Exogenous strigolactones impact metabolic profiles and phosphate starvation signalling in roots
Funding information: Consejo Superior de Investigaciones Científicas, Grant/Award Number: 201640I040; European Regional Development Fund, Grant/Award Numbers: AGL2017-88-083-R, RTI2018-094350-B-C31; Generalitat Valenciana, Grant/Award Number: CDEIGENT/2018/015; Ministerio de Ciencia, Innovación y Universidades, Grant/Award Numbers: AGL2017-88-083-R, RTI2018-094350-B-C31, FJCI-2015-23575; National Network of Carotenoids, Grant/Award Number: BIO2017-90877-REDT
Abstract
Strigolactones (SLs) are important ex-planta signalling molecules in the rhizosphere, promoting the association with beneficial microorganisms, but also affecting plant interactions with harmful organisms. They are also plant hormones in-planta, acting as modulators of plant responses under nutrient-deficient conditions, mainly phosphate (Pi) starvation. In the present work, we investigate the potential role of SLs as regulators of early Pi starvation signalling in plants. A short-term pulse of the synthetic SL analogue 2′-epi-GR24 promoted SL accumulation and the expression of Pi starvation markers in tomato and wheat under Pi deprivation. 2′-epi-GR24 application also increased SL production and the expression of Pi starvation markers under normal Pi conditions, being its effect dependent on the endogenous SL levels. Remarkably, 2′-epi-GR24 also impacted the root metabolic profile under these conditions, promoting the levels of metabolites associated to plant responses to Pi limitation, thus partially mimicking the pattern observed under Pi deprivation. The results suggest an endogenous role for SLs as Pi starvation signals. In agreement with this idea, SL-deficient plants were less sensitive to this stress. Based on the results, we propose that SLs may act as early modulators of plant responses to P starvation.
1 INTRODUCTION
Phosphorus (P) is an essential nutrient for plants, as it is a structural component of many biomolecules, including nucleic acids, lipids and proteins, and it is involved in many cellular processes such as primary metabolism, protein activation, energy transfer and signal transduction cascades (Ham, Chen, Yan, & Lucas, 2018; Scheible & Rojas-Triana, 2015). However, despite its relevance, P is one of the less-abundant macronutrients present in soils. It is mainly acquired by plants in the form of inorganic phosphate (Pi), which has high-affinity to mineral particles and organic matter, thus reducing its bioavailability markedly, and limiting plant growth and development (Lynch, 2011; Raghothama, 2000). Along evolution, plants have developed a set of complex physiological, biochemical, metabolic and molecular modifications to cope with Pi limitation in the soil, collectively known as Pi starvation responses (PSRs) (Ham et al., 2018; Puga et al., 2017). PSRs include alterations in shoot and root morphology, the regulation of high-affinity Pi transporters (PHTs), modifications in the primary and secondary metabolism, as well as the exudation into the rhizosphere of Pi-releasing enzymes, organic acids and signalling molecules to associate with beneficial soil microorganisms that can improve Pi uptake (Andreo-Jiménez, Ruyter-Spira, Bouwmeester, & López-Ráez, 2015; Campos et al., 2018; Lambers, Martinoia, & Renton, 2015; Puga et al., 2017). Overall, PSRs aim to improve P-use efficiency by affecting both Pi acquisition and reallocation and remobilization of internal P.
Establishment and regulation of PSRs require a fine-tuned coordination and integration of local and systemic signalling pathways, which are mediated by a number of genes and signalling molecules (Ham et al., 2018; Lan, Li, & Schmidt, 2015; Puga et al., 2017; Scheible & Rojas-Triana, 2015). It is well established that the transcriptional activator PHOSPHATE STARVATION RESPONSE 1 (PHR1), and related transcription factors, play a central role by regulating the expression of many Pi starvation-induced genes (Bustos et al., 2010; Rubio et al., 2001; Zhou et al., 2008). PHR1 is constitutively expressed, but its activity is regulated by the plant Pi status. Indeed, PHR1 activity is negatively regulated by SYG1/Pho81/XPR1 (SPX)-domain proteins (Figure 1), which sense inositol phosphates (InsP) as a Pi signal (Puga et al., 2017; Secco et al., 2012). Under Pi limitation, InsP concentration drops, making the complex SPX-PHR1 no longer stable releasing PHR1. Then, PHR1 induces the expression of certain high-affinity transporters of the PHT1 family (Figure 1), facilitating Pi-acquisition and translocation in-planta (Huang et al., 2013; Liu et al., 2012; Puga et al., 2017). PHR1 also promotes the expression of the microRNA miR399, whose levels are highly induced soon upon Pi limitation (Pant, Buhtz, Kehr, & Scheible, 2008). miR399 modulates the activity of PHO2, encoding a ubiquitin-conjugating E2 enzyme involved in protein degradation (Lin et al., 2008). Subsequently, down-regulation of PHO2 prevents the degradation of PHO1, a Pi transporter involved in Pi loading into the xylem (Liu et al., 2012). In parallel to miR399, PHR1 also promotes the expression of the non-protein coding gene IPS1 (Franco-Zorrilla et al., 2007). IPS1 sequesters free miR399 through a target mimicry mechanism, preventing the interaction miR399-PHO2 and the degradation of PHO2 transcripts (Figure 1) (Franco-Zorrilla et al., 2007). Thus, plant Pi acquisition and homeostasis is finely regulated mainly by the interaction of the triad IPS1-miR399-PHO2.
