Association of Cytokine Gene Polymorphisms in Patients with Tuberculosis and Their Household Contacts
Abstract
Cytokine gene polymorphisms are known to be associated with functional differences in cytokine regulation and may affect host susceptibility to tuberculosis (TB). Contacts are important group in developing tuberculosis infection and are 10–60 times more likely to develop TB than general population. The present study was carried out in patients with TB (N = 176), their household contacts (HHC; N = 155) from Free Chest TB Clinic PPM DOTS (1TU) covering 500,000 population at Mahavir Hospital and Research Centre, Hyderabad, and healthy controls (HC; N = 170) also included. The association of single-nucleotide polymorphisms (SNPs) in the promoter region of TNF-α (−308G/A), IL-2 (−330T/G), IL-4 (−589C/T) and in exon region of TGF-β1 (+869T/C) genes was assessed by ARMS & PCR-RFLP using specific primers in the above-mentioned subjects. The differences in allelic or genotypic frequencies of TNF-α (−308G/A) between patients, their HHC and HC were not statistically significant (P > 0.05). IL-2 (−330T/G) TG genotype was significantly different between patients, HHC compared to HC (P < 0.002, OR-1.997, 95%CI-1.260-3.168, P < 0.03, OR-1.602, 955CI-1.003-2.561).IL-4 (−589C/T) CC genotype was significantly different between patients and HC (P < 0.03, OR-1.791, 95%CI-1.009-3.189) as well as between HHC and HC at P < 0.0001, OR-2.56, 95%CI-1.448-4.545. In addition, the TGF-β 1 (+869T/C) TC genotype was significantly associated with susceptibility to tuberculosis in patients when compared against HC(P < 0.0001, OR-3.416, 95%CI-2.063-5.670) and HHC (P < 0.0001, OR-2.357, 95%CI-1.439-3.868), respectively.MDR analysis indicated that TT genotype of TGF-β1 with TT and CT genotypes of IL-4 showed high risk with GA, TT genotypes of TNF-α, IL-2, respectively. Our results suggest that IL-2 (-330T/G), IL-4 (-589 C/T) and TGF-β1 (+869T/C) gene polymorphisms may be associated with TB susceptibility.
Introduction
Tuberculosis (TB), primarily caused by Mycobacterium tuberculosis (M.tb), continues to be one of the most important infectious causes of death 1. In 2011, there were an estimated 9.2 million new cases of TB, while India alone shows over 3.4 million cases 2. Although, one-third of the World's population is infected with M.tb; only 10% of those infected develop active disease during their lifetime 3.
Household contacts of patients with tuberculosis comprise a high-risk group for tuberculosis, and their examination carries great importance for prevention and control of active disease 4. Studies have reported around 80–100% of the contacts may have TB infection, but on an average 20% of them are found to develop disease 5. The risk of developing disease may be associated with genetic variability at various cytokine gene loci 6. Cytokines influence immune responses by activating a network of pro-inflammatory and downregulatory molecules derived from both T cells and macrophages, which determines the disease outcome 7, 8. Functional single-nucleotide polymorphisms (SNPs) in the promoter and protein coding regions of key genes have been associated with susceptibility and/or resistance to disease 9.
The tumour necrosis factor (TNF)-α gene is located within the major histocompatibility complex class III region at the short arm of chromosome 6, and its production plays a pivotal role in macrophage activation for controlling M. tuberculosis infection 10. Genomic polymorphism resulting in substitution of the nucleotide adenine (A) for guanine (G) at position −308 (−308G > A, relative to the transcription start site) was discovered within a promoter region of the TNF-α locus (rs1800629 G/A) has been the most common target of the association studies 11, 12. The presence of allele A substitution leads to increase in the binding of nuclear factors and enhanced transcription of the gene 13, 14. Thus, −308 polymorphism could potentially affect the cell type and specific regulation of TNF-α synthesis at the transcriptional level 15. Interleukin (IL-2) gene located on the long arm of chromosome 4 plays a pivotal role in generating an immune response and can influence the course of mycobacterial infections, either alone or in combination with other cytokines 16. The −330T/G promoter (rs2069762) polymorphism is implicated in the increased susceptibility to inflammatory diseases and cancers 17, making it an ideal marker for genetic association studies 18.
Polymorphisms in the IL-4 promoter (rs2243250 C/T) could influence the transcription levels of the gene. Specifically, a functional SNP, located 589 bp upstream of the transcriptional site, has been associated with increased promoter strength, stronger binding of transcription factors to the promoter and also with different levels of IL-4 activity 19, 20.
