Drought responsiveness in black pepper (Piper nigrum L.): Genes associated and development of a web-genomic resource
Funding information: Indian Council of Agricultural Research, Grant/Award Number: F. no. Agril. Edn.4–1/2013-A&P; Indian Council of Agricultural Research, Ministry of Agriculture and Farmers' Welfare, Government of India; CABin Grant
Abstract
Black pepper (Piper nigrum L.; 2n = 52; Piperaceae), the king of spices, is a perennial, trailing woody flowering vine and has global importance with widespread dietary, medicinal, and preservative uses. It is an economically important germplasm cultivated for its fruit and the major cash crop in >30 tropical countries. Crop production is mainly affected by drought stress. The present study deals with the candidate gene identification from drought-affected black pepper leaf transcriptome generated by Illumina Hiseq2000. It also aims to mine putative molecular markers (namely SSRs, SNPs, and InDels) and generate primers for them. The identification of transcription factors and pathways involved in drought tolerance is also reported here. De novo transcriptome assembly was performed with trinity assembler. In total, 4914 differential expressed genes, 2110 transcriptional factors, 786 domains and 1137 families, 20,124 putative SSR markers, and 259,236 variants were identified. At2g30105 (unidentified gene containing leucine-rich repeats and ubiquitin-like domain), serine threonine protein kinase, Mitogen-activated protein kinase, Nucleotide Binding Site-Leucine Rich Repeat, Myeloblastosis-related proteins, basic helix–loop–helix are all found upregulated and are reported to be associated with plant tolerance against drought condition. All these information are catalogued in the Black Pepper Drought Transcriptome Database (BPDRTDb), freely accessible for academic use at http://webtom.cabgrid.res.in/bpdrtdb/. This database is a good foundation for the genetic improvement of pepper plants, breeding programmes, and mapping population of this crop. Putative markers can also be a reliable genomic resource to develop drought-tolerant variety for better black pepper productivity.
Abbreviations
-
- bHLH
-
- basic helix–loop–helix
-
- BWA
-
- Burrows-Wheeler Aligner
-
- CFA
-
- cyclization of fatty acid acyl
-
- CPC
-
- Coding Potential Calculator
-
- DEG
-
- differentially expressed gene
-
- FC
-
- fold change
-
- FDR
-
- fase discovery rate
-
- GO
-
- Gene Ontology
-
- INDELs
-
- INnsertion and DELetion
-
- lncRNAs
-
- long non-coding RNAs
-
- PDRTDb
-
- Black Pepper Drought Transcriptome Database
-
- SNP
-
- Single Nucleotide Polymorphism
-
- SRA
-
- Sequence Retrieval Archive
-
- SSR
-
- simple sequence repeat
-
- SSR-FDM
-
- SSR-functional domain markers
-
- WGD
-
- whole-genome duplication
1 INTRODUCTION
Black pepper (Piper nigrum L.) is a trailing woody flowering vine belonging to the family Piperaceae, also commonly known as white pepper, green pepper, peppercorn, and Madagascar pepper. It is predominantly a self-pollinated plant and is cultivated commercially by the orthotropic stem-cutting method. P. nigrum L. is a diploid species with 2n = 52 chromosomes (Sasikumar, 1993). Due to its broader applications in dietary, medicine, preservatives, and global trade, it is known as the “King of spices” (Quijano-Abril et al., 2006). It has great nutritional and agricultural significance and is used as a preservative, perfume, and insecticide due to its antioxidant, anti-inflammatory, and anticancerous properties. It is also used for upset stomach, bronchitis, malaria, and cholera (Gulcin, 2005; Hu et al., 2015; Raja & Sethuraman, 2008; Vijayan & Thampuran, 2000).
Black pepper originates from tropical evergreen forests of the Indian Western Ghats. It is one of the most highly traded spice germplasm in the world. Globally the crop is grown in an area of 586,078 ha, with India covering 130,870 ha area. It is cultivated as a major cash crop in more than 30 tropical countries, like Vietnam, India, Indonesia, China, Malaysia, and Brazil (Ahmad et al., 2010; Tian et al., 2006). Vietnam is the world's leader country: the largest producer (35% of the world's production) and exporter of pepper (FAO 2017). India had an estimated production of 67,427 tonnes out of the total global production of 732,524 tonnes in the year 2018 (FAO, 2017). The global demand for spices, including pepper, is around 3.19% per year, which is more than the population growth rate, and thus there will be greater than fivefold demand by 2050. However, the use of black pepper as an immune booster in a post-COVID-19 world is going to increase the demand more than what was expected (Rastogi et al., 2020; http://spices.res.in/sites/default/files/Vision-IISR-2050.pdf). Unfortunately, pepper plants are sensitive to water deficit due to their large leaf area with high stomatal conductance (Campos et al., 2014; Gonzalez-Dugo et al., 2010). The recent climatic changes have inflated the length and occurrence of abiotic stresses affecting crop development (Haider et al., 2018). There is significant economic loss reported, as high as 70% (Pascale et al., 2003), due to reduced yield and fruit size under water stress. The most critical moisture stress-prone stage of black pepper is the stage prior to blossoming (Penella & Calatayud, 2018). Lack of moisture results in spike shedding causing yield loss. The productivity of black pepper is affected by drought or water deficit stress (George et al., 2017) leading to major economic losses.