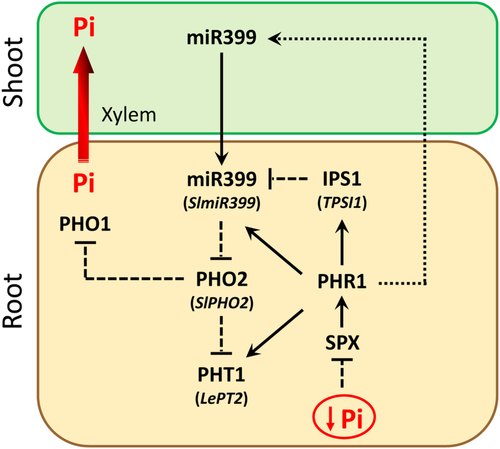
Plant responses to environmental challenges, including Pi starvation, are also mediated by phytohormones. Thus, it has been shown that Pi deficiency response is associated with downregulation of gibberellins and cytokinins, while other phytohormones such as auxin, ethylene, abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA) and strigolactones (SLs) are up-regulated (Chiou & Lin, 2011; Khan, Vogiatzaki, Glauser, & Poirier, 2016; López-Ráez et al., 2008; Pérez-Torres et al., 2008; Prerostova et al., 2018; Song et al., 2016). SLs are the latest plant hormones described, acting as modulators of plant responses under nutrient-deficient conditions, mainly Pi starvation, and other abiotic stresses such as drought and salinity (Al-Babili & Bouwmeester, 2015; Andreo-Jiménez et al., 2015). They are carotenoid-derived, belonging to the apocarotenoid class, as ABA. They are produced by the action of a β-carotene isomerase (D27) and sequential oxidative cleavage by two carotenoid cleavage dioxygenases (CCD7 and CCD8), giving rise to carlactone, the precursor of all canonical SLs, including strigol- and orobanchol-type (Al-Babili & Bouwmeester, 2015; Waters, Gutjahr, Bennett, & Nelson, 2017). Under Pi limitation, they hinder shoot growth and promote root system development – inhibiting primary root growth, promoting lateral root formation, and root hair number and elongation –, thus increasing soil exploration capacity and improving minerals and/or water acquisition under stress (Kapulnik et al., 2011; Ruyter-Spira et al., 2011). In addition to their role as phytohormones, they are important ex-planta signalling molecules in the rhizosphere, promoting the association with beneficial microorganisms, such as arbuscular mycorrhizal fungi and rhizobia, also to improve nutrients (mainly Pi and nitrogen) and water acquisition (López-Ráez, Shirasu, & Foo, 2017). Despite the key role of SLs under Pi starvation, how they modulate plant responses and whether they are also involved in P signalling remain unclear. We have previously proposed that higher Pi acquisition efficiency in a commercial wheat cultivar might be related to its improved SLs-P signalling, which modulates PHO2 activity (de Souza Campos et al., 2019). To further investigate the potential role of SLs as regulators of early Pi starvation signalling, here we explore the transcriptional and metabolic responses of the plant to a short-term pulse of the synthetic SL analogue 2′-epi-GR24, both under normal and Pi limitation conditions. Moreover, the expression pattern of Pi starvation signalling maker genes was assessed in the tomato SL-deficient line SlCCD8-RNAi L04. Our results suggest that SLs can act as modulators of plant responses during Pi limitation. Improving our understanding of Pi starvation signalling is essential to develop new agricultural strategies in order to optimize plant Pi uptake and reduce the use of P fertilizers.
2 MATERIALS AND METHODS
2.1 Plant growth conditions and treatments
Tomato (Solanum lycopersicum L. cv. MoneyMaker) and wheat seeds (Triticum aestivum cv. Tukan) were surfaced-sterilized in 4% sodium hypochlorite containing 0.02% (v/v) Tween 20, rinsed thoroughly with sterile water and germinated for 2 d in a plate on moistened filter paper at 25°C in darkness. Subsequently, seedlings were grown hydroponically in 3 L plastic containers with modified Long Ashton nutrient solution (Hewitt, 1966) containing 800 μM of Pi with constant aeration in a greenhouse for 4 weeks. After that, half of the plants were transferred to a modified nutrient solution without Pi (−P) and were let to grow for another week before applying 2′-epi-GR24 treatments. The other half was maintained under normal Pi (+P) conditions. Nutrient solution was replaced twice in a week. The active diasteroisomer 2′-epi-GR24 (orobanchol-type) (Figure S1) was applied to the nutrient solution (with and without Pi) at 4 different concentrations (0, 10, 100 and 1000 nM) for 1 h. The SL analogue 2′-epi-GR24 was kindly provided by Dr. Xie (Utsunomiya University, Japan). To prepare 2′-epi-GR24, 1 mg of the compound was dissolved in 330 μL of pure acetone to obtain a stock solution of 1 M. The stock was serially diluted in sterile demiwater to obtain the desired final concentrations. Then, the corresponding nutrient solution was replaced without 2′-epi-GR24, and plants were grown for an additional 24 h. Six seedlings per treatment were grown. Shoots and roots were collected, weighed, frozen in liquid nitrogen and kept at −80 °C until use.
Plants of the SL-deficient tomato line SlCCD8-RNAi line L04 and its corresponding wild-type cv. Craigella (LA3247) were grown in pots as described in López-Ráez et al. (2008). Seeds were surfaced-sterilized and germinated as described above. The seedlings were sown and grown in 0.5 L pots with sand/vermiculate (1:1) for 4 weeks in a greenhouse at 21/18 °C with 16/8 h photoperiod and 70% humidity. Plants were watered twice a week with modified Long Ashton nutrient solution (Hewitt, 1966). Half of the plants (Craigella and SlCCD8-RNAi) were watered with standard Pi levels (800 μM), whereas the other half was watered with 25% Pi of the standard solution (200 μM), to subject the plants to Pi limitation and induce the characteristic SL-deficient phenotype. Six seedlings per cultivar and treatment were grown. Roots from each pot were collected separately, frozen in liquid nitrogen and stored at −80 °C until use.
