Crop genome-wide association study: a harvest of biological relevance
Summary
With the advent of rapid genotyping and next-generation sequencing technologies, genome-wide association study (GWAS) has become a routine strategy for decoding genotype–phenotype associations in many species. More than 1000 such studies over the last decade have revealed substantial genotype–phenotype associations in crops and provided unparalleled opportunities to probe functional genomics. Beyond the many ‘hits’ obtained, this review summarizes recent efforts to increase our understanding of the genetic architecture of complex traits by focusing on non-main effects including epistasis, pleiotropy, and phenotypic plasticity. We also discuss how these achievements and the remaining gaps in our knowledge will guide future studies. Synthetic association is highlighted as leading to false causality, which is prevalent but largely underestimated. Furthermore, validation evidence is appealing for future GWAS, especially in the context of emerging genome-editing technologies.
Introduction
Since the concept was first applied in maize in 2001 (Thornsberry et al., 2001), association mapping studies in crop species have revealed links between tens of thousands of genomic regions and various traits. Association mapping is a quantitative approach for determining if a genomic variant is associated with a trait of interest using a natural population or a collection of diverse individuals. The main hypothesis states that a particular phenotype shared by a subset of individuals will be highly linked to neighboring genetic variations (linkage disequilibrium, LD; Glossary Box) in their recent ancestor, where the causal mutation and corresponding phenotype arose. Recent advances in high-throughput genotyping technologies and increases in computational power have made it possible to carry out association studies on genome-wide sets of genetic variants, an approach known as genome-wide association study (GWAS), thus greatly changing the mapping of quantitative traits. While relatively low-resolution and time-consuming methods such as linkage mapping are typically used for mapping in biparental populations, the emergence of GWAS provides an opportunity to discover genes or regions associated with given traits in a relatively high resolution and unbiased manner in broad-based and diverse populations. GWAS can also reveal the global landscape of a trait, known as its genetic architecture (Figure 1), a term used to describe the genetic basis of a trait based on information regarding the number of causative genes or alleles, their interactions, and the distribution and patterns of their effects (Hansen, 2006).
Sequencing-based GWAS has become a routine tool in crop genetics over the last decade, making outstanding achievements in two major ways. The first is in redefining the concept of a ‘trait’, from conventional developmental traits (e.g. Buckler et al., 2009; for maize flowering; Huang et al., 2010; for rice agronomic traits), to individual responses to environmental factors (biotic or abiotic stress tolerance) (e.g. Li et al., 2017 for rice blast resistance; Wang et al., 2016 and Guo et al., 2018 for drought tolerance in maize and rice, respectively; Kuroha et al., 2018 for periodic flooding adaptation), to large-scale molecular-level quantification (e.g. Fu et al., 2013 and Chen et al., 2018 for determination of the structural transcriptome of the maize kernel, respectively; Chen et al., 2014 and Wen et al., 2014a with respect to metabolism in rice and maize), to more complex phenotypic variations as long as they are heritable and measurable, such as rice heterosis (Huang et al., 2015) and maize haploid male fertility (Ma et al., 2018a). The level of precision is expected to extend further in the next decade, from single-trait variables to a variable vector depicting a dynamic developmental process with the recent development of plant phenomics techniques (Yang et al., 2013b; Tardieu et al., 2017; Singh et al., 2018). The second impressive achievement is that as increasing numbers of crop species enter the ’omics era, GWAS is being performed not only in cereals (in particular rice and maize) but also in a broad range of crops, including soybean (i.e., Hwang et al., 2014; Wen et al., 2014b; Zhang et al., 2015a; Fang et al., 2017a; Leamy et al., 2017), cotton (Fang et al., 2017b; Wang et al., 2017b; Du et al., 2018; Ma et al., 2018b), tomato (Lin et al., 2014; Tieman et al., 2017; Zhu et al., 2018), cucumber (Shang et al., 2014; Zhang et al., 2015c), sesame (Wei et al., 2015), peanut (Pandey et al., 2014; Zhang et al., 2017b), peach (Cao et al., 2016), and lettuce (Zhang et al., 2017a). These studies, together with purpose-developed populations, catalogs of allelic variants, and corresponding genotype–phenotype associations, provide unprecedented resources for understanding crop functional genomics. These studies have not only validated known trait associations, but also identified new favorable haplotypes or, in some cases, revealed previously unknown pathways.
