543 | 65 |
| S. pyogenes | 19615 | 85 |
| S. pneumoniae | 6303 | 125 |
| E. cloacae | 13047 | 30 |
| E. aerogenes | 15038 | 25 |
| S. lugdunensis | 43809 | 25 |
| S. enterica | 700720 | 60 |
| E. faecalis | 51299 | 25 |
| S. bovis | 35034 | 35 |
| S. warneri | 27839 | 50 |
| C. flavescens | 10340 | 26 |
| S. agalactiae | 13813 | 33 |
| M. luteus | 381 | 15 |
| A. baumannii | 15308 | 50 |
| E. faecium | 19434 | 50 |
We also tested the ability of the ETGA-PC assay to detect an additional 22 clinically relevant bacterial species from PC aliquots spiked with microbes at levels close to 200 CFUs/mL. As shown in Table 3, the ETGA-PC assay was able to detect all 22 bacterial species close to, or below this level.
Monitoring of bacterial growth within PC units
Next, we set out to determine whether the ETGA-PC assay could monitor the growth of seven different bacterial species within PC units. To start, we determined the approximate CFUs/mL within each of the spiked PC units by dividing the number of CFUs spiked (obtained from plating analysis) by the volume of PC contained within each unit. The resultant CFUs/mL values for each of the bacterially spiked units are shown in the bottom right portion of the graph in Fig. 5 and are all less than 1 CFU/mL. In Fig. 5, the qPCR-Ct values obtained from ETGA-PC assay analysis of PC aliquots were plotted versus the time for which each unit was incubated. The ETGA-PC assay results and CFUs/mL values (determined by plating analysis) are also presented in table format directly below the line plots (Fig. 5).
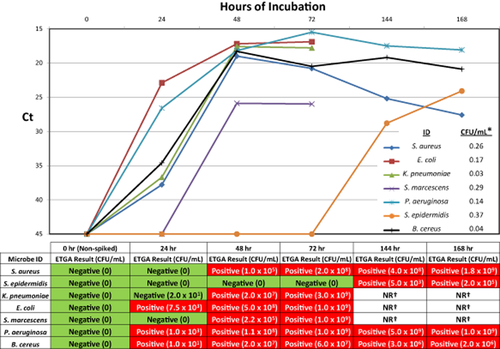
Time course analysis of bacterially spiked PC units. PC units were spiked with seven different bacterial species and ETGA-PC assay and CFU plating analysis were performed on aliquots in a time course fashion. The resultant ETGA assay Ct values were plotted versus the time that each unit spent in a PC incubator before aliquot removal. In the bottom right portion of the time course plot, the starting CFUs/mL values are shown for each of the spiked PC units. The ETGA-PC assay and CFU plating results from the time course experiments are also presented in table format directly below the time course plots. *The starting CFUs/mL within the respective PC units as determined by plating analysis. †Samples were not removed from PC unit due to massive aggregation or coagulation.
Discussion
In this report we assessed the feasibility of a novel approach toward sensitive and universal detection of bacterially contaminated PCs via measurement of bacterially derived DNA polymerase activity (Fig. 1). Since anucleated cells such as PLTs have been established to contain mitochondrial DNA polymerase,22 we hypothesized that PCs might yield ETGA assay background signal. As suspected, early feasibility experiments presented herein showed that PCs contribute significant DNA polymerase–dependent ETGA assay background signal and therefore need to be reduced to enable detection of DNA polymerase activity from spiked bacteria. To this end, we demonstrated that a differential cell lysis procedure could effectively reduce the ETGA assay background signal derived from three different types of PCs. We then compiled the background data from additional PC units and generated an experimental positivity threshold. In subsequent experiments, ETGA-PC assay signals derived from PC aliquots containing serial dilutions of four clinically relevant bacteria were distinguishable from donor-matched PC aliquots that had not been spiked with bacteria. In fact, when using the experimental positivity threshold as an objective measure, these preliminary experiments demonstrated that the ETGA-PC assay can detect as little as 30 CFUs/mL bacteria within several of the spiked PC aliquots. As shown in Tables 1 and 2, we did notice that some bacterial species were not consistently detected at less than 100 CFUs/mL when spiked into PC units derived from different donors. This inconsistent detection at low levels is likely reflective of PC donor–dependent bacterial recovery differences associated with the ETGA-PC assay sample preparation procedure. Considering this, more replicates are certainly needed to determine a true limit-of-detection (LOD) value for each bacterial species; however, when spiked into six aliquots of PC (representing three independent ALR and ALNR units), our data suggest that the ETGA-PC assay is able to consistently detect close to, or less than 200 CFUs/mL E. coli, K. pneumoniae, S. aureus, and S. epidermidis. When compared to the claimed LOD values for current FDA-approved POI tests,19, 24, 25 the ETGA-PC assay could represent approximately a 50- to 300-fold improvement in sensitivity. Furthermore, independent studies have shown that one of the FDA-approved POI assays is prone to failure when challenged with clinically relevant Gram-negative bacterial species that were spiked into PCs at levels well above the claimed LOD, thus raising concerns regarding its reliability.26-28
In addition to some commonly encountered contaminants, a wide range of bacterial species have been isolated from contaminated PC units.8, 9, 29, 30 It is therefore imperative that a POI test is not only sensitive, but also universal in nature. To this end, we also set out to verify that the ETGA-PC assay is able to detect a range of clinically relevant bacterial species. Our experiments showed that the ETGA-PC assay is also able to sensitively detect the presence of an additional 22 clinically relevant bacterial species. These data further support the notion that measurement of DNA polymerase activity represents a bona fide universal detection approach.
We felt that it was also important to assess the ability of the ETGA-PC assay to monitor the growth of bacteria within PC units incubated under standard conditions. These experiments effectively demonstrated that the ETGA-PC assay can sensitively detect the presence of bacteria during incubation of spiked PC units. Similar to previous studies,26, 31 S. epidermidis is prone to slow growth within PLT units, and in our experiments, it was not detected by plating or ETGA until after 144 hours of incubation. Except for the 24-hour time point taken from the PC unit spiked with K. pneumoniae, all of the plating results are in agreement with the ETGA-PC assay results. The subsequent 48-hour time point for this unit yielded a strong ETGA-PC assay positive signal, which was consistent with the high levels of CFUs determined by parallel plating analysis.
Considering the feasibility data presented herein, we feel that the ETGA-PC assay has potential to be used as a POI test for rapid detection of bacterially contaminated PC units close to the time of transfusion. To our knowledge, this is first time that microbes have been detected from PCs via measurement of their endogenous DNA polymerase activity and therefore represents a truly novel approach. In its current manual form, the ETGA-PC assay requires approximately 2 hours to obtain results for a batch of six samples, 90 minutes of which are spent in sample incubations and PCR thermocycling. Experiments are under way to further simplify the assay, reduce the overall time to result, and/or make the procedure more amenable to automation.
Acknowledgments
We thank Kevin Cochrane and the rest of the St Luke's University Hospital blood bank team for all of their help with supplying freshly expired apheresis PLT units.
Conflict of Interest
DRZ, NMR, MJK, and SMO are employees of ZEUS Scientific Incorporated and have no other conflicts of interest to declare.