2.2 Extraction and quantification of strigolactones from roots
SL quantification was performed as described in Rial, Varela, Molinillo, López Ráez, and Macías (2009). Fifty milligram of tomato root extracts from each treatment were ground in a mortar with liquid nitrogen. Root material was extracted with 1 mL of ethyl acetate in an ultrasonic bath for 10 min, centrifuged for 10 min at 5000 rpm, concentrated in a rotary evaporator, and stored at −80 °C. Before the analysis, the extracts were dissolved with MeOH to achieve a ratio 1:1 g · L−1 (w/v). (±)-GR24 (racGR24), used as internal standard, was dissolved in MeOH to 10 mg · L−1 and added to all samples at 10 μg · L−1. The samples were analysed on a Bruker EVOQ Triple Quadrupole Mass Spectrometer (Bruker, Madrid, Spain), using as ionization source an electrospray (ESI+). The samples were injected and separated using an ACE Excel 1.7 C18 (100 mm × 2.1 mm, 1.7 μm particle size) (Advanced Chromatography Technologies Ltd., Aberdeen, Scotland) maintained at 40 °C. The mobile phases were solvent A (water, 0.1% formic acid) and solvent B (MeOH, 0.1% formic acid), with a flow rate set to 0.3 mL min−1. The linear gradient was: 0–0.5 min, 50% B; 0.5–5 min, to 100% B; 5–7 min, 100% B; 7–7.5 min, to 50% B and 7.5–10.5 min, 50% B. The injection volume was 5 μL. The instrument parameters were: spray voltage +4500 V, cone temperature 300 °C, cone gas flow 15 psi, heated probe temperature 400 °C, heated probe gas flow 15 psi, nebulizer gas flow 55 psi and collision pressure 2.0 mTorr. The compound-dependent parameters for orobanchol, solanacol and the IS, the parent or precursor ions, the fragments obtained by MRM analysis and the collision energy to achieve each fragmentation are provided in Table S1. Orobanchol and solanacol were kindly supplied by Professor Xiaonan Xie and Professor Koichi Yoneyama (Weed Science Center, Utsunomiya University, Japan), and racGR24 was provided by Professor Binne Zwanenburg (Department of Organic Chemistry, Radboud University, Nijmegen, Netherlands).
2.3 RNA isolation and gene expression analysis by quantitative real-time RT-PCR (qPCR)
Total RNA was extracted using TRIsure reagent (Bioline, Barcelona, Spain) according to the manufacturer's instructions. Subsequently, the RNA was treated with RQ1 DNase (Promega, Madrid, Spain) and purified through a silica column using the RNA Clean & Concentrator kit (Zymo Research, Madrid, Spain). RNA was quantified using a Nanodrop (Thermo Fisher Scientific, Madrid, Spain), and its integrity checked by gel electrophoresis before stored at −80 °C. The first-strand cDNA was synthesized with 1 μg of purified total RNA using the PrimeScript RT Master Mix kit (Takara, Saint-Germain-en-Laye, France) according to the manufacturer's instructions. Real-time quantitative PCR (qPCR) was performed in a StepOnePlus real-time PCR system (Thermo Fisher Scientific, Madrid, Spain), using the TB Green Premix ExTaq kit (Takara, Saint-Germain-en-Laye, France) and specific primers (Table S2). Five independent biological replicates were analysed per treatment. Relative quantification of specific mRNA levels was performed using the comparative 2-Δ(ΔCt) method (Livak & Schmittgen, 2001). Expression values were normalized using the housekeeping gene SlActin for tomato and TahnRNPQ (the heterogeneous nuclear ribonucleoprotein Q) for wheat (Grün, Buchner, Broadley, & Hawkesford, 2018).
2.4 Metabolic analyses
Untargeted metabolic profiles of tomato roots were performed by liquid chromatography and electrospray ionization (LC-ESI) full scan mass spectrometry, as described in Rivero, Álvarez, Flors, Azcón-Aguilar, and Pozo (2018). Briefly, 50 mg of freeze-dried root material was extracted at 4 °C with 1 mL of MeOH:H2O (10:90, v:v) containing 0.01% of HCOOH. After the centrifugation at full speed at 4 °C for 15 min, the supernatant was filtered through 0.2 μm cellulose filters (Regenerated Cellulose Filter, 0.20 μm, 13 mm D. pk/100; Teknokroma, Barcelona, Spain). Subsequently, 20 μL were injected into an Acquity UPLC system (Waters, Mildford, MA, USA) interfaced with a hybrid quadrupole time-of-flight instrument (QTOF MS Premier). Subsequently, a second fragmentation function was introduced into the TOF analyser to identify the signals detected. This function was programmed in a t-wave ranging from 5 to 45 eV to obtain a fragmentation spectrum of each analyte (Gamir, Pastor, Cerezo, & Flors, 2012). Positive and negative electrospray signals were analysed independently to obtain a global view of the data conduct. To elute the analytes, a gradient of methanol and water containing 0.01% HCOOH was used. Six independent biological samples were randomly injected. The LC separation was performed using a UPLC Kinetex 2.6 μm particle size EVO C18 100 A, 50 x 2.1 mm (Phenomenex, Madrid, Spain). Chromatographic conditions and solvent gradients and further were established as described by Rivero et al. (2018).