Despite great success, current GWAS analysis has clear limitations, especially issues of population structure correlation and low-frequency causal alleles leading to false-negative results (Korte and Farlow, 2013). For example, only one gene (ZmCCT) was revealed for flowering time using a diverse association mapping panel consisting of 500 inbred lines (Yang et al., 2013a), as flowering time is a typical adaptive trait and is always confounded (i.e., highly correlated) with population structure. It has been widely accepted that many false negatives occur for such confounded traits when correcting for population structure in GWAS (Huang and Han, 2014). Another example showed only five inbred lines (<1%) possessing functionally alternative alleles at the Brachytic2 locus for plant height among 527 lines (Xing et al., 2015); it is thus not possible to identify this locus using routine association mapping analysis. A similar phenomenon is also seen in rice, where (putative) causal alleles within most of the cloned yield-related quantitative trait loci (QTLs) are at low frequency in diverse germplasms (2% for Ghd7, Xue et al., 2008; Lu et al., 2012; 1% for GS3, Fan et al., 2006; Mao et al., 2010; 2% for qGL3, Zhang et al., 2012; 6% for TGW6, Ishimaru et al., 2013). Solving these issues by developing novel statistical models to explore rare functional alleles (Zhu et al., 2011; Listgarten et al., 2013; Kaakinen et al., 2017) or employing artificially designed populations to balance allelic frequencies and control population structure (Buckler et al., 2009; Dell'Acqua et al., 2015; Romero Navarro et al., 2017; Wen et al., 2018) are of great importance for GWAS. As these matters have been extensively discussed previously (Gibson, 2012; Korte and Farlow, 2013; Xiao et al., 2017; Cockram and Mackay, 2018), they will not be the further focus of the present review.
For the foreseeable future, GWAS will continue to be a common research tool for providing a bird's-eye view of the genetic structure of any trait of interest. The generation of various ’omics data will increase and enable OWAS (’omics-wide association study; Glossary Box) (Xiao et al., 2017). As the number of GWAS-based candidate gene regions has greatly increased in recent years, a review of new insights into crop genetic architecture is required. We summarize here (Box 1) how recent GWAS results have led to an updated view of several important issues of genetic architecture, including epistasis, pleiotropy, and phenotypic plasticity. We also discuss the likely causes of synthetic association, an increasingly common GWAS artifact. We examine the impact of these new biological insights and problems on the practice of GWAS and emphasize the need to validate GWAS results, especially by using the increasingly popular genome-editing technology.
Box 1. Highlights
- Crop GWAS has ushered a transition to ’omics-wide association mapping (OWAS), promising a better understanding of genetic architecture of complex traits.
- The large number of studies provides an unprecedented opportunity to increase in-depth understanding of the classical concepts of epistasis and pleiotropy.
- Phenotypic plasticity is largely ignored and requires intensive data collection and general statistical modeling.
- Synthetic association exists frequently in GWAS and is considered to result from the presence of multiple independent alleles within a locus.
- Emerging novel technologies such as genome editing can be used for further GWAS validation.
Epistasis: Negligible or neglected?