In response to the functional role, this SNP has been investigated for genetic susceptibility in several immune related diseases such as asthma, pulmonary tuberculosis and cancers 21-23. TGF-β 1 (rs9282871T/C) gene located on long arm of chromosome 19; a potent macrophage-deactivating molecule reduces the effectiveness of macrophages to contain Mycobacterium tuberculosis 24. A T/C substitution at position +869 (codon 10) in exon 1 of the gene has been associated with high serum levels of TGF-β1 25, 26. T869C TGF-β1 polymorphisms have been reported to influence the susceptibility to various diseases 27.
Hence, the present study was designed to investigate the association of single-nucleotide polymorphisms in the promoter region of TNF-α (−308G/A), IL-2 (−330T/G), IL-4 (−589C/T) and in exon region of TGF-β1 (+869T/C) genes in patients, their household contacts (HHC) and healthy controls (HC) in relation to tuberculosis susceptibility.
Materials and methods
Subjects
Patients with tuberculosis (N = 176), HHC (N = 155) who attended Free Chest TB Clinic PPM DOTS (1TU) 5,00,000 population at Mahavir Hospital and Research Centre, Hyderabad, and healthy controls (HC) (N = 170) of same region with no history of tuberculosis and other diseases were studied.
Patients were categorized based upon sputum, smear examination for acid-fast bacilli, sputum culture, chest X-ray and histological examinations as per Revised National Tuberculosis Control Program (RNTCP) guidelines. HIV positive, patients with comorbid conditions like hypertension, diabetes mellitus, renal failure and cancer were excluded from the study. HHC are the family members who are exposed to index patient and living in the same household. Informed consents were obtained from all the subjects. Body mass index has been calculated. Tuberculin skin test (TST) positivity was assessed by administering five tuberculin units (TU) subcutaneously on the left arm. An induration of >10 mm in between 48–72 h was considered positive (TST+) and was not performed in healthy controls (HC). The study has been approved by Institutional ethical committee (Institutional Ethics Committee for BioMedical Research at Bhagwan Mahavir Medical Research Centre, India).
DNA extraction
Genomic DNA from peripheral blood (1–2 ml) was extracted using Flexi gene kit according to the manufacturer's protocol (QIAGEN, Hilden, Germany). Quantity of DNA was confirmed by Nano Drop and was stored at −20 °C.
TNF-α (−308G/A) genotyping
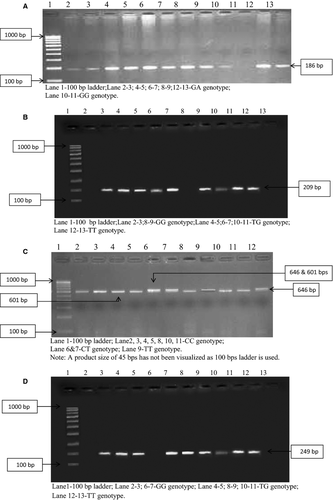
Amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) was carried out for TNF- α (−308G/A) genotyping using common primer 5′-tct cgg ttt ctt ctc cat cg-3′, G allele primer 5′-ata ggt ttt gag ggg cat gg -3′ and A allele primer 5′- ata ggt ttt gag ggg cat ga-3′, with cycling conditions of 95 °C for 8 min followed by 30 cycles at 95 °C for 90 s, 60 °C for 90 s, 72 °C for 1 min and finally for 10 min extension at 72 °C. A product size of 186 bp was detected (Fig. 1A).
IL-2 (−330T/G) genotyping
A 209 bp of IL-2 (−330T/G) was detected by ARMS-PCR using common primer 5′-aag agt cat cag aag agg aa-3′, T allele primer 5′-ttg tac att gtg gca gga gtt-3′ and G allele 5′-ttg tac att gtg gca gga gtg-3′, with cycling conditions of 95 °C for 2 min followed by 30 cycles of denaturation at 95 °C for 30 s, 59 °C for 45 s, 72 °C for 30 s and final extension at 72 °C for 5 min (Fig. 1B).
IL-4 (−589C/T) genotyping
Restriction fragment length polymorphism (PCR-RFLP) was performed for IL-4 (−589C/T) genotyping using forward primer 5′-agg tgt cga ttt gca gtg ac-3′ and reverse primer 5′-act agg cct cac ctg ata cg-3′, with cycling conditions of 94 °C for 2 min followed by 35 cycles at 94 °C for 30 s, 62 °C for 30 s, 72 °C for 30 s and finally for 7 min extension at 72 °C. The resulting amplicon after overnight digestion with BsmF1 enzyme (purchased from NEB enzymes, supplied by Bionova, Hyderabad) at 65 °C produced two fragments of 601 and 45 bp. In heterozygote (C/T) three fragments (646,601 and 45 bp), in homozygote (C/C) two fragments (601 and 45 bp) and in homozygote (T/T) undigested product of 646 bp were observed (Fig. 1C).