2.5 Full scan data analysis
Full scan data files were acquired with the Masslynx 4.1 software (Masslynx 4.1; Waters, Barcelona, Spain) and were transformed from .raw format into .cdf with Databridge tool provided by Masslynx software. The software R (http://www.r-project.org/) was used to process chromatographic data file using the XCMS algorithm (www.bioconductor.org) to obtain the peak peaking, grouping signals and signal corrections. Peak area was normalized relative to the dry weight. To test the metabolic differences between treatments, a non-parametric Kruskal–Wallis test (P < 0.05) was performed. Partial least square discriminant analysis and heat map analysis were performed with the metaboAnalyst 4.0 (Chong & Xia, 2018). Adduct and isotope correction, filtering, clustering, exact mass mapping and metabolic pathway exploration was carried out with the packages MarVis filter, MarVis cluster and MarVis pathway that are integrated in the Marvis suit 2.0 (Kaever et al., 2015). Metabolite identification was carried out based on exact mass accuracy and fragmentation spectra matching with different online database. The database kegg (https://www.genome.jp/kegg/) was used for exact mass identity and for fragmentation spectrum analysis, the Massbank and the Metlin databases were used (www.massbank.jp; www.masspec.scripps.edu).
2.6 Statistical analyses
Data were subjected to one-way analysis of variance (ANOVA) using the software SPSS Statistics v. 20 for Windows (SPSS Inc., Chicago, IL, USA). Duncan's multiple range test was applied when suited. Full scan data was subjected to Kruskal–Wallis test and signals with P-value ≤0.05 between treatments were used for identification.
3 RESULTS
3.1 Low doses of 2′-epi-GR24 stimulate strigolactone biosynthesis in roots under normal and low Pi conditions
It is well known that SL biosynthesis is promoted under Pi deficiency (López-Ráez, Charnikhova, Gómez-Roldán, et al., 2008; Yoneyama et al., 2012). Here, we explore the potential feedback in SL biosynthesis under both optimal and deficient Pi conditions. Tomato plants were grown hydroponically under normal Pi conditions (+Pi) or subjected to Pi limitation for a week (−Pi). Then, half of the plants of each condition were given a 1 h-pulse with different concentrations (0, 10, 100 or 1000 nM) of 2′-epi-GR24. The SL analogue racGR24 is a racemic mixture of four diastereoisomers, where some of the enantiomers do not present SL activity (Scaffidi et al., 2014). Here, to avoid side-effects of the non-active molecules, the active diasteroisomer 2′-epi-GR24 (orobanchol-type) (Figure S1) was applied to the nutrient solution. Upon the 1 h-pulse, plants grew for additional 24 h with the corresponding nutrient solution (with or without Pi) without 2′-epi-GR24 to evaluate the response to the treatment.
As expected, the levels of the characterized tomato SLs orobanchol and solanacol (Figure S1) (López-Ráez, Charnikhova, Gómez-Roldán, et al., 2008) were promoted after 1 week Pi starvation (Figure 2a,b). In addition, the levels of these two SLs were further enhanced by the exogenous application of 2′-epi-GR24. Remarkably, this effect was dose-dependent, being most pronounced at the lowest dose (10 nM) and disappearing at higher doses (100 and 1000 nM) (Figure 2a,b). The same pattern as for the analytical quantification was observed by qPCR when using molecular markers for the SL biosynthesis pathway. The two genes studied were SlD27, encoding for a β-carotenoid isomerase which converts all-trans-β-carotene to 9-cis-β-carotene and SlCCD8, which encodes a carotenoid cleavage enzyme catalysing the production of carlactone, the precursor of all canonical SLs (Al-Babili & Bouwmeester, 2015; Waters et al., 2017). The expression of both genes was induced about 3 times under Pi starvation, and was further promoted up to 8 and 6 times, respectively, by 10 nM of 2′-epi-GR24 (Figure 2c,d).
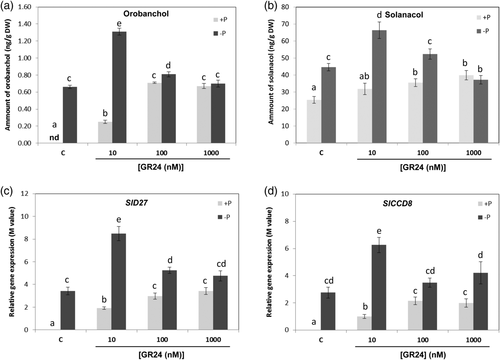
Orobanchol and solanacol levels also increased by 2′-epi-GR24 in plants grown under normal Pi conditions, somehow resembling those observed in Pi limitation. Here, the effect was also dose-dependent, but the highest levels were observed at higher concentrations of 2′-epi-GR24 (Figure 2a,b). A very similar pattern was found regarding the expression of SlD27 and SlCCD8. They were also induced under these conditions (Figure 2c,d). The expression of these two SL biosynthesis genes was analysed in another important agricultural crop as wheat. Interestingly, a similar trend was observed in the expression levels of the wheat TaD27 and TaCCD8 genes, being both induced by Pi deprivation and by 10 nM 2-epi-GR24 under normal Pi conditions (Figure S2).