The recent advances in NGS-based transcriptomic studies pave the way for the identification of novel genes involved in stress adaptation and post-transcriptional regulation by miRNAs, which can be useful for the genetic improvement of abiotic stress resistance in crops (Abdelrahman et al., 2018). In previous studies, de novo transcriptomic study has been done in leaf and fruit tissues of black pepper (P. nigrum L.) to identify polymorphic simple sequence repeat (SSR) and primers (Hu et al., 2015; Joy et al., 2013), but no attempt has been made to study the transcriptome profiling upon drought stress in this crop. This study aims to identify drought-responsive genes from the leaf transcriptome of black pepper. The identification of transcriptional factors, domains and families, mining of putative SSR markers, variants, long non-coding RNAs has been carried out for the development of web-genomic resources.
2 MATERIALS AND METHODS
2.1 Collection and maintenance of leaf tissue of black pepper genotype
Black pepper genotype (P. nigrum accession 4226) was cultivated at ICAR-Indian Institute of Spices Research, Calicut, India (Latitude: 11° 15′ 0.00″ N; Longitude: 75° 46’ 12.00″ E). This genotype was previously identified as drought tolerant (Krishnamurthy et al., 2016). The plants were maintained as rooted cuttings under greenhouse conditions with temperature above 30°C, relative humidity >75 and day length of 12 h. The plants were grown in pot (one plant per pot) using soil, cow dung, and sand in the ratio of 1:1:1 and watered once a day for 100 days. Abiotic water stress was induced by withdrawal of irrigation on the 15th day, before the 4–5 nodes stage, mimicking drought conditions (Vijayakumari & Puthur, 2014). The control plants were watered regularly.
2.2 RNA isolation and sequencing
The RNA was extracted by pooling leaf tissues from uniform-sized leaves randomly collected from 10 different plants just before wilting, both for control and drought-affected samples. To minimize the across-sample variability, pooling approach of 10 biological replicates was employed (Zou et al., 2016). Total RNA was isolated from the Liquid N2 frozen leaves of drought-induced and control plants using Spectrum Plant Total RNA Kit (Sigma-Aldrich) and On–column DNAse I digestion set (Merck) was used for elimination of DNA from the total RNA preparations, following manufacturer's protocol. The RNA quality was checked by both gel electrophoresis (1% agarose gel) and Bioanalyzer (Agilent 2100, Average RIN 6.4). Paired-end sequencing from the drought-induced and control RNA samples was done using Illumina HiSeq 2000 platform and utilising TruSeq RNA Sample Prep Kit v2 for the library preparation to get a read length of 101 bp. The paired reads fastq files from all the samples used in transcriptome analysis are available in the Sequence Retrieval Archive (SRA) at National Center for Biotechnology Information (NCBI) with BioProject: PRJNA515366 and BioSamples: (SAMN10754251, SAMN10754252).
2.3 Pre-processing and de novo assembly
The obtained 17.5 GB was pre-processed before assembly. The visualization for accessing the raw reads was done using FASTQC. Trimmomatic (v0.36) was used for the removal of low-quality reads, adaptors, and overrepresented sequences (Bolger et al., 2014). Reads with phred-score ≥ 30 and read length ≥ 36 bp were taken for downstream analysis. Trinity (Haas et al., 2013) and SOAPdenovo-Trans (Xie et al., 2014) assembler was used for de novo transcriptome assembly of control and drought samples of P. nigrum. Based on the N50 value, trinity assembly was used for further analysis. CAP3 assembler was employed to remove redundant reads from the trinity assembly (Huang & Madan, 1999).
2.4 Abundance estimation and identification of differentially expressed genes
The contigs generated by de novo assembly of P. nigrum transcriptome were considered as reference on which the trimmed reads of control and drought-affected samples were aligned using Bowtie tool (Langmead et al., 2009). The calculation of expression values in the form of fragments per kilo base of exon per million mapped reads (FPKM) was performed using RNA-Seq by Expectation–Maximization (RSEM) tool (Li & Dewey, 2011). For the identification of differential expressed genes, EdgeR (Empirical analysis of Digital Gene Expression in R; Robinson et al., 2010) of the R Bioconductor and NOIseq (Tarazona et al., 2011) tools were used to reduce the noise and enhanced the computational accuracy. EdgeR package was employed for the identification of the differentially expressed genes (DEGs) in drought-stressed samples while comparing it with control samples using stringent parameters of FDR 0.01 and log2 fold change (FC) value as five. A total of 4914 differentially expressed genes were found, out of which 2862 and 2052 were upregulated and downregulated, respectively. The results of edgeR tool were further validated by NOIseq tool, which is comparatively better as it can handle non-replicate data with its data-adaptive and non-parametric approaches. For the identification of significant DEGs, the threshold P-value and variance were set to 0.05 (Jaiswal et al., 2018). To check the reliability of results, a comparative analysis of the EdgeR results with q = 0.9 and q = 0.95 was performed.
Our de novo assembly of black pepper leaf transcriptome was mapped on the reference genome of Hu et al., 2019 (available at NCBI: BioProject accession PRJNA529758, PRJNA529760 and genome assembly at http://cotton.hzau.edu.cn/EN/download.php). This was compared at three different thresholds of DEGs (75, 90, 95 of % similarity) to have an idea of accuracy and uniformity of our transcriptome assembly.
