Activation of PIF4/5 by the Transcription Factor TCP4 Promotes Hypocotyl Elongation in Arabidopsis
Funding: This work was supported by National Natural Science Foundation of China (NSFC) (No. 32370405) and Natural Science Foundation of Yunnan Province (2017FB050).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors is: Xudong Sun ([email protected]).
ABSTRACT
Transcription factor TEOSINTE BRANCHED1, CYCLODEA, PROLIFERATING CELL FACTORS 4 (TCP4) plays essential roles in plant development processes. The role of TCP4 in the regulation of hypocotyl growth remains a matter of debate. Our results cast doubt on the previous claims that TCP4 directly activates YUC5 expression to promote hypocotyl elongation, and clearly show that TCP4 promotes hypocotyl elongation dependent on PIF4/5.
Transcription factors TEOSINTE BRANCHED1, CYCLODEA, PROLIFERATING CELL FACTORS 4 (TCP4) play essential roles in plant development processes, such as leaf morphogenesis (Bresso et al. 2017; Li et al. 2012; Palatnik et al. 2003; Schommer et al. 2008), jasmonic acid biosynthesis (Schommer et al. 2008), cell proliferation (Schommer et al. 2014), flower development and flowering time (Kubota et al. 2017; Nag et al. 2009), and secondary cell wall synthesis (Sun et al. 2017). Overexpression of miR319-resistant TCP4 (rTCP4) results in long hypocotyls, hyponastic cotyledons, and smaller rosette leaves. Jaw-D, a miR319a overexpressing line, has opposite phenotypes (Schommer et al. 2008; Sun et al. 2017). We recently identified that a UVR8-TCP4-LOX2 module regulates UV-B tolerance dependent on the JA signaling pathway in Arabidopsis thaliana, and TCP4 regulates UV-B tolerance independently of ELONGATED HYPOCOTYL 5 (HY5) signaling (Li et al. 2024). HY5 is the central regulator of light signal transduction for hypocotyl growth and anthocyanin homeostasis (Contreras-Avilés et al. 2024). These results also indicated that TCP4 regulates hypocotyl elongation independent of HY5. Challa et al. (2016) reported that TCP4 directly activates YUCCA5 (YUC5) expression to promote Arabidopsis hypocotyl elongation. Although overexpression of YUC5 causes elongated hypocotyls and narrow leaves, the cotyledons are not hyponastic (Woodward et al. 2005). Similarly, hypocotyl lengths did not show obvious differences in the yuc5 mutant compared to WT (Müller-Moulé et al. 2016). The role of TCP4, therefore, in the regulation of hypocotyl growth remains a matter of debate.
To test the genetic relationship between TCP4 and YUC5, we crossed rTCP4 with yucQ (a yuc3/5/7/8/9 quintuple mutant). Our results showed that whereas the yucQ mutant had a WT-like hypocotyl length, rTCP4-3 × yucQ had a similar phenotype to rTCP4-3 (Figure 1A,B). A previous study showed that YUC3, YUC5, YUC7, YUC8, and YUC9 genes are expressed in seedling roots and that the loss-of-function yucQ mutant generates a short and agravitropic primary root (Chen et al. 2014). Although overexpression of YUC5 resulted in elongated hypocotyls, the loss-of-function of the yuc5 mutant did not reduce hypocotyl length (Müller-Moulé et al. 2016). In the present study, we found no differences in the relative expression of YUC5 between rTCP4-3 and WT hypocotyls (Figure 1C). Together, these results strongly suggested that YUC5 is not the target gene of TCP4 in hypocotyl elongation processes.
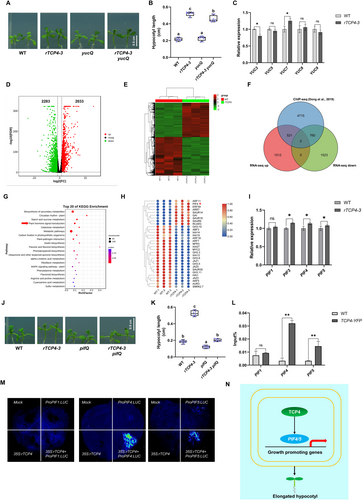
We undertook comparative transcriptome analysis to elucidate the candidate target genes for TCP4's regulation of hypocotyl elongation. A total of 4316 differentially expressed genes (DEGs) were identified (FDR < 0.05 and log2|FoldChange| > 0.8), 2033 genes of which were significantly up-regulated and 2283 genes which were significantly down-regulated (Figure 1D,E and Table S1). Correlation heatmap and Principal Component Analysis (PCA) indicated that rTCP4-3 groups and WT groups had distinct gene expression profiles, and our analysis was repeatable and stable (Figure S1). By combining RNA-seq with ChIP-seq data of TCP4 (Dong et al. 2019), an overlap was found for 521 up-regulated and 760 down-regulated DEGs between RNA-seq and ChIP-seq experiments (Figure 1F, Table S2). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed enrichment of pathways associated with circadian rhythm, starch and sucrose metabolism, and “Plant hormone signal transduction” (Figure 1G). An expression heatmap identified the key components of “Plant hormone signal transduction” and, coupled with qRT-PCR, revealed that PIF4/5 relative expression levels were strongly up-regulated in the hypocotyls of rTCP4 lines (Figure 1H,I).
To test the genetic relationship between TCP4 and PIF4/5, we obtained rTCP4 x pifQ (a pif1/3/4/5 quadruple mutant). The resultant hypocotyl phenotype of rTCP4-3 x pifQ was similar to pifQ, with a length significantly shorter than WT (Figure 1J,K), further confirming that the PIFs may be involved in TCP4-mediated hypocotyl elongation. The ChIP assay revealed that TCP4 could directly bind to the promoter of PIF4/5 (Figure 1L). A previous study also revealed that TCP4 could bind to PIF4/5 in the genome (Dong et al. 2019). To support this, we undertook a luciferase assay, which revealed that TCP4 could positively regulate the promoter activity of PIF4/5 (Figure 1M). Combining the above results, we hypothesize that PIF4/5 is the target genes for TCP4-mediated hypocotyl elongation in Arabidopsis.
In conclusion, we propose a working model for a TCP4-PIF4/5 regulatory module that governs hypocotyl elongation (Figure 1N). Our results cast doubt on the previous claims that TCP4 directly activates YUC5 expression to promote hypocotyl elongation and clearly show that TCP4 promotes hypocotyl elongation dependent on PIF4/5.
Author Contributions
Y.Z., C.C.C.C., and X.S. wrote the article; Y.Z. and Q.L. collected and processed the data; Y.D., Y.Y., and X.S. designed the research.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (NSFC) (No. 32370405), Natural Science Foundation of Yunnan Province (2017FB050).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
All relevant data are available from the corresponding authors upon request. There are no restrictions on data availability. RNA-seq related data were submitted at https://ngdc.cncb.ac.cn/gsa/, with the GSA number CRA018400.