3.2 Short-term application of strigolactones enhance plant Pi starvation signalling
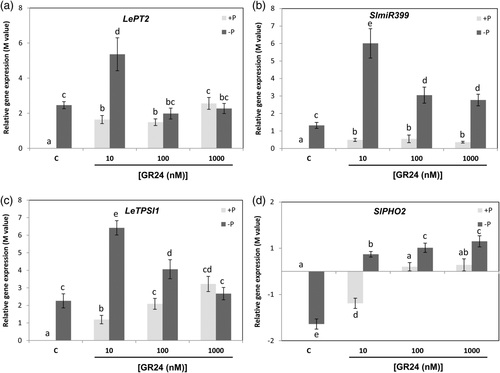
The influence of SLs on P-related signalling was assessed by analysing the expression of key genes involved in plant P responses, such as the triad IPS1-miR399-PHO2, and the PHT LePT2. LePT2 belongs to the PHT1 family, and its expression is strongly dependent on the plant Pi status (Franco-Zorrilla et al., 2007; Lin et al., 2008; Nagy et al., 2005; Pant et al., 2008). The expression of LePT2 was induced in the roots more than 2 times under Pi limitation. Interestingly, its expression was further increased up to 5 times when 10 nM 2′-epi-GR24 was applied, while higher concentrations had no effect in its expression under this Pi limiting conditions (Figure 3a). The application of 2′-epi-GR24 under normal Pi conditions induced an increase in the expression of LePT2, reaching at all SL concentrations similar expression levels to those observed for Pi starvation (Figure 3a). A very similar expression profile was observed for the genes of triad IPS1-miR399-PHO2. Transcript levels of LeTPSI1, the tomato homolog to IPS1 (Liu, Muchhal, & Raghothama, 1997), and SlmiR399 were promoted by Pi deprivation, and this induction potentiated by SL addition (Figure 3b,c). Moreover, under normal Pi conditions gene expression was enhanced by all 2′-epi-GR24 treatments (Figure 3b,c). A different behaviour was detected for the other key gene in Pi response. Transcript levels of PHO2 were almost 2 times down-regulated by Pi starvation, levels that were recovered upon application of 2′-epi-GR24 (Figure 3d). Nevertheless, the application of 10 nM 2′-epi-GR24 under normal Pi conditions repressed the expression of PHO2 1.5 times (Figure 3d), resembling the effect of Pi starvation.
The expression pattern of IPS1, miR399 and PHO2 and of a high-affinity Pi transporter (TaPht2) was also analysed in wheat. As for tomato, transcript levels of TaPht2, taemiR399 and TaIPS1 were clearly induced by Pi starvation and by 10 nM 2′-epi-GR24 application under normal Pi conditions (Figure S3). In the case of TaPHO2, Pi starvation did not induce any significant change. However, 2′-epi-GR24 increased its transcript levels only under Pi limiting conditions (Figure S3d).
3.3 SL-deficient plants are less sensitive to Pi starvation
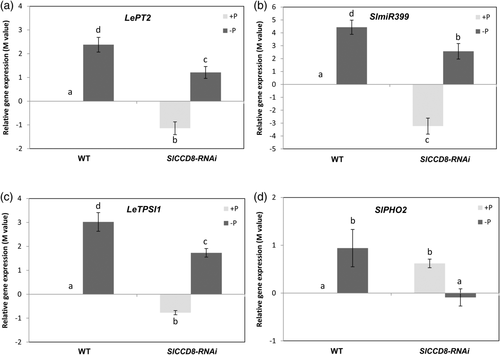
We previously generated and characterized knock-down lines for the SL biosynthesis gene SlCCD8 in tomato (Kohlen et al., 2012). One of these transgenic lines – SlCCD8-RNAi L04 – presented a 92% reduction on SLs levels. Here, we analysed the response of this SL-deficient line to Pi starvation by checking the expression of Pi marker genes. Basal expression of the Pi transporter LePT2 was about twofold lower in the SlCCD8-RNAi line compared to the corresponding wild-type under normal Pi conditions. An increase in LePT2 transcript levels was observed under Pi limitation both in the wild-type and the transgenic line. However, the final value reached in SlCCD8-RNAi was lower because of their reduced basal levels (Figure 4a). The same behaviour was observed for SlmiR399 and LeTPSI1, showing an induction by Pi starvation both in the wild-type and the transgenic line (Figure 4b,c). As for LePT2, basal transcriptional levels of these genes were also lower in SlCCD8-RNAi, therefore reaching lower final values. In the case of SlPHO2, under normal Pi conditions, the basal levels in the SlCCD8-RNAi were higher than in the wild-type, in contrast to the pattern observed for SlmiR399 and LeTPSI1 (Figure 4d). On the other hand, an induction for SlPHO2 was detected in the wild-type in Pi starvation compared to normal Pi conditions, while in SlCCD8-RNAi a slight reduction was observed (Figure 4d). Thus, Pi starvation had opposite effects in the wild-type and in the SL-deficient line in the expression of SlPHO2, supporting the regulatory role of SLs in Pi responses.
3.4 Low doses of 2-epi-GR24 impact metabolic profiles in the roots, resembling those of Pi starvation
The previous data evidence a parallelism between the plant response to Pi starvation and to low doses of SLs (2′-epi-GR24). In addition, altered basal levels of Pi marker genes and response to Pi limitation were observed in the SL-deficient line SlCCD8-RNAi. To investigate a potential direct relationship between SLs and Pi signalling, the reprogramming of tomato root metabolism associated with responses to Pi starvation and exogenous application of 2′-epi-GR24 were compared. Since major effects at transcriptional and SL levels were observed at low doses of 2′-epi-GR24 (10 nM), root metabolic profiles upon application of this concentration of 2′-epi-GR24 under normal and limited Pi conditions were analysed. Untargeted metabolomics analyses of extracts via HPLC coupled with a quadrupole time-of-flight mass spectrometer were performed. Following the chromatographic analyses, a bioinformatics processing of the detected signals was performed using the MarVis Suit 2.0 software tool for clustering and visualization of the metabolic markers (Kaever et al., 2015). Clustering and functional pathway (KEGG Solanum lycopersicum pathway Database) analyses were further performed to obtain potential biological information of the metabolic reprogramming.