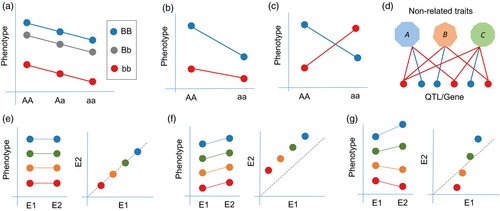
Epistasis represents a non-linear interaction between two or more segregating loci with different alleles across genetic backgrounds. This type of interaction between segregating loci is expected to contribute to phenotypes by biologically plausible mechanisms (Mackay, 2014). However, most studies have focused on additive genetic variance (Figure 1a), with relatively little attention paid to epistasis (Figure 1b, c), the importance of which is still being established in different biological systems. While its prominent role in heterosis has been widely recognized in various crops (Yu et al., 1997; Li et al., 2001; Melchinger et al., 2007; Garcia et al., 2008; Shen et al., 2014; Jiang et al., 2017), the prevalence of epistasis in maize trait architecture is thought to be small (Buckler et al., 2009; Tian et al., 2011; Xiao et al., 2016), or results in a large effect only at specific loci (Studer and Doebley, 2011; Durand et al., 2012). However, many studies indicate that epistasis is pervasive in contributing to various quantitative trait phenotypes (Manicacci et al., 2009; Würschum et al., 2011; Zhang et al., 2015a; Wen et al., 2016; He et al., 2017; Luo et al., 2017; Mathew et al., 2018) and can be further used to improve the accuracy of trait prediction for both inbreds and hybrids (Maurer et al., 2015; Santos et al., 2015; Luo et al., 2017). These results were observed in a variety of populations, such as a recombinant inbred line (RIL) population, a multi-parent advanced-generation inter-cross (MAGIC) population, and diversity panels, and for different traits, such as morphological characteristics, resistance to disease, and cellular metabolite levels.
Even though the importance of epistasis is increasingly recognized, the detection of epistatic effects is difficult, especially in GWAS using populations of unrelated individuals. Large numbers of variants, each present at a low frequency, create a major challenge, leading to low statistical confidence in the level of epistasis. Thanks to recently proposed efficient computing algorithms (Hemani et al., 2011; Gyenesei et al., 2012; Lishout et al., 2015; Zhang et al., 2016; Cowman and Koyuturk, 2017) and alternative, non-exhaustive modeling approaches (Guo et al., 2014; Leem et al., 2014; Karkkainen et al., 2015; Wang et al., 2015a; Zhang et al., 2016; Mathew et al., 2018), both the computational cost and multiple-testing burden can be effectively addressed and even high-order interactions can be uncovered, provided there is a sufficient number of individuals in the population under study. Still, because of the need to test alleles pairwise, those with relatively high frequency will provide the greatest probability of discovering epistasis, and low-frequency alleles at either locus will reduce the statistical power. This could be why epistatic effects appear to be population dependent. For a panel with diverse lines, a large population size is needed to reach the high statistical power necessary to uncover QTLs with moderate or subtle epistatic effects. However, artificially designed populations with balanced allele frequency, such as MAGIC (Mathew et al., 2018), promise to greatly facilitate epistasis discovery. Interestingly, Wei et al. (2018) found that loci identified using genotypic-variability-based GWAS can be used to evaluate potential epistatic interactions.
In general, integrating improved algorithms with experimental crop populations will improve the accuracy of future interaction studies. These insights will enable future crop engineering, as demonstrated by a pioneering study in tomato that optimized inflorescence architecture and high yield by eliminating undesirable epistasis (Soyk et al., 2017).
Pleiotropy: What is the promise of GWAS?
Understanding pleiotropy, a well known phenomenon identified a century ago (Stearns, 2010) in which one allele or gene affects multiple phenotypes (Figure 1d), is crucial for understanding genetic mechanisms and for simultaneous breeding of multiple complex traits. While the presence of pleiotropy is often mistakenly assumed when a locus is found to be associated with two or more traits, there are at least two additional assumptions that need to be emphasized. The first is that the association with two or more traits comes from the same causal gene within the locus, which is particularly important as the mapping resolution in crop GWAS limits the ability to discriminate between multiple candidates in high LD with each other. The second assumption is that the associated traits should be uncorrelated or, more precisely, they should be independently affected by the same causal gene rather than affected by confounding between the phenotypes (Solovieff et al., 2013). This makes the identification of pleiotropy more difficult in both plant species and human beings.