TGF-β1 (+869T/C) genotyping
For TGF-β1 + 869 T/C genotyping, ARMS-PCR was conducted using antisense primer 5′-tcc gtg gga tac tga gac ac- 3′, T sense primer 5′-agc agc ggt agc agc agc a-3′ and C sense primer 5′-gca gcg gta gca gca gcg-3′ with cycling conditions of 95 °C for 1 min followed by 30 cycles at 95 °C for 20 s, 62 °C for 1 min, 72 °C for 50 s and final extension at 72 °C for 5 min. A product size of 241 bp was detected (Fig. 1D).
All the primers were designed by using primer BLAST (NCBI) and were commercially obtained from Eurofins (MWG operon). PCR mix (buffer, dntps, Taq polymerase) was purchased from Genei Bangalore. Amplification was carried out using Bio-Rad Thermal cycler.
Electrophoresis of PCR products and digested fragments was carried out on 2% agarose gel stained with ethidium bromide, and presence or absence of fragments was visualized by UV transilluminator.
Statistical analysis
Statistical analysis was performed by spss version 20.0 (IBM, White Plains, NY, USA). Differences in the distribution of clinical characteristics (selected variables) between the patients, HHC and HC were evaluated by using the Student's t-test (for continuous variables) or χ2-test (for categorical variables). Hardy–Weinberg equilibrium (HWE) was tested using a goodness-of-fit χ2-test. Allele and genotype frequencies were compared by Pearson chi-square test between the groups; odds ratio (OR) with respective confidence intervals (95% CI) was also calculated. P < 0.05 was considered significant. Multifactor dimensionality reduction (MDR) analysis was carried out to detect gene–gene interactions by using mdr software 2.0beta8.4 (Prof. Jason H. Moore, Dartmouth Medical School, Lebanon, NH, USA).
Results
The clinical characteristics of patients with TB, their HHC and HC are summarized in Table 1. Age-matched patients and HC were studied. Mean age of the patients, HHC and HC was 28.37 ± 11.61 (94 men and 82 women), 32.9 ± 10.44 (68 men and 87 women) and 30.28 ± 9.29 (110 men and 60 women), respectively. HHC age group was found to be significant from that of patients (P < 0.0001) and HC (P < 0.016). Gender difference was also observed among all the subjects (P < 0.001).
| Variable | Pts (N = 176) | HHC (N = 155) | HC(N = 170) | P-value |
|---|---|---|---|---|
| Age(y) | 28.37 ± 11.61 | 32.9 ± 10.44 | 30.28 ± 9.29 |
Pts versus HHC (<0.0001)* HHC versus HC (<0.016)* |
| Gender M/F | 94 (53.5%)/82 (46.5%) | 68 (43.8%)/87 (56.2%) | 110 (64.7%)/60 (35.3%) | 0.001+ |
| TST P/N | 127 (72.1%)/49 (27.9%) | 110 (70.9%)/45 (29.1%) | – | 0.81+ |
| BMI (kg/m2) | 16.7 ± 2.95 | 22.98 ± 4.54 | 24.7 ± 4.37 |
Pts versus HHC and HC (<0.0001)* |
- Pts, patients; HHC, household contacts, HC, healthy controls.
- a Mean ± SD independent samples t-test; + χ2 test; M/F-male/female; P/N-positive/negative; BMI, body mass index.
Of 176 patients, 127 were tuberculin skin test (TST) positive and 49 were negative. Among 155 HHC, 110 were TST positive and 45 were negative. Mean BMI of patients, HHC and HC was 16.7 ± 2.95, 22.98 ± 4.54 and 24.7 ± 4.37, respectively. Significant difference was observed between patients compared to HHC and HC at P < 0.0001.