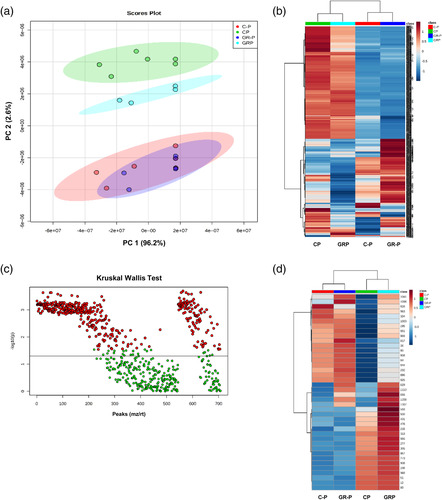
Metabolic analysis yielded a total of 1180 signals, 298 in ESI− mode (Table S3) and 882 in ESI+ mode (Table S4). A combined principal component analysis (PCA) (P < 0.05) of the signals obtained from the ESI+ and ESI− modes showed that the principal source of variation resulted from Pi starvation [Control -P (C-P) and GR24-treated -P (GR-P)]. Plant samples subjected to one week of Pi deprivation grouped together in the PCA, and clearly separated from those of plants grown under normal Pi conditions [Control +P (CP)], explaining 2.6% of the variation (component 2) (Figure 5a). Under Pi starvation, exogenous application of 2′-epi-GR24 did not induce significant changes in the plant (GR-P vs C-P), showing a priority effect of Pi starvation in the plant metabolome. However, under normal Pi conditions, 2′-epi-GR24 application [GR24-treated +P (GRP) vs Control +P (CP)] induced plant metabolic responses, revealing some SL-derived metabolic responses leading to profiles closer to those observed in plants grown under Pi limitation (Figure 5a). Hierarchical cluster analysis of the different groups confirmed the observations of the PCA analysis, supporting that the main source of variability is the plant Pi status. Remarkably, the heatmap analysis showed that rather than inducing, Pi starvation repressed the biosynthesis of most secondary metabolites detected (Figure 5b). The Kruskal–Wallis test revealed 408 significant (P < 0.05) features, of which 166 showed differential signals when comparing control plants (CP) with 2-epi-GR24 treated plants (GRP and GR-P) (Figure 5c). Out of the 166 features, 40 signals were increased by 2-epi-GR24 (Figure 5d).
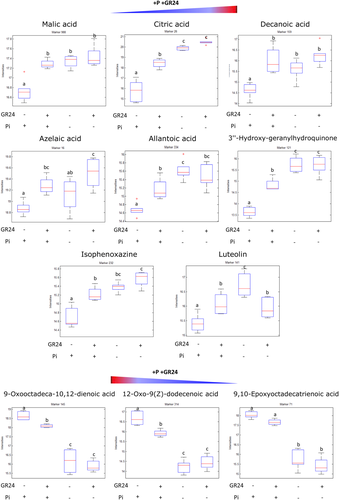
The major impact took place at the primary metabolism, including signals associated to carboxylic acids, fatty acids and purine metabolism, but also at the secondary metabolism, mainly associated to phenylpropanoids (Figure 6), changes already reported to be associated to PSRs (Pant et al., 2015; Ziegler et al., 2016). Among the identified compounds, we found the carboxylic acids malic and citric acids, whose levels were increased by Pi starvation and by 2-epi-GR24 in normal Pi conditions. The same pattern was observed for the fatty acids decanoic and azelaic acids, allantoic acid (purine metabolism), 3′′-Hydroxy-geranylhydroquinone (ubiquinone and other terpenoid-quinone biosynthesis), isophenoxazine (tryptophan metabolism) and the flavonoid luteolin, all of them induced by Pi starvation, but also by 2-epi-GR24 under normal Pi conditions. Among the compounds that showed reduced accumulation under Pi starvation, several also showed a reduction with the application of 2-epi-GR24 under normal Pi conditions, as is the case of some fatty acids, especially compounds associated to the linoleic and alpha-linolenic acids metabolism, including 9-Oxooctadeca-10, 12-Oxo-9(z)-dodecenoic acid and 9,10-Epoxyoctadecatrienoic acid (Figure 6).
4 DISCUSSION
P is one of the less-abundant macronutrients in soils, which negatively impacts plant growth and development, and therefore agricultural production. In intensive agriculture, the abuse of P-fertilizers originates considerable costs and environmental damage, as soil and groundwater contamination. Therefore, understanding how plants sense, signal and respond to low Pi availability is essential to optimize the use of these fertilizers, alleviating agricultural costs and the excessive consumption of this non-renewable resource.