Pleiotropic effects have largely not been systematically explored in crops, even though a number of pleiotropic genes have been implicated in specific case studies. For example, Ghd7 controls heading date, grain number, plant height, and flag leaf area in rice (Xue et al., 2008; Tan et al., 2012; Weng et al., 2014), and its ortholog in maize, ZmCCT, affects both flowering time and disease resistance (Hung et al., 2012; Yang et al., 2013a; Wang et al., 2017a). Several recent studies have attempted to evaluate the shared genetic basis for crop genotype−phenotype links on a genome-wide basis. Schulthess et al. (2017) identified several potential pleiotropic loci for wheat yield-related traits using multiple-trait-based association mapping. This study also presented a simulation analysis of factors, including minor allele frequency and QTL effect sizes and distances for different traits, affecting the power to identify pleiotropy, which can be used as reference for future study designs. However, the population diversity and low genotypic density in this study limited the differentiation of true pleiotropy from the effects of closely linked genes or spurious pleiotropy (Glossary Box). By integrating a large and diverse soybean collection with whole-genome sequencing, Fang et al. (2017a) found that genetic sharing for different traits is widespread and, interestingly, the E2 locus exhibits pleiotropy for both yield and seed quality. Pleiotropy has also been identified for maize carbon and nitrogen metabolism (Zhang et al., 2015b).
Beyond the accumulative GWAS of various traits for crop species, improved methods provide an unprecedented opportunity to effectively dissect the contribution of pleiotropy to crop trait variation. Commonly applied multi-trait-based methods have been comprehensively reviewed (Solovieff et al., 2013; Hackinger and Zeggini, 2017) and can be applied to crop studies. We emphasize that whole-genome gene expression QTL (eQTL) results can also be used to analyze pleiotropy, and Zhu et al. (2016) have proposed a strategy to integrate eQTL data with knowledge of QTL resulting in observable phenotypes to identify the variants displaying pleiotropic effects for both trait and gene expression. This method was further applied by Hannon et al. (2017) to study the pleiotropic variants associated with quantitative traits and DNA methylation.
Pleiotropy seems common and important for increasing trait prediction accuracy for complex human traits (Maier et al., 2018), but our understanding of this phenomenon is still greatly limited in crop genetics. As the number of genotype–phenotype links grows, pleiotropy will draw greater attention from crop researchers.
Phenotypic plasticity: A power to nurture the nature
Most current efforts focus on mapping the genetics of complex trait variance in populations; however, the phenotypic performance of an individual can change with fluctuating environment. The ability to respond to environmental change by expressing variable phenotypes without genotypic change is called phenotypic plasticity (Figure 1e–g). The phenomenon of different alleles displaying varied plastic responses is described as genotype–environment interaction (G × E). The significance of phenotypic plasticity, which was considered to be genetically heritable, has been realized for several decades (Bradshaw, 1965; Weaver and Ingram, 1969; Schlichting, 1986; Gavrilets and Scheiner, 1993; Dewitt et al., 1998). These preliminary studies described morphological changes mainly by performing theoretical or simulation investigations. Various quantitative genetic models controlling phenotypic plasticity have been proposed, including the allelic sensitivity model and the over-dominance model (Scheiner, 1993). Current genetic mapping studies provide an unparalleled chance to explore the quantitative architecture of changing phenotypic responses and possibly even uncover the underlying genes (Wang et al., 2013; Zhai et al., 2014). Phenotypic plasticity adds a significantly complex layer to the genetic architecture of complex traits.
Phenotypic measures of plasticity include two aspects: the degree of change in the phenotypic mean across environments and the pattern of such change (Schlichting and Levin, 1984). The coefficient estimated from regression analysis, and summary statistics including range, standard deviation, and coefficient of variation are simple measures. However, these only describe the amount of change without indexing the pattern of change. The coefficient obtained from regression along an environmental gradient can be specified as the reaction norm to describe both the degree and pattern of plastic change (Eberhart and Russell, 1966; Freeman, 1973); however, the environmental gradient is trait and genotype dependent and therefore difficult to characterize when numbers of individuals are measured. After obtaining the measurements, phenotypic plasticity can be mapped genetically and compared in the same way as phenotypic mean, or modeled along specific frameworks (Wang et al., 2013; Zhai et al., 2014).