Gene polymorphisms in promoter region of TNF-α (−308G/A), IL-2 (−330T/G), IL-4 (−589C/T) and in exon region of TGF-β1 (+869T/C) were studied. The comparison of the allele and genotype frequencies for TNF-α (−308G/A) showed no significant differences among the patients, HHC and HC (data not shown) where as for IL-2 (−330T/G) TG (P < 0.002, P < 0.037) and GG (P < 0.01, P < 0.02) genotypes were significant in patients and HHC compared to HC (Table 2). IL-4 (−589C/T) CC genotype was associated in patients (P < 0.03) where as in HHC, TT (P < 0.01) and CC (P < 0.0001) genotypes and both the alleles (P < 0.0001) were significant when compared to HC (Table 3). TGF-β 1 (+869T/C) TT, TC genotypes and alleles were significantly associated in patients and HHC compared to HC at P < 0.0001 (Table 4). The genotype distributions of IL-2 (−330T/G), IL-4 (−589C/T) and TGF-β1 (+869T/C) were in Hardy–Weinberg equilibrium (HWE) (χ2-1.05, 3.49 and 3.3 at P > 0.05) for HC but not for patients and HHC. However, TNF-α (−308G/A) deviates from HWE.
| IL-2 (−330 T/G) | |||||
|---|---|---|---|---|---|
| HC | Patients | OR P-value CI | HHC | OR P-value CI | |
| Genotypes | N = 170 (%) | N = 176 (%) | N = 155 (%) | ||
| TT | 58 (34.11) | 45 (25.5) |
0.663 <0.082 0.406–1.083 |
47 (30.3) |
0.84 <0.465 0.513–1.376 |
| TG | 88 (51.7) | 120 (68.2) |
1.997 <0.002** 1.260–3.168 |
98 (63.25) |
1.602 <0.037* 1.003–2.561 |
| GG | 24 (14.11) | 11 (6.3) |
0.406 <0.01* 0.179–0.902 |
10 (6.45) |
0.42 <0.02* 0.180–0.959 |
| Alleles | N = 340 (%) | N = 352 (%) | N = 310 (%) | ||
| T | 204 (60) | 210 (59.6) |
0.986 <0.927 0.719–1.352 |
192 (61.9) |
1.085 <0.613 0.781–1.506 |
| G | 136 (40) | 142 (40.4) |
1.014 <0.927 0.740–1.391 |
118 (38.1) |
0.922 <0.613 0.664–1.280 |
- *Significant, **highly significant, bold value indicates risk (P < 0.05), 95% CI, confidence interval; OR, odds ratio.
| IL-4 (−589C/T) | |||||
|---|---|---|---|---|---|
| HC | Patients | OR P-value CI | HHC | OR P-value CI | |
| Genotypes | N = 170 (%) | N = 176 (%) | N = 155 (%) | ||
| CC | 26 (15.2) | 43 (24.4) |
1.791 <0.03* 1.009–3.189 |
49 (31.6) |
2.56 <0.0001 ** 1.448–4.545 |
| CT | 66 (38.8) | 71 (40.4) |
1.066 <0.773 0.677–1.678 |
56 (36.2) |
0.891 <0.616 0.554–1.433 |
| TT | 78 (45.8) | 62 (35.2) |
0.641 <0.04* 0.407–1.011 |
50 (32.2) |
0.562 <0.01* 0.348–0.905 |
| Alleles | N = 340 (%) | N = 352 (%) | N = 310 (%) | ||
| C | 118 (34.7) | 142 (40.4) |
1.272 <0.126 0.923–1.753 |
154 (49.7) |
1.857 <0.0001** 1.338–2.578 |
| T | 222 (65.2) | 210 (59.6) |
0.786 <0.126 0.570–1.083 |
156 (50.3) |
0.538 <0.0001** 0.388–0.747 |
- *Significant, **highly significant, bold value indicates risk (P < 0.05), 95% CI, confidence interval; OR, odds ratio.
| TGF-β 1 (+869 T/C) | |||||
|---|---|---|---|---|---|
| HC | Patients | OR P value CI | HHC | OR P value CI | |
| Genotypes | N = 170 (%) | N = 176 (%) | N = 155 (%) | ||
| TT | 58 (34.11) | 12 (6.81) |
0.141 <0.0001* 0.068–0.286 |
10 (6.45) |
0.133 <0.0001* 0.061–0.284 |
| TC | 92 (54.11) | 141 (80.11) |
3.416 <0.0001* 2.063–5.670 |
119 (76.78) |
2.357 <0.0001* 1.439–3.868 |
| CC | 20 (11.76) | 23 (13.08) |
1.127 <0.713 0.568–2.242 |
26 (16.77) |
1.512 <0.196 0.772–2.969 |
| Alleles | N = 340 (%) | N = 352 (%) | N = 310 (%) | ||
| T | 208 (61.17) | 165 (46.87) |
0.560 <0.0001* 0.409–0.766 |
139 (44.83) |
0.516 <0.0001* 0.373–0.714 |
| C | 132 (38.8) | 187 (53.13) |
1.786 <0.0001* 1.305–2.445 |
171 (55.17) |
1.931 <0.0001* 1.401–2.683 |
- a Highly significant, bold value indicates risk (P < 0.05), 95% CI, confidence interval; OR, odds ratio.