2.5 Homology search, annotation, and functional characterization
The homology search of DEGs from P. nigrum L. transcriptome assembly was performed with NCBI non-redundant database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) as reference using Blastx algorithm. The standalone local ncbi-blast-2.2.31+ with threshold E-value 1e-3 (Altschul et al., 1990) was considered for the same. For further analyzes, Blast2GO Pro tool (https://www.blast2go.com/; Conesa et al., 2005) was used for mapping and annotation. The functional characterization of the identified candidate genes along with the involved pathways and the interproscan of DEGs were identified. The functional classification was done using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases for a broader overview of the crop species and the pathways involved. These genes were functionally categorized into three sub-categories: biological processes, cellular components, and molecular functions. Further, Blastx tool was performed against PlantTFDB 4.0 (http://planttfdb.cbi.pku.edu.cn/download.php; Jin et al., 2016) for finding out transcriptional factors in the differentially expressed genes. The KEGG pathways were also identified using Blast2GO tool.
2.6 Putative molecular markers identification
Mining putative SSR markers was done from de novo transcriptome assembly of P. nigrum L. using perl script of MISA (Micro SAtellite identification tool; http://pgrc.ipk-gatersleben.de/misa/; Thiel et al., 2003). For identification of SSRs, default parameters were considered, i.e., 10 repeating units in case of mononucleotides, six repeating units in case of dinucleotides, and five repeating units in case of trinucleotides, tetranucleotides, pentanucleotides, and hexanucleotides. The maximum difference between two SSRs markers was kept as 100 bp. Primers were generated for the desired marker using PRIMER3 tool (http://sourceforge.net/projects/primer3/; Untergasser et al., 2012).
For variant detection, alignment and mapping were done, where the reads were mapped over P. nigrum L de novo transcriptome assembly. This was performed using Burrows-Wheeler Aligner (BWA) tool (http://bio-bwa.sourceforge.net/; Li & Durbin, 2009). For findins SNPs (Single Nucleotide Polymorphism) and INDELs (INnsertion and DELetion), the SAM tools package was used (http://samtools.sourceforge.net/; Li, 2011). Filtering of SNPs was done using SNP-EFF tool. The critera appliers were: read depth ≥ 4, quality score ≥ 20, and flanking regions of 50 on both the sides.
2.7 Identification of long non-coding RNAs and their annotation
Long non-coding RNAs (lncRNAs), usually >200 nucleotides, are involved in various developmental process and stress response. For the identification of lncRNAs in the present study, assembled contigs were filtered based on transcript length (> 200 nt). The transcripts with open reading frame (ORF) length longer than 100 amino acids were discarded using ORF Predictor (Min et al., 2005) (https://bioinformatics.ysu.edu/tools/OrfPredictor.html). The remaining transcripts were filtered based on the coding potential using CPC (Kong et al., 2007; http://cpc.gao-lab.org/) and PLEK (Li et al., 2014). Transcripts with coding-potential score ≥ 0.5 were discarded; PLEK is based on k-mer and SVM-based approach identifying lncRNAs from mRNAs sequences. The remaining transcripts were searched with the help of BLASTx against the NCBI-nr (E-value 1e-03), Swissprot database (with e-value: 1e-4, coverage: 35%, alignment length: 40 aa, % sequence identity: 35%), and HMMER search against the Pfam database (Finn et al., 2016) with default parameters. This was done to filter the transcripts that matched to protein coding regions or to protein family domains. To remove the possibility of getting housekeeping genes (including tRNA, rRNA, snRNAs, and snoRNAs etc.), the transcripts were also cross-checked against the Rfam database (http://rfam.xfam.org/: using INFERNAL) and rRNA database (https://www.arb-silva.de/; Kang & Liu, 2015, Kumar et al., 2019).
2.8 Development of transcriptome-based web resource of black pepper
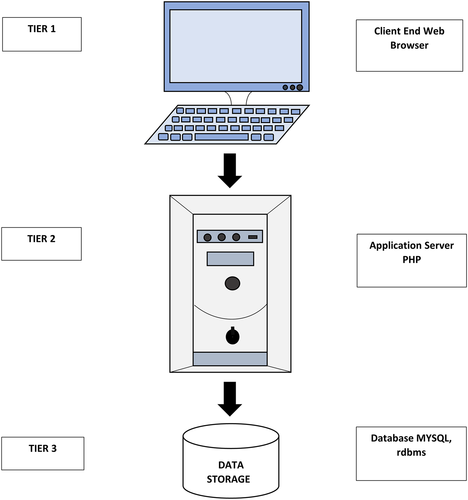
The Black Pepper Drought Transcriptome Database (BPDRTDb) is based on “three-tier architecture,” also called as client–server architecture, where functional and logical processes, data access, user interface, and computer data storage were developed as independent modules and maintained on separate platforms. The client–server architecture comprises of client tier or presentation tier, middle tier or logical and database tier or physical tier (Figure 1). The top most level (i.e., client tier) is the user interface for which web pages were developed using HTML (Hypertext markup language) and Javascript for defining the queries and browsing. It translates tasks resulting to something that user can recognize. In the middle tier or logical tier, PHP (Hypertext Pre-processor) language has been used for writing codes for the server to process, define queries, fetch data, and create and maintain database connectivity. The last tier (i.e., database tier) was developed using MySQL, the information is stored and retrieved from the database for storing various data in the form of tables such as contigs ID, sequence, length, assembly, blast result file, DEGs, expression values, markers like SSRs, SNPs, INDELs, transcription factors, pathways. All the tables are interlinked among themselves and to contigs ID. It also provides provision for users to blast transcript IDs against NCBI non-redundant database.