SLs are key modulators of plant responses to Pi limitation in the soil, significantly altering plant physiology and development to optimize Pi uptake and use (reviewed in Waters et al., 2017). Indeed, their biosynthesis is highly promoted under Pi limiting conditions (López-Ráez, Charnikhova, Gómez-Roldán, et al., 2008; Yoneyama et al., 2012). They have the capacity of inhibiting bud outgrowth under Pi shortage in order to reduce shoot biomass and minimize Pi demand. Actually, SL-deficient plants show a typical dwarf and bushy phenotype in different species, which is restored upon exogenous application of racGR24 (Gómez-Roldán et al., 2008; Umehara et al., 2008). In the aerial part, they also promote internode elongation, secondary growth of the stem and early leaf senescence to facilitate Pi reallocation, and inhibit adventitious rooting. In the roots, where they are mainly produced and accumulated, SLs repress the growth of the primary root, while stimulating the outgrowth of lateral roots, root hair number and elongation (Kapulnik et al., 2011; Ruyter-Spira et al., 2011). All these morphological modifications are well documented to be associated to plant responses to Pi starvation, and they are oriented to increase the root surface area to facilitate Pi uptake in the soil (Lynch, 2011; Raghothama, 2000). More recently, it has been reported that exogenous racGR24 application promoted anthocyanin accumulation and the activation of acid phosphatases, typical early PSRs in plants (Ito et al., 2015). However, how they modulate plant Pi responses and whether they are involved in P signalling is not clear. We show here that a 1 h-pulse of the active SL analogue 2′-epi-GR24 was able to further promote the biosynthesis of endogenous SLs, already induced by Pi limitation, both in tomato and wheat (Figure 2), supporting a positive feedback loop in SL biosynthesis. Interestingly, 2′-epi-GR24 application increased SL levels also under optimal Pi conditions, where SL levels are usually low and even undetectable (López-Ráez, Charnikhova, Gómez-Roldán, et al., 2008; Yoneyama et al., 2012), suggesting that SLs could act as signals triggering plant responses to Pi deficiency. In agreement with their potential regulatory role in P signalling, the positive effect of GR24 was mainly observed at low doses (10 nM) in plants grown under Pi starvation, with higher basal levels of endogenous SLs, while the main effect under optimal Pi conditions (with low endogenous SL levels) was detected at higher doses of GR24 (100 and 1000 mM). Other dose-dependent effects for SL action have been previously reported. We previously showed that racGR24 application under normal Pi conditions suppressed lateral root formation, while it was promoted at Pi limitation. Therefore, it was proposed that endogenous SLs are important for the final output in lateral root development (Ruyter-Spira et al., 2011). Similarly, De Cuyper and co-workers showed that in the interaction Sinorhizobium meliloti-Medicago truncatula racGR24 treatment differentially affected nodulation, depending on its concentration. The authors showed that low doses were able to promote the number of nodules, whereas high doses reduced it (De Cuyper et al., 2015). Therefore, it seems that the net effect of SLs depends on the P nutritional conditions and on their optimum endogenous levels, as described for most plant hormones.
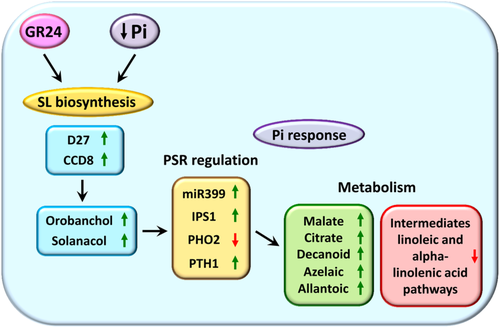
As mentioned, plant P responses are finely regulated by PHR1 and the triad IPS1-miR399-PHO2 (Figure 1) (Franco-Zorrilla et al., 2007; Ham et al., 2018; Puga et al., 2017). We have recently proposed that higher basal levels of SLs in a commercial wheat cultivar would act as a priming signal to boost plant responses to Pi starvation, regulating the expression levels of the three genes of the triad IPS1, miR399 and PHO2 (de Souza Campos et al., 2019). The fine-tuning modulation of PHO2 activity would reduce shoot Pi loading and favour the development of the root system, thus improving Pi acquisition efficiency and use (de Souza Campos et al., 2019). Here, we show that the short-term application of 2′-epi-GR24 also affected the expression of IPS1-miR399-PHO2 and that of the high-affinity transporter LePT2 genes, both in tomato and wheat. Low doses of GR24 boosted the expression of these genes, already promoted by Pi starvation, both in tomato and wheat. In addition, in the case of PHO2, which expression was reduced under Pi limitation, it was promoted by 2′-epi-GR24. This fact could be explained as an effect of timing and/or endogenous concentration of SLs. Indeed, a time-dependent increase of PHO2 transcripts has been shown in wheat, which would coincide with a progressive increase in SL levels (de Souza Campos et al., 2019). 2′-epi-GR24 also induced the expression of these Pi marker genes even at optimal Pi conditions, partially mimicking the effect observed in Pi starvation. A promoter effect of racGR24 in the expression of the high-affinity Pi transporter Pht1;7 under Pi deprivation has also been observed in Arabidopsis (Prerostova et al., 2018). Conversely, a down-regulation of Pi transporters from the Pht1 family in the SL-deficient mutant max1-1 was shown (Ito et al., 2015). Altogether, the results point to an involvement of SLs in regulating early P signalling in plants (Figure 7). In agreement with this idea, the SL-deficient line SlCCD8-RNAi showed altered levels of these key Pi response regulatory elements, being also less sensitive to Pi starvation. A similar effect was observed in Arabidopsis, where the SL-deficient max1-1 and the SL-signalling max2-1 mutants were less sensitive to Pi limitation, producing less root hairs and anthocyanins under stress. This mutant showed reduced transcript levels of IPS1, whereas those of PHO2 were up-regulated (Ito et al., 2015). We report here the same pattern for tomato. Interestingly, the application of racGR24 partially rescued the phenotype in the SL-deficient mutant, but not in the SL-signalling mutant (Ito et al., 2015; Kapulnik et al., 2011). Thus, our results support an important role of SLs in the plant response to P levels. Further research is required to decipher how plants perceive Pi stress and how the relationship SL-P signalling is regulated.