In a recent pioneering study, Kusmec et al. (2017) analyzed 23 agronomic traits in 4−11 environments using a nested association mapping (NAM) population consisting of about 5000 RILs. By partitioning the trait into phenotypic mean, linear, and non-linear plasticities with the Bayesian Finlay−Wilkinson Regression (FWR) (Su et al., 2006), structurally and functionally distinct candidate genes were found in association with mean and plastic phenotypes. This distinct genetic architecture provides a further opportunity to simultaneously manage trait mean and plasticity for a given environment or for a changing climate (Nicotra et al., 2010). Flowering time is a classic example used in model and crop plants to study the genetic control of both mean and plastic phenotypes (Ungerer et al., 2003; Anderson et al., 2012; Brachi et al., 2013; Mendez-Vigo et al., 2016; Mao et al., 2017). Another recent study on sorghum flowering time (Li et al., 2018) indicated that modeling plasticity can not only explain the genetic response to different environments, but also enable highly accurate prediction of trait performance in new environments. Beyond such studies on individual traits, a new model has been proposed (Zhou et al., 2015) to explore the genetic architecture of phenotypic plasticity in multiple correlative traits.
Even though the statistical framework (Wang et al., 2013; Zhai et al., 2014; Zhou et al., 2015) has been established for years and representative studies (Kusmec et al., 2017; Li et al., 2018) have been performed, our understanding of the mechanisms of plasticity and its effect on shaping crop diversity along environmental gradients is still limited. The lack of deep insights into this common and important issue may be because of the massive data sets needed, including environmental measurements in addition to standard genotypic and phenotypic variations. Particularly, multiple environmental conditions along a gradient of variation are necessary to unravel the quantitative attributes of phenotypic responses. When describing the interaction between genome and environment, understanding phenotypic plasticity helps interpret the evolutionary and environmental forces modifying genetic architectures (Josephs, 2018). Therefore, investigating phenotypic plasticity will also contribute to crop improvement, when integrated with breeding programs that are facing climate change and instability.
Synthetic association: Misleading for causality
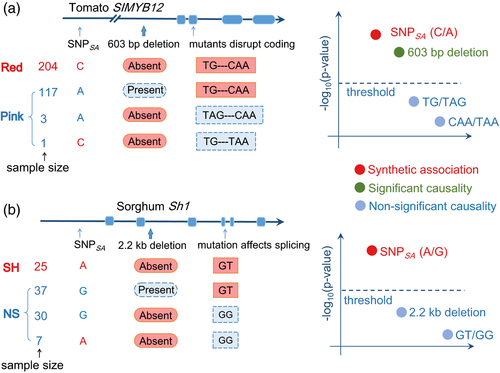
Confusingly, sometimes the non-causative loci show more significant associations in GWAS than the causative ones. In other words, the causative genes are sometimes located away from the GWAS peaks. This has been observed in a number of association studies in plants including Arabidopsis (Atwell et al., 2010; Kerdaffrec et al., 2016), rice (Huang et al., 2010, 2011; Yano et al., 2016), sorghum (Lin et al., 2012), and tomato (Lin et al., 2014). This misleading association is called synthetic association (or ‘ghost association’), which is presumed to be caused by LD between common tagged markers and rare causative variants (Dickson et al., 2010; Chang and Keinan, 2012). The rare-allele hypothesis can be understood such that the common variant would show significant association signal with a given trait when it is linked to a low frequency but large-effect causative variant. This can also explain the ‘missing heritability’ issue, because the common markers (identified with greater significance) can explain only a limited fraction of trait variance as a result of their imperfect association with the causative low-frequency variant. However, some cases do not follow the simple rare-allele assumption and can be explained by another phenomenon, that the trait variation is caused by multiple alleles within one gene (Lin et al., 2012; Yano et al., 2016).
We tend to interpret the synthetic association issue as the ‘presence of multiple causative alleles’, in which the standard single-variant-based GWAS has insufficient power to detect any of these because of the genetic interference between different alleles (Figure 2). Given that mutation constantly generates new variants, multiple independent alleles within one gene leading to the same phenotype could be common. Haplotype- or gene-based methodologies have good potential for identifying such situations, although current haplotype-based association mapping is still imperfect (Hayes, 2013) and both inferring accurate haplotypes and incorporating haplotypes into association mapping remain particularly challenging in plants. Additionally, the multiple functional alleles in such cases of synthetic association with high LD are unlikely to be captured by haplotype phasing. A better understanding of the underlying causes of synthetic associations would help in the design of future studies to detect causative genetic variants while avoiding artifacts.