3 RESULTS
3.1 Pre-processing and de novo assembly
The reads' quality assessment was done using FASTQC tool based on several parameters, namely basic statistics, per sequence quality scores, per base sequence content, adapter content, per sequence GC content, sequence length distribution, per base N content, sequence duplication levels, overrepresented sequences, per tile sequence quality, and k-mer content. The higher the score, the better is the base call. The median score was found to be approximately 41, which indicates good quality of reads. Further, quality trimming was performed using Trimmomatic, where reads were truncated on the basis of their average quality.
A total of 16,396,607 and 19,883,219 paired-end reads in control and drought-stressed samples, respectively, were generated with reads length of 101 bp. After removal of 22,587 and 28,596 low-quality reads from the control and drought stress drought-stressed samples, respectively, the remaining reads were further used for downstream analysis. These cleaned and high-quality reads were further used for de novo transcriptome assembly of P. nigrum L. We got a total of 149,678 transcripts generated by Trinity with 42.75% GC content. The total Trinity genes were 90,064 with N50 value of 1409. The average contig length was 843.72 bp (Table 1). Further, the redundant sequences were removed using Cap3 assembly. A total of 114,598 transcripts were generated showing 42.49% GC content and N50 value as 1481 bp. The range of transcripts varied from 201 bp to 12,166 bp. The minimum length of transcripts was 201 bp, while the largest transcript of the length was 12,166 bp. The maximum numbers of transcripts (i.e. 31,719 transcripts) ranged between 200 to 299, followed by 16,632 and 9317 transcripts for the lengths ranging between 300 to 399 and 400 to 499, respectively. Also, the comparison of three different thresholds of reads (75, 90, 95% similarity) showed a variation < 5% when mapped to the reference genome (Table 2). This low variation is indicative of a high accuracy of the transcriptome assembly, uniformity of coverage, and read depth.
| Total trinity transcripts | 149,678 |
| Total trinity genes | 90,064 |
| Percent GC | 42.75% |
| Contig N50 | 1409 bp |
| Median contig length | 487 bp |
| Average contig length | 843.72 bp |
| Total assembled bases | 126,285,990 |
| Total number of transcripts | Total number of mapped transcripts over reference genome | Percentage mapped transcripts | Percentage similarity threshold |
|---|---|---|---|
| 114,598 | 112,630 | 98.2827 | 75 |
| 114,598 | 111,435 | 97.2399 | 90 |
| 114,598 | 104,274 | 90.9911 | 95 |
3.2 Abundance estimation and identification of differentially expressed genes
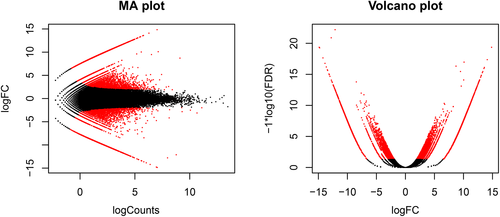
The paired-end reads of both control and drought-affected samples were mapped and aligned to the de novo transcriptome assembly of P. nigrum L. This was done to calculate the expression values in the form of Fragments Per Kilo base of exon per Million mapped reads (FPKM) values. These expression values were used for getting the differentially expressed genes (DEGs). A total of 4914 DEGs were found, out of which 2862 and 2052 were upregulated and downregulated, respectively. MA plot visualizes the differences between the data in two samples, i.e., control and drought affected. In MA plot, the horizontal x-axis represents A, which is the log2 transformed mean expression level and the vertical y-axis represents the transformed data onto M (log 2 transformed fold change). The horizontal axis in the Volcano plot is the fold change (FC) on log scale so that upregulated and downregulated genes appear symmetric. The vertical axis represents the mean expression value of −log10 (False discovery rate). The light black dots indicate the up and downregulated DEGs, while the pink dots represent non DEGs (Figure 2).
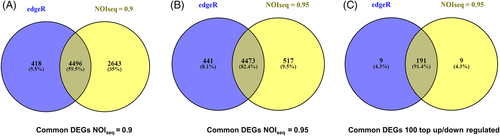
A total of 4914 DEGs were obtained from edgeR tool, out of which 2862 and 2052 were upregulated and downregulated, respectively. Whereas we found 7139 DEGs in q = 0.9 (4670 up and 2469 down [± 7 fold change value]) and 4990 DEGs in q = 0.95 (3130 up and 1860 downregulated [± 8 fold change value]). Out of the total 4914 DEGs from edgeR, 4496 and 4473 DEGs were commonly found with NOIseq at q = 0.9 and q = 0.95, respectively, as evident in the Venn diagram in Figure 3.
Also, the comparison of three different thresholds of DEGs (75, 90, 95% similarity) mapped to the reference genome showed < 5% variation, indicating the accuracy of transcriptome assembly, uniformity of coverage and read depth (Table 3).
