The Arabidopsis U-box E3 ubiquitin ligase PUB30 negatively regulates salt tolerance by facilitating BRI1 kinase inhibitor 1 (BKI1) degradation
Abstract
The Arabidopsis U-box E3 ubiquitin ligases play an important role in the ubiquitin/26S proteasome-mediated protein degradation pathway. Recently, PUB30 has been reported to participate in the salt stress response during seed germination stage in abscisic acid (ABA)-independent manner, but the molecular mechanism remains to be elucidated. Here, we displayed that the pub30 mutant was more tolerant to salt stress during seed germination, whereas the mutant of its closest homologue PUB31 showed mild sensitivity to salt stress. PUB30 exhibited E3 ubiquitin ligase activity in vitro. PUB30 specifically interacted with BRI1 kinase inhibitor 1 (BKI1), a regulator playing dual roles in brassinosteroids signaling, in vitro and in vivo. We found that BKI1 protein was ubiquitinated and degraded by the 26S proteasome. The degradation of BKI1 was slowed down in the pub30-1 mutant compared with that in the wild type. The bki1 mutant was sensitive to salt, whereas the transgenic plants overexpressing BKI1 showed salt tolerant phenotype. All these results indicate that PUB30 negatively regulates salt tolerance probably through regulating the degradation of BKI1 and brassinosteroids signaling in Arabidopsis.
1 INTRODUCTION
Salinity is one of the major threats affecting land use capability and crop yield worldwide. Understanding the molecular mechanism governing the salt stress response will be a great asset to develop salt resistant crop plants (Ji et al., 2016). Salinity causes both ion toxicity and osmotic stress, and plants have developed a very complex salt tolerance mechanism during the course of evolution (Zhu, 2002). The salt overly sensitive (SOS) signaling pathway is crucial for maintaining ion homeostasis under salt stress. SOS1, SOS2, SOS3, and SOS3-like calcium binding protein8 (SCaBP8) are major components in this pathway. SOS1 encodes a plasma membrane-localized Na+/H+ antiporter that functions in maintaining the ion homeostasis in the cell (Qiu, Guo, Dietrich, Schumaker, & Zhu, 2002; Shi, Ishitani, Kim, & Zhu, 2000). SOS2 is a serine/threonine protein kinase that phosphorylates and activates SOS1 (Liu, Ishitani, Halfter, Kim, & Zhu, 2000; Qiu et al., 2002). SOS3 encodes a calcium-binding protein that interacts with SOS2, recruiting it to the plasma membrane and activating it in calcium-dependent manner that further activates SOS1 in plasma membrane. (Albrecht, Ritz, Linder, Harter, & Kudla, 2001; Guo, Halfter, Ishitani, & Zhu, 2001; Halfter, Ishitani, & Zhu, 2000; Quintero et al., 2011; Quintero, Ohta, Shi, Zhu, & Pardo, 2002). SCaBP8/Calcineurin B-like 10 is a homologous protein of SOS3 and also crucial for SOS2 activation. SCaBP8 mainly functions in the shoot, whereas SOS3 functions in the root. These two proteins share the same properties in the regulation of SOS2 (B. G. Kim et al., 2007; Quan et al., 2007).
The ubiquitin/26S proteasome-mediated pathway is involved in selective degradation of proteins in cells where E3 ubiquitin ligases are responsible for substrate specificity. According to the mode of action and the presence of specific domains, E3 ligases are categorized into four classes: RING, HECT, F-box, and U-box (Yee & Goring, 2009). In the Arabidopsis genome, 64 plant U-box (PUB) genes have been predicted, and they function in diverse biological processes (Yee & Goring, 2009). PUB22 and its homologous protein PUB23 coordinately regulate plant response to drought by ubiquitinating cytosolic RPN12a, a subunit of the 19S regulatory particle in the 26S proteasome (Cho, Ryu, Song, Kwak, & Kim, 2008). Additionally, PUB22 was reported to attenuate the pathogen-associated molecular patterns (PAMPs)-induced signaling by mediating degradation of Exo70B2, a subunit of the exocyst complex that regulates vesicle tethering during exocytosis (Stegmann et al., 2012). PUB12 and PUB13 positively regulate ABA signaling by ubiquitinating ABI1 only when it interacts with ABA receptors (Kong et al., 2015). The pub18 pub19 double mutant was found less sensitive to ABA and sodium chloride (NaCl) stress compared to the wild type during seed germination stage (Bergler & Hoth, 2011). Recently, PUB30 was reported to be induced specifically by high salinity and participated in the salt stress response as a negative factor in seed germination, but the molecular mechanism is yet to be completely elucidated (Hwang, Seo, Kang, Kwak, & Kim, 2015).