Validation: A necessary adoption for future GWAS
Even with a strong theoretical foundation and efforts to remove undesired noise (i.e., population structure) and employ strict probability cut-offs, false-positive associations will still occur due to the enormous number of statistical inferences and other unaccounted factors, such as low-accuracy genotype calling at some loci (Browning and Yu, 2009), small population size (Finno et al., 2014), and synthetic associations introduced above. This calls for an independent validation process, which has seldom been incorporated into GWAS design. Validation includes at least two methodologies: one validating candidates of interest in different populations, the associations would be assumed to be more likely when being detected in independent studies; and the other using laboratory experiments, such as candidate gene knock-out, over-expression, or genetic complementation. Cross-population validation is currently achieved by integrating association mapping in diverse panels or linkage mapping in RIL population(s) or F2 populations. Taking as an example the recent study on cloning ZmCCT9, which affects maize flowering time (Huang et al., 2018), this locus was simultaneously identified by NAM (Buckler et al., 2009) and maize-teosinte RIL populations under association and linkage mapping. Furthermore, the causal allele, an InDel of a harbinger-like transposon, has been identified in an association panel containing 513 diverse maize inbred lines (Li et al., 2013) and validated in the above two populations used to map the locus. Another model case is the association mapping of rice chlorophyll content in a diverse panel of 529 individuals followed by three customized F2 populations to validate GWAS signals (Wang et al., 2015b). With increasing numbers of populations and corresponding genotypic and phenotypic variations, association mapping in multiple populations is becoming possible and should make the observed hits much more effective and reliable. At this point, cross-population analysis could be extended to cross-species analysis for similar or homologous traits. A better understanding of the conserved, differentiated, and/or dynamic genetic architecture for any trait of interest will be valuable, and such cross-species analysis has already been implemented in rice (Huang et al., 2011) and cereals (Chen et al., 2016; Liu et al., 2017).
Beyond statistical inference, molecular and genetic experimentation is a reliable way to validate GWAS hits. This is still difficult because of the low throughput and case dependence of most wet experiments. Fortunately, the emerging genome-editing technologies promise an effective and high-throughput approach. Recently, two Chinese teams have simultaneously created genome-wide targeted mutant libraries using the CRISPR/Cas9 technique in rice (Lu et al., 2017; Meng et al., 2017). Obtaining the functional gene within each GWAS peak should be quicker than ever expected by combining high-throughput forward- and reverse-genetic techniques.
Glossary Box
- Linkage disequilibrium (LD): is a phenomenon in which co-occurrence of alleles at different loci is non-random in a given population, being either higher of lower than expected if they were independent. The presence of genetic linkage makes it is unnecessary for association studies to examine every polymorphism since linked variants are strongly correlated.
- Genome-wide association study (GWAS): is a statistical approach of mapping quantitative trait loci to link phenotypes of interest to whole-genomic genotypes by taking advantage of historic linkage disequilibrium.
- ’Omics-wide association study (OWAS): extends genome-wide association studies to multiple ’omic variations, including genomics, transcriptomics, proteomics, and metabolomics, with the aim of characterizing a full molecular functional and dynamic picture of variations in phenotype.
- Quantitative trait locus (QTL): is a genomic confidence interval associated with a trait of interest, which varies in degree of effect size and physical length, and includes at least one causal gene or other functional element. QTLs exert main, epistatic, and interaction with the environment effects, while the main effects can be additive or dominant.
- Epistasis: describes an interaction relationship between genes (or loci) in which the effect of one gene can vary (in size or even direction) among different alleles of the other gene, leading to non-linear consequences for phenotypic variation. Epistasis can modify effects in an additive and/or dominant manner at the interacting loci, with epistatic effect models described as additive-by-additive, additive-by-dominance, and dominance-by-dominance. Additive-by-additive interactions are the most studied as only a small population is required.