| Total number of DEGs | Total number of mapped DEGs over reference genome | Percentage mapped DEGs | Percentage similarity threshold |
|---|---|---|---|
| Upregulated DEGs | |||
| 2862 | 2772 | 96.8553 | 90 |
| 2862 | 2521 | 88.0852 | 95 |
| Downregulated DEGs | |||
| 2052 | 1960 | 95.5166 | 90 |
| 2052 | 1791 | 87.2807 | 95 |
3.3 Homology search, annotation, and functional characterization
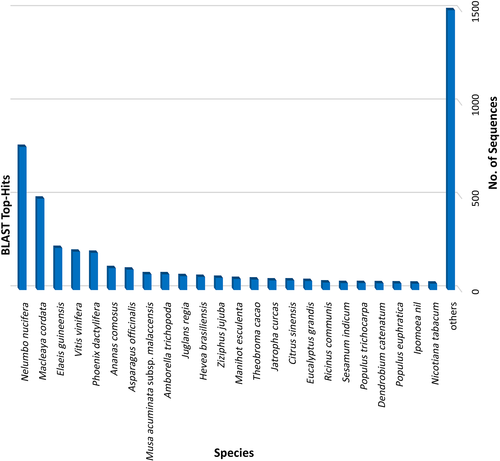
The homology search of DEGs was performed using BlastX to identify similar genes present in the database. Out of 4914 transcripts, we found that 4161 transcripts showed similarity with other genes present in the database, while 529 transcripts were novel as they did not show any similarity or were without hits. A total of 46 transcripts were used in mapping and 148 transcripts for the GO annotation process. The top hit species distribution revealed that maximum hits were found with Nelumbo nucifera (i.e. 766 transcripts), followed by Macleaya cordata and Elaeis guineensis in 489 and 225 transcripts, respectively (Figure 4).
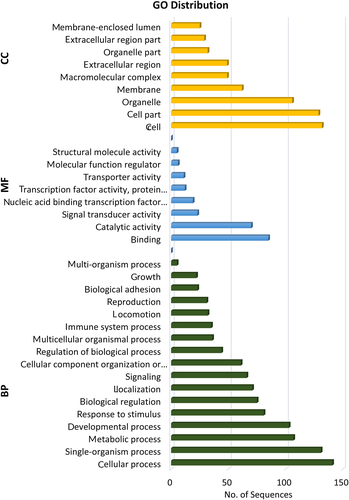
Blast2GO annotation results revealed that these differentially expressed genes are into three sub-categories: biological process, molecular functions, and cellular components. In biological process, molecular function and cellular component, a total of 1064, 231, and 613 transcripts were found, respectively. Under biological process, there were 141 transcripts involved in cellular process, 107 in metabolic process, 103 in development process, 75 in biological regulation, 66 in signalling, and 22 in growth. In molecular functions, 70 transcripts were involved in catalytic activity, 85 in binding, 5 in structural molecule activity, and 6 in molecular function regulation. In cellular component, 132 transcripts were involved in cell, 129 in cell part, 106 in organelle, 62 in membrane, and 29 in extracellular part (Figure 5).
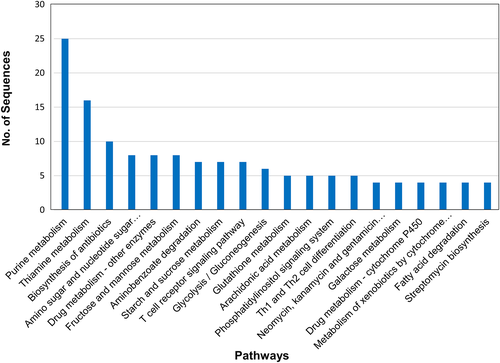
To study the pathways involved in stress mechanism, KEGG pathway analysis was done. A total of 4914 transcripts were involved in 187 pathways. The highest number of transcripts (i.e. 25) was involved in purine metabolism. This was followed by 16 transcripts in thiamine metabolism and 10 in biosynthesis of antibiotics (Figure 6).
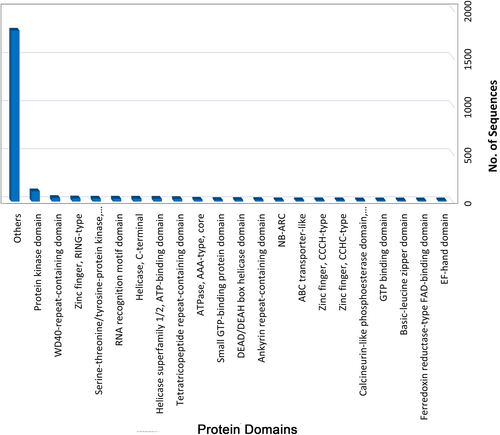
The interproscan was used to identify domains and families in DEGs. A total of 786 domains in 2298 transcripts were observed. The highest number of transcripts belonged to the protein kinase domain (IPR000719; 107 transcripts) followed by the WD40-repeat-containing domain (IPR017986; 38 transcripts) (Figure 7).
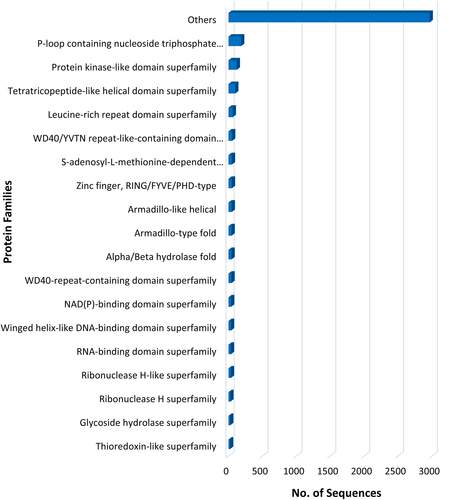
In InterProScan family search, a total of 3999 transcripts were found to be involved in 1137 families. Many transcripts were present in the family of P-loop containing nucleoside triphosphate hydrolase ((IPR027417; 186 transcripts) followed by the protein kinase-like domain superfamily (IPR011009; 122 transcripts) (Figure 8).