Genotype combinations for four cytokine genes were assessed by MDR analysis where TT genotype of TGF-β 1 shows high risk in combination with TT and CT genotypes of IL-4 and also with GA, TG genotypes of TNF-α, IL-2, respectively. TC genotype of TGF-β1 shows high risk with CC genotype of IL-4 and also with GA, TT genotypes of TNF-α, IL-2. Similarly, CC genotype of TGF-β1 shows high risk with TT genotype of IL-4 and also with GA, TG genotypes of TNF-α, IL-2 in patients compared to HC (Fig. 2). These SNPs had a training accuracy of 0.7022 and testing accuracy of 0.6226.

In HHC and HC, TT genotype of TGF-β 1 shows high risk with CT genotype of IL-4 with GA, TG genotypes of TNF-α, IL-2, respectively (Fig. 1). These SNPs had a training accuracy of 0.7011 and testing accuracy of 0.6053. The cross-validation consistency for all the groups was 10/10. The statistical significance for the combinations shown P < 0.0001 between patients and HHC compared against HC.
Discussion
TNF-α is an important mediator in the inflammatory response against TB 28. In the present study, no significant association of TNF-308G/A polymorphisms was observed, similar to the other studies in Asians, that is, South Indian population 29, North Indian population 30, Turkish 31 and Thai population 32. However, Bikmaeva et al. 33 have shown an association of ‘A’ allele with risk of pulmonary TB in Russian population. Scola et al. 34 demonstrated a reduction in low-producer GG homozygous individuals in TB affected group among Caucasians.
Polymorphisms in cytokine genes might regulate cytokine production as shown in human IL-2 for -330 promoter polymorphism 35, 36. We observed significant association of TG, GG genotypes in patients and HHC compared to HC, where as no significant association was shown in Macedonian population by Dejan Trajkov et al. 37. Selvaraj et al. 18 in south Indian population observed significantly decreased TT genotype frequency in patients compared to controls, which was not similar with the present study. Naslednikova et al. 38 study revealed a positive correlation of the G allele, TG genotype and GG genotype with pulmonary tuberculosis in Caucasian population, which seems to be similar with the present study.
IL-4 is a pleiotropic cytokine produced by Th2-cells, eosinophils and mast cells, which mediates the humoral immune response and exerts anti-inflammatory effects by inhibiting the production of pro-inflammatory cytokines and by changing the expression profile of macrophages 39. Our study reports that TT genotype frequency was higher in healthy controls (45.8%) and CT genotype frequency in patients and HHC (40.4% & 36.2%), even though the homozygous' CC' genotype shows significantly lower frequency (24.4%) in patients, but based on OR suggesting that CC genotype may be associated with susceptibility for TB, which was consistent with Dejan Trajkov et al. 37. Other studies Naslednikova et al. 38 in Russian population observed significance for CC, CT genotypes in patients and controls where as we found TT & CC genotypes significant between patients compared to HC and CC genotype compared to HHC. Hence, IL-4-589C/T gene polymorphisms may be associated with Tuberculosis, where as studies by Amirzargar et al. 40 in Iranian population found negative association with IL-4-590T/T in patients with TB.
Transforming growth factor-beta (TGF-β) is a multifunctional cytokine, which plays a central immunomodulatory role and TGF-β1 together with other cytokines such as TNF-α may be responsible for the tissue destruction in tuberculosis 41. We observed TT and TC genotypes significant in patients and HHC compared to HC where as no significance was shown in Chinese study by Judith et al. 42. Similarly, a significant negative association with TT genotype was reported in Iranian population by Amirzargar et al. 40. However, Henao et al. 43 did not confirm any association with TGFβ 1 in tuberculosis.
These divergent findings may be due to ethnical differences in various populations. Therefore, we conclude that TG of IL-2, CC of IL-4 and TC of TGF-β1 genotypes and also their combination with TNF- α shown high risk towards the disease for identifying susceptible individuals (HHC). This can be confirmed by further studies in large number of samples.
Acknowledgment
We thank Staff of the Free Chest Clinic Mahavir PPM DOTS, tuberculosis unit (1T.U), Bhagwan Mahavir Trust. Financial support was provided by DBT-RGYI (Sanction no: 102/IFD/PR/2029/2007-2008 dated 18/01/2008) and COE (Sanction No: BT/01/COE/07/02, dated 30/12/08).
Conflict of interest
Authors have no conflict of interest.