Brassinosteroids (BRs) are a group of important plant hormones that control a variety of important plant growth and developmental processes, including cell division/elongation, photomorphogenesis, and adaptation to biotic/abiotic stresses (Abraham et al., 2003; Clouse & Sasse, 1998; Krishna, 2003). BRs biosynthesis and signal transduction play an important role in the plant response to salt stress (Krishna, 2003; Steber & McCourt, 2001). BR-deficient det2-1 and BR-insensitive bin2-1 mutants displayed salt sensitive phenotype in seed germination and seedling growth compared with the wild-type plants (Zeng, Tang, & Hua, 2010). The exogenous application of BR significantly increases the salt stress tolerance during seed germination and in postgerminative growth stages (Kagale, Divi, Krochko, Keller, & Krishna, 2007; Ozdemir, Bor, Demiral, & Turkan, 2004). BRs signaling pathways were extensively studied in Arabidopsis (Clouse, 2011; Clouse & Sasse, 1998; T. W. Kim & Wang, 2010). BRI1 kinase inhibitor 1 (BKI1), a membrane-associated protein, functions as a negative regulator of the BR receptor, BR insensitive 1 (BRI1). In the presence of BR, BKI1 could be phosphorylated by BRI1 that results in delocalization of BKI1 from plasma membrane into the cytoplasm and release of BRI1 from the inhibition by BKI1 (X. Wang & Chory, 2006). The plasma membrane-dissociated and phosphorylated BKI1 also plays positive roles in BR signaling by interacting with a subset of 14-3-3 proteins. The cytosolic BKI1 antagonizes the 14-3-3 s and enhances the BRI1 EMS suppressor 1 (BES1)/brassinazole resistant 1 (BZR1) accumulation in the nucleus to activate the BR responsive gene expression (H. Wang et al., 2011).
In the present study, we reported that the pub30-1 and its allelic mutant pub30-3 are more tolerant to salinity stress in the seed germination stage. PUB30 plays an antagonistic role with its homologous protein PUB31 in seed germination although they share 81% identity. We showed that PUB30 possessed E3 ubiquitin ligase activity in vitro and interacted with BKI1 in the yeast two-hybrid assay. Pull-down and co-immunoprecipitation assays further confirmed the specificity of PUB30 over PUB31 for the interaction with BKI1. The accumulation of BKI1 protein was observed after treatment with the proteasome inhibitor MG132. BKI1 protein was ubiquitinated and degraded by the proteasome. We found that the cell-free degradation of BKI1 was slowed down in the pub30-1 mutant compared with the wild type. The transgenic seedlings overexpressing BKI1 showed salt-tolerance phenotype, whereas the bki1 mutant displayed hypersensitivity to salt stress, suggesting that BKI1 is a positive regulator in salt stress tolerance mechanism. Overall, these results suggest that PUB30 negatively regulates salt tolerance through facilitating BKI1 degradation.
2 MATERIALS AND METHODS
2.1 Plant materials, growth conditions and stress treatments
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild type. T-DNA inserted mutants, pub30-1 (Salk_012549), pub30-3 (Salk_073739), and pub31 (Salk_054774), were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH, USA). Seeds were sterilized with 20% bleach and 0.1% Triton-X100 for 10 min and washed 5 times with sterilized water, and then sown on plastic Petri plates containing half-strength Murashige–Skoog (MS) medium (Duchefa Biochemie, Haarlem, The Netherlands) including 1% sucrose and 0.3% phytogel (Sigma-Aldrich, St Louis, MO, USA) with or without 150 mM NaCl. The seeds were laid in the refrigerator at 4 °C for 3 days and transferred to the incubator at 23 °C with continuous light with an intensity of 120 μmol m−2 s−1.
2.2 Real-time quantitative reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was isolated from 10-day-old seedlings sown on 1/2 MS medium using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) following the manufacturer's instructions. Total RNA (2 μg) was reverse-transcribed using Transcript All-in-one First-Strand cDNA Synthesis Supermix for qPCR Kit (Transgene, Beijing, China) and equal quantities of reverse-transcribed products were used for the real-time quantitative RT-PCR reactions. Quantitative RT-PCRs were performed using a SYBR Premix Ex Taq™ RT-PCR kit (Takara, Shiga, Japan) following the manufacturer's instructions with the primers listed in Table S1.
2.3 Yeast two-hybrid assay
In the initial library screening step, full-length PUB30 was used as a bait to screen a cDNA library made from 12-day-old Arabidopsis seedlings with the Matchmaker Library Construction & Screening Kit (Clontech). To confirm the interaction between PUB30 and BKI1, the full-length coding sequence of PUB30 and PUB31 were amplified and cloned into the pGBKT7 (BD-PUB30 and BD-PUB31) and BKI1 was inserted into pGADT7 (AD-BKI1). The polymerase chain reaction (PCR) primers used for plasmid construction are listed in Table S2. The combinations of BD-PUB30 and AD-BKI1, BD-PUB31 and AD-BKI1, BD-PUB30 and AD, BD-PUB31 and AD, and BD and AD-BKI1 were transformed into yeast strain AH109. Yeast synthetic drop-out media lacking leucine (Leu) and tryptophan (Trp; Coolaber, Beijing, China) was used to identify transformed colonies. Three days after transformation, selective colonies were grown in –Leu and −Trp liquid media for 24 h. Then, cell suspension was pipetted onto yeast synthetic drop-out media lacking histidine (His), Leu, and Trp (Coolaber, Beijing, China) with 2.5 mM 3-amino-1,2,4-triazole (3-AT, Sigma-Aldrich, St Louis, MO, USA).