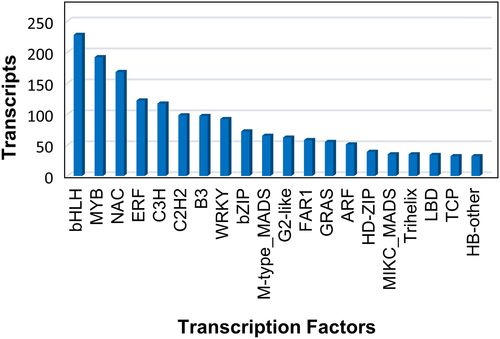
The transcription factors (TF) were identified by performing blast in the PlantTFDB 4.0 using BlastX tool. A total of 2110 transcription factors among the 4914 transcripts with an expected e-value 1e-3 were observed. The transcripts corresponding to bHLH, MYB, and NAC transcription factors occurred a maximum number of times, i.e., 228, 192 and 168, respectively (Figure 9).
3.4 Identification of putative molecular markers
3.4.1 Putative simple sequence repeats (SSRs)
A total of 20,124 genic region markers were mined from the differentially expressed genes of the de novo assembled transcriptome of P. nigrum L. The number of SSRs present in simple formation was 93.6%, while they were 6.4% in compound formation. Around 2639 sequences contained more than one SSR. The highest numbers of SSRs obtained were mono-nucleotide, followed by tri-nucleotide and di-nucleotide. The mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides reported were 58.77%, 13.84%, 26.28%, 0.83%, 0.13%, and 0.15%, respectively, as described in Table 4. A total of 14,742 primers were generated for these markers using PRIMER3 tool.
| Information regarding SSRs | De novo assembly |
|---|---|
| Mono-nucleotide | 11,827 |
| Di-nucleotide | 2785 |
| Tri-nucleotide | 5288 |
| Tetra-nucleotide | 167 |
| Penta-nucleotide | 26 |
| Hexa-nucleotide | 31 |
3.4.2 Mining of single nucleotide polymorphism
Using the various filtering criteria (i.e., read depth ≥ 4, quality score ≥ 20 and flanking regions of 50 on both sides), a total of 259,236 variants were mined. Out of these variants, 246,458 were SNPs and 12,778 were Indels. The maximum number of variants were found in contig 4105 and contig 4169 (i.e., 112 variants), followed by contig 3995, 4066, and 10,346 with 105, 95, and 93 unique variants, respectively. Contig 4105 was predicted as acetyl-CoA carboxylase 1, contig 4169 was predicted as the hypothetical protein CRG98_045342, contig 3995 was predicted as phosphotidylinositol3/4 kinase, and contig 4066 was predicted as E3 ubiquitin protein ligase UPL3.
3.5 Identification of lncRNAs and their annotation
A total of 114,598 assembled transcripts had their lengths greater than 200 bp and they were subsequently used for downstream analysis. We retained 70,608 transcripts (~61.61%) with ORF length less than 100 amino acids as predicted by ORF Predictor. After filtering mRNAs using PLEK, a total of 70,364 sequences were found to have a coding-potential score less than 0.5 using CPC (Coding Potential Calculator). A total of 46,702 transcripts were removed on the account of similarity with known proteins or protein domains when BLAST against NCBI-nr, Swissprot, Pfam and housekeeping genes (tRNAs, snRNAs, and snoRNAs). Finally, a total of 23,662 transcripts were found as high confidence lncRNAs in the black pepper transcriptome. It was observed that the maximum number of sequence (17,006 corresponding to 71.87%) had a length ranging between 200 and 400 bp, followed by 4177 (17.65%) of the range 400–600, and only 0.74% of sequence with a size above 1200 bp (Table 5).
| Sequence length range | No. of sequences |
|---|---|
| 200–400 | 17,006 |
| 400–600 | 4177 |
| 600–800 | 1502 |
| 800–1000 | 577 |
| 1000–1200 | 226 |
| >1200 | 174 |
| Total | 23,662 |
3.6 Black Pepper Transcriptome Database: BPDRTDb
The Black Pepper Drought Transcriptome Database based on three-tier architecture contains the information of candidate genes, putative genic region markers (SSRs, SNPs, Indels), transcriptional factors, pathways, domains, families, etc. It houses 114,598 transcripts, 4914 differential expressed genes, 20,124 putative markers, 14,742 primers, and 259,236 variants, which contains 246,458 SNPs and 12,778 Indels identified from de novo assembly. A total of 2110 transcriptional factors, 786 domains and 1137 families are also catalogued in this resource. It contains five tabs: Home, Transcripts, Markers, Candidate genes, and Supplements (Figure 10).

The first page, namely “Home page,” shows general information regarding the crop (P. nigrum L.) data and database. The “Transcripts” tab is further categorized into Expression profile, Transcription factor families, Domain and family, and Pathways. The expression values are expressed in the form of FPKM values of control and drought-affected samples with BLAST results under the Expression profile tab. The transcriptional factors identified in DEGs have hyperlinks to PlantTFDB, which was used as a reference to BLAST DEGs under the Transcription Factor families tab. Under the tab, Domain and family search, one can access the domain and families and hyperlinks are provided for direct link to Interpro database website. The “Pathways” tab has the details of pathways identified from DEGs directly linked to KEGG database.
The “Markers” tab has detailed information on the molecular markers identified from the black pepper transcriptome analyzed in this study. This tab contains three sub-tabs, namely SSRs, SNPs, and Indels. SSRs tab provides the information of repeats detailing the three sets of primers, while the SNPs and Indels tabs provide the variants, insertion and deletion with 50 bp flanking regions on both sides. For SSRs, three sets of primers will be generated for each particular marker, searchable using a transcript ID. The result displays the start and end positions of primers and primer length.