In addition to promoting SL biosynthesis and increasing the gene expression of P signalling marker genes under low and optimal Pi conditions, low doses of 2′-epi-GR24 altered root metabolic profiles in plants grown under optimal Pi conditions, partially resembling those observed under Pi starvation (Figure 5a). Among the identified compounds, an increase in malate and citrate was observed under both Pi starvation and 2′-epi-GR24 application. An accumulation and exudation into the rhizosphere of these carboxylic acids are generally observed in plants exposed to Pi shortage (Pant et al., 2015). It is suggested that they can improve Pi availability by mobilizing different P forms from the soil through the chelation of metal ions such as Fe, Al or Ca. Moreover, a key role for malate in the characteristic changes triggered by Pi starvation in root system architecture has been recently described (Mora-Macías et al., 2017). Another important metabolite associated to plant responses to Pi availability which levels were accumulated by Pi starvation and 2′-epi-GR24 under optimal Pi conditions was allantoic acid. The accumulation of nitrogen-rich compounds, including the ureides allantoin and its degradation product allantoate, is related to increased nucleotide degradation and the consequent Pi mobilization under this stress condition, and the crosstalk between P and nitrogen metabolism (Medici et al., 2019; Pant et al., 2015). Overall, the accumulation of these compounds would help the plant to cope with low Pi availability by optimizing P use and internal mobilization.
Another metabolite specifically accumulated under Pi limitation and 2′-epi-GR24 application was the saturated dicarboxylic acid azelaic acid. This compound has been associated to priming of plant immunity (systemic acquired resistance, SAR), conferring local and systemic resistance against bacterial pathogens by inducing the production of SA (Jung, Tschaplinski, Wang, Glazebrook, & Greenberg, 2009). Increased levels of SA in roots subjected to Pi starvation have been shown (Prerostova et al., 2018) (López-Ráez et al. unpublished data), whereas a reduced accumulation of this hormone was observed in the tomato SL-deficient SlCCD8-RNAi line L9 (Torres-Vera, García, Pozo, & López-Ráez, 2014). Interestingly, this SL-deficient line was more susceptible to the fungal pathogen Botrytis cinerea. Similarly, Arabidopsis SL-deficient plants were hypersensitive to the actinomycete Rhodococcus fascians, whereas the application of racGR24 to wild-type plants induced resistance against this pathogen (Stes et al., 2015). A connection between Pi starvation and SAR through the stimulation of SA by the Pi transporter PHT4;1 was made in Arabidopsis (Wang et al., 2011), where the authors proposed a critical role of this Pi transporter in regulating innate immunity in Arabidopsis. Parallelism between Pi starvation and 2′-epi-GR24 application was also found in the reduction of compounds associated to the linoleic and alpha-linolenic acids metabolism was observed (Figure 6). These pathways are related to the biosynthesis of essential fatty acids and oxylipins metabolism, which includes the biosynthesis of JA and derivatives. The reduction of these intermediate compounds could indicate an increase of the final products of these pathways such as JA, among others. Indeed, a promotion of JA content by Pi starvation has been reported (Khan et al., 2016; Prerostova et al., 2018). Interestingly, Khan and co-workers showed that the accumulation of JA in Pi-starved plants was mediated by PHR1, and that it was associated with resistance to insect herbivores (Khan et al., 2016). As for SA, reduced levels of JA were observed in the SL-deficient SlCCD8-RNAi line L9 (Torres-Vera et al., 2014), pointing to a role of SLs in defence responses. A cross-talk between Pi starvation and signalling pathways regulating plant responses to other environmental stresses, including biotic stresses, has been suggested, opening up a broad field of research. SLs, through its interaction with other phytohormones, might be regulating these plant stress responses in a dose- and likely tissue-dependent manner.
Summarizing, we provide experimental evidences supporting that SLs are early modulators of plant responses to low Pi availability, promoting the expression of key regulatory genes and that of PHTs associated to this stress, and altering metabolic profiles to cope with Pi limitation (Figure 7). A short-term pulse of low doses of the SL analogue 2′-epi-GR24 at optimal Pi conditions was able to partially mimic the plant response to Pi starvation, supporting the role of SLs in Pi-related signalling. The results presented here could be extrapolated to crop varieties with higher endogenous SLs' levels or with increased sensitivity to this plant hormone. This knowledge may help to develop new strategies to optimize plant Pi acquisition efficiency and use, thus reducing the excessive use of P fertilizers for a more sustainable agriculture.
ACKNOWLEDGMENTS
This work was supported by grants RTI2018-094350-B-C31 and AGL2017-88-083-R from the Spanish National R&D Plan of the Ministry of Science, Innovation and Universities Economy and Competitiveness (MICIU) and the European Regional Development Fund (ERDF), and 201640I040 from the Spanish National Research Council (CSIC). J.G. was supported by two Postdoctoral fellowships: FJCI-2015-23575 from Ministry of Science, Innovation and Universities, and CDEIGENT/2018/015 from Generalitat Valenciana. The laboratory participates in the National Network of Carotenoids (CaRed) (BIO2017-90877-REDT). We thank Dr. Koichi Yoneyama and Dr. Xiaoan Xie for kindly providing the natural strigolactones used to set up their analytical quantification and Dr. Binne Zwanenburg for providing racGR24. We also thank the SCIC of the University Jaume I for technical support in the metabolomic analysis. We declare that we do not have any conflict of interest.