- Pleiotropy: is a phenomenon in which one gene directly contributes to more than one seemingly unrelated phenotypic trait. The likely underlying mechanism is that the product of a given gene can be either used by various cells or function in cascade-like signaling to various targets. As genes usually function as networks and developmental phenotypes are interactive, it is very hard to distinguish true biological pleiotropy from mediated pleiotropy and spurious pleiotropy. Mediated pleiotropy occurs when phenotype 1 lies on a causal path to phenotype 2; an association between gene(s) with phenotype 1 will thus also occur with phenotype 2. Spurious pleiotropy is reflected by both genotypic and phenotypic aspects: the identified variant is in high linkage disequilibrium with two causal variants in distinct genes that contribute to different phenotypes; or different phenotypes are misclassified into one phenotype such that any causal variant/gene affects one of them and a spurious association occurs for the other.
- Phenotypic plasticity: is used to describe all kinds of phenotypic responses to environmental change without any change of genome sequence. In theory, not all phenotypic plasticity is exactly the same, but it covers the concept of genotype–environment interaction (G × E), which presents a narrower situation that different alleles respond to environmental change to different degrees. However, in practice, G × E is almost equivalent to phenotypic plasticity with only a slight difference in emphasis. Phenotypic plasticity is considered more important for plants than animals due to their immobility and has been revealed to be highly relevant to plant traits including flowering timing, leaf shape variations, allocation of soil nutrients, and size of seeds.
- CRISPR-based genome editing: is a genetic engineering technology targeting specific genome locations with operations including insertion, deletion, modification, or replacement. CRISPR is an abbreviation of Clustered Regularly Interspaced Short Palindromic Repeats, a family of DNA sequences in Bacteria and Archaea that play key roles in the prokaryotic defense system. CRISPR associated proteins (Cas) process these sequences and cut matching viral DNA sequences. CRISPR/Cas form the basis of the emerging highly efficient and specific CRISPR-based genome-editing technology.
Conclusions: Prospects and challenges
The power of crop GWAS to explore the genetic architecture of complex traits has been demonstrated in multiple species, and this number will continue to increase rapidly. However, most studies are of limited scope to the main (additive) effect of genetic architecture. This is why the present review attempts to restate the complexity of the concept of genetic architecture, exploring the architecture beyond additive effects and underscoring the importance of understanding trait variability (Box 2). This complexity is not only the result of differences in gene action, but also determined by ontogenic gene networks or even epigenetic effects, and the interaction with surroundings, including living neighbors and greatly changing environmental conditions. Compared with unprecedented achievements in the study of main effects, the application of GWAS to non-linear effects has been limited, providing only a rudimentary view of the comprehensive picture of genetic architecture.
Sun and Wu (2015) draw a complete picture of genetic architecture and propose a conceptual framework. They consider the current genetic mapping, i.e., linking genomic variants to individual phenotypes, as the first stage, followed by functional mapping, systems mapping, network and ecosystem mapping, and consideration of biological mechanisms, treating each trait as a dynamic vector, integrating full ’omics variations, and incorporating the role of ecological interactions in the formation of complex traits. Such complexity makes future mapping a great challenge for data collection, integration, and statistical modeling.
Box 2. Open questions
- What is the pervasiveness of the non-additive effect, is this effect dominant for certain traits, and what is the underlying mechanism?
- Is it possible to map causal genes (or variants) with ultra-high resolution to understand the generality of pleiotropy and how to apply it in future genetic improvement?
- Which genes and mechanisms contribute to phenotypic plasticity, and will identify these help to predict crop responses to climate change?
- Is synthetic association widespread, and how can it be addressed effectively?
- How far are we away from the precise design of new cultivars by integrating natural and created variations?
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2016YFD0100803, 2016YFD0101003), National Natural Science Foundation of China (31525017), the Hubei Provincial Natural Science Foundation of China (2015CFA008), and the Postdoctoral Talent Innovation Program of China (BX201700092).
Conflict of Interest
The authors declare no competing interests.