The “candidate genes” tab results into transcript IDs, log Fold Change value, log Counts Per Million reads value, log P-value, False Discovery Rate value. It also allows user to blast transcript IDs against the NCBI non-redundant database.
4 DISCUSSION
Being one of the expensive spice germplasms, molecular studies on black pepper (P. nigrum L.) are necessary. As drought is a major abiotic stress, studies on transcriptome assembly and analysis can help in the implementation of genetic improvement and breeding programmes to improve black pepper yield in the long run.
Our transcriptome assembly statistics having GC content 42.75% and N50 as 1409 is supported by the earlier de novo transcriptome assembly of Piper species where GC content and N50 are reported to be ~45% and ~ 1300 bp, respectively. Also, the maximum numbers of transcripts fall in the range of 200 to 299 bp as in a similar study (Hao et al., 2016). The comparison of reads mapped at 75, 90, and 95% similarity thresholds with < 5% variation align with previous studies, indicating that our transcriptome assembly is precise and the coverage/depth is uniform (Jaiswal et al., 2019). Black pepper is reported to have 63,466 protein-coding genes (Hu et al., 2019). Our transcriptome assembly counted 114,598 transcripts, this over-representation may be due to polyploidization or whole-genome duplication (WGD) events that are a major driving force of evolution, especially in plants (Adams, 2013; Hu et al., 2019; Jiao et al., 2011).
A total of 4914 differentially expressed genes were found, out of which 2862 were upregulated and 2052 downregulated. The differentially expressed genes give an idea of the genes playing an important role in the survival of the crop in drought conditions and can also be further used for introgression into varieties for drought- or stress-responsiveness. Genetic engineering is another approach to produce modified plants with better responsiveness towards survival in drought condition. With the help of information related to the upregulated genes, it would be possible to produce plants with better drought-tolerant capacity. Also, when the DEGs were mapped to the reference genome at 70%, 90%, and 95% similarity, low variation (<5%) was observed, supporting the accuracy and uniformity of genome coverage.
The homology search of DEGs would give the clue of similar genes present in the database. In the present study, 4161 transcripts having similarity with other genes is due to the already reported drought-responsive genes in the existing databases. Piper nigrum showed maximum hits (similar sequences) with plants like Nelumbo nucifera, Macleaya cordata, and Elaeis guineensis. The highest hit with Nelumbo nucifera indicates the commonality between these two crops. Both black pepper and Nelumbo nucifera are dicot and have common polyol-alkaloid pathways, potent aldose reductase inhibitory activity is found in their alkaloidal extracts (Gupta et al., 2014). Both are medicinal plants and have larvicidal and repellent activity in the methanol and ethyl acetate extracts (Kamaraj et al., 2011). Also, both crops exhibit similar antioxidant and clastogenic activity (Archana et al., 2015). However, 529 DEG transcripts did not show any similarity or had no hits in any of the databases, which indicates that these may be novel genes.
Transcription factors are reported to play critical roles in the regulation of various abiotic stress responses as molecular switches that control stress-responsive gene expression. (Khan et al., 2018). The involvement of NAC transcription factor in drought response has been evoked (Jogaiah et al., 2013). The presence of NAC transcription factors in our study shows enhanced tolerance and diverse role in drought stress regulation as previously reported in pepper (Guo et al., 2015; Zhang et al., 2020). The bHLH gene found in our analysis has been earlier reported to improve tolerance to multiple abiotic stressors in transgenic plants in wheat (Zhai et al., 2016), Arabidopsis (Le et al., 2017), and Populus (Dong et al., 2014). A novel bHLH transcription factor, PebHLH35, confers drought tolerance by reducing stomatal density with reduced photosynthesis efficiency and promoting growth of roots, over shoots, as reported in Populus, Arabidopsis, and maize (Bai & Settles, 2015; Ludwig et al., 1989).
The regulation of stress-related genes and signalling networks to counteract abiotic stress like drought is a common strategy in plants. Transcription factors like MYB plays a significant role in regulating drought stress in crops like maize (Guangsheng et al., 2012), wheat (Olowe et al., 2015), and Arabidopsis (Baldoni et al., 2015; Javed et al., 2020).
The Gene Ontology classification of DEGs was done into three major categories, namely biological processes, cellular components, and molecular function. It was observed that cellular/single-organism/metabolic processes were majorly represented by the gene categories under biological processes. Similarly, binding and catalytic activity were distributed higher under molecular functions. The cellular components category annotated cell, cell parts, and organelle as the major phenomena. This is in concordance with the result of the fruit transcriptome report of black pepper (Hu et al., 2015). These functional annotations can be source of information for specific biochemical processes or developmental processes in black pepper.
We found nearly 5000 transcripts representing 187 pathways. Many belonged to the purine metabolism, followed by thiamine metabolism and antibiotics biosynthesis. The over-representation of purine metabolism was also reported in the fruit transcriptome analysis of black pepper (Khew et al., 2020). This confirmed the role of purine metabolites in drought tolerance as reported in Arabidopsis (Watanabe et al., 2010) and almond-peach rootstock (Bielsa et al., 2018).
The dominance of protein kinase domain is validating our result as they are reported to be the regulatory drought-responsive genes. The role of kinase in the activation of ABF results in the mediation of ABA-responsive genes responsible for stomatal closure during drought stress (Bielsa et al., 2018). The presence of WD40-repeat-containing domains in our study has also been reported to mediate the response to drought stress in wheat (Hu et al., 2018; Kong et al., 2015) and are important findings ad those repeats mediate the response to drought stress.
The higher number of occurrences of bHLH, MYB and NAC transcription factors is due to their significant roles in drought tolerance and adaptation. They regulate the elongation of root hair, to absorb water more efficiently, and the vacuolar acidification required to maintain the internal water balance in response to water deficit (Lloyd et al., 2017). BHLH and MYB1R1 are phosphorylated and are involved in stress defence, water stress signalling and proteins like glutathione peroxidase, heat shock factor, glycine-rich factor have been involved in better stress responsiveness (Kosova et al., 2018). NAC transcription factors play an important role in root growth, which provide drought tolerance in wheat, cotton, and Arabidopsis (Iquebal et al., 2019) but they are reported to produce multiple stress tolerance in plants (Shao et al., 2015).
The DEGs in the study are found to have homology with patatin, which is a potato tuber storage protein that encodes for lipolytic acyl hydrolase enzyme. The corresponding gene is highly expressed in water deficit conditions and induces defence responses in plants (Matos et al., 2001). We also found DEGs sharing homology with genes encoding mycolic acid cyclopropane synthase, formed from the cyclization of fatty acid acyl (CFA) chains, that decreases the level of unsaturated fatty acids in membranes. This reduces the membrane permeability and fluidity, thus providing rigidity to the lipid bilayers, which helps in tolerance to drought and adverse conditions (Yu et al., 2011). Purine metabolism is involved in ABA accumulation, hormones important in the abiotic response in plants (Jaiswal et al., 2018). The protein kinase domain is associated with genes like SNF1 and exhibits osmotic stress tolerance in plants like Arabidopsis thaliana (Umezawa et al., 2004).
Among the total 20,124 putative markers generated from de novo assembly of Piper nigrum L., the mononucleotide repeats were the most abundant with 11,827 SSRs (58.77%), followed by 5288 (26.27%) trinucleotide repeat motifs and 2785 (13.83%) dinucleotide repeats. This higher abundance of mononucleotide may be due to the inherent limitations of the NGS technology that adds mono-type leading to sequencing error (Haseneyer et al., 2011). Based on the SSR containing sequences, we successfully designed 14,742 primers. These SSR primers can be a valuable and useful biomarker resource of P. nigrum (Hu et al., 2015).
SSR markers have broad use, for example, in variety identification, development, improvement, mapping, and molecular breeding. They can also be used in product traceability, phylogenetic, and taxonomic comparison (Jasrotia et al., 2019). The SSR markers mined from the transcriptome are commonly referred to as SSR-functional domain markers (SSR-FDM) (Liu et al., 2013). The SSR-FDM in our study can be used in linkage mapping, genetic variability, and functional diversity analysis (Kujur et al., 2013). The genic region putative SSR markers catalogued in the Black Pepper Drought Transcriptome Database (BPDRTDb) can be a valuable resource for variety differentiation. Applications of such types of SSR-FDM have been reported in Solanaceae crops (Yu et al., 2010), sugarcane (Parida et al., 2010), tulsi (Gupta et al., 2010), brinzal (Stàgel et al., 2008), oil palm (Tranbarger et al., 2012), etc.
SNP mining results showed a total of 259,236 variants, which contains 246,458 SNPs and 12,778 Indels. SNPs mining is important for studying linkage mapping, genetic variation, map-based gene isolation, population structure analysis, and plant breeding and association genetics. Mining of SNPs and Indels could possibly lead to the development of improved black pepper varieties and such type of similar studies have been reported in various other crops such as Seasme indicum and Camellia sinensis (Sahu et al., 2012). A total of 23,662 transcripts reported in the black pepper transcriptome were high-confident lncRNAs. Such lncRNAs are associated with drought stress resistance in crops like cassava (Xiao et al., 2019) and cotton (Lu et al., 2016).
In our study, we have catalogued in the Black Pepper Drought Transcriptome Database (BPDRTDb) the genomic information obtained from drought-stressed leaf transcriptome analysis of black pepper, namely differentially expressed genes, transcriptional factors, pathways, domains, families, SSR marker, and variants. This is freely accessible to researchers for academic use and can be utilized in the implementation of genetic improvement and breeding programmes to improve the crop yield. There does not exist any genomic resource of the drought transcriptome of black pepper. The developed genomic resources are useful for finding candidate gene based on discovery program and trait-based association studies, for mapping studies and cultivar identification in endeavour of black pepper germplasm improvement.
ACKNOWLEDGEMENT
The authors are thankful to Indian Council of Agricultural Research, Ministry of Agriculture and Farmers' Welfare, Government of India for creation of Advanced Super Computing Hub for Omics Knowledge in Agriculture (ASHOKA) facility where the work was carried out. The authors further acknowledge with thanks the supportive role of Directors of ICAR-IASRI, New Delhi, and ICAR-IISR, Kozhikode.
AUTHOR CONTRIBUTIONS
Johnson George K, Dinesh Kumar, and Sarika Jaiswal conceived and designed the experiments; Johnson George K, Soumya Madhavan, Manju KP, and Umadevi P performed the experiments; Ankita Negi, Rahul S. Jasrotia, Sarika Jaiswal, U.B. Angadi, M.A. Iquebal, and Anil Rai analyzed the data; Anil Rai, Rahul S. Jasrotia, M.A. Iquebal, Sarika Jaiswal, and Dinesh Kumar wrote the paper. All the authors read and approved the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
Data Availability:Data generated has been submitted to SRA of NCBI with BioProject: PRJNA515366 and BioSamples: (SAMN10754251, SAMN10754252).