2.4 Pull-down assay
All of the PCR primers used for plasmid construction are listed in Table S2. In order to express the PUB30, PUB31, and BKI1 proteins in Escherichia coli DE3 cells, the coding sequence of PUB30, PUB31, and BKI1 were amplified and individually cloned into pCold-MBP, pCold-MBP, and pCold-GST vectors with infusion cloning kit (Takara, Shiga, Japan). The glutathione S-transferase (GST) and maltose binding protein (MBP) tags were fused to the N-terminus of target proteins (pCold-MBP-PUB30, pCold-MBP-PUB31, and pCold-GST-BKI1).
In pull-down assay, 2 μg of GST-BKI1 protein was incubated with 10 μg of MBP-PUB30 or MBP-PUB31 that was attached onto the agarose beads for 30 min at room temperature in 100 μl of binding buffer (20 mM Tris–HCl, pH 7.2, 10 mM MgCl2, 2 mM DTT, and 5% NP40). After 5 times washing with binding buffer, the beads were boiled with sodium dodecyl sulfate loading buffer, and samples were separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and detected with 1:3,000 GST or 1:5,000 MBP antibody (CWBIO, Beijing, China). The horseradish peroxidase-conjugated anti-mouse IgG (H + L; 1:3,000 dilution, CWBIO, Beijing, China) was used as the second antibody. The western blots were developed with enhanced chemiluminescence reagent (Coolaber, Beijing, China).
2.5 Co-immunoprecipitation assay
The full-length coding sequence of PUB30, PUB31, and BKI1 were amplified and individually cloned into the vector 1307-FLAG and 1307-Myc (Zhao et al., 2016). The primers used for plasmid construction are listed in Table S2. The combinations of 1307-FLAG-PUB30 and 1307-Myc-BKI1 and 1307-FLAG-PUB31 and 1307-Myc-BKI1 were transformed to Arabidopsis mesophyll protoplast according to the standard protocols (Sheen, 2001). The transfected protoplasts were incubated overnight at 23 °C and homogenized in immunoprecipitation buffer containing 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM DTT, 0.1% Triton, and 1% protease inhibitor cocktail (Roche, Indianapolis, IN, USA). The sample was centrifuged at 12,000 rpm for 10 min at 4 °C, and the supernatant was incubated with agarose-conjugated anti-Myc antibody (Sigma-Aldrich, St Louis, MO, USA) for 3 hr at 4 °C. After 5 times washing with immunoprecipitation buffer, the protein were separated on 10% SDS-PAGE gel and subjected to immunoblotting using anti-Myc antibody (CWBIO, Beijing, China) and anti-FLAG antibody (CWBIO, Beijing, China).
2.6 In vitro self-ubiquitination assay
In vitro self-ubiquitination assay was carried out according to the method reported by Xie et al. (2002). MBP-PUB30 (500 ng) was immobilized on amylose resin beads. UBA1, UBC8, and UBQ14 were individually cloned into the pET28a vector to express E1, E2, and ubiquitin. The primers used for plasmid construction are listed in Table S2. Ubiquitination assays were carried out by adding 200 ng UBA1 (E1), 100 ng UBC8 (E2), and 2 μg ubiquitin in the buffer containing 50 mM Tris–HCl, pH 7.4, 2 mM ATP, 5 mM MgCl2, and 2 mM DTT. The reaction mix (30 μl) was incubated for 2 hr at 30 °C. The reaction products were separated on 6% SDS-PAGE gel and subjected to immunoblot using anti-MBP antibody (1:10,000 dilution, CWBIO, Beijing, China) and anti-ubiquitin (anti-Ub) antibody (1:3,000 dilution, Santa Cruz, CA, USA).
2.7 In vivo ubiquitination assay
The in vivo ubiquitination assay was carried out according to the reported method (Irigoyen et al., 2014). Briefly, the wild-type and transgenic plants overexpressing 1307-Myc-BKI1 were grown on MS medium for 10 days and then treated with 50 μM MG132 for 6 hr. Fresh seedlings (0.2 g) were grinded in liquid nitrogen, and the powder was solubilized in 600 μl BI buffer containing 50 mM Tris–HCl, pH 7.5, 20 mM NaCl, 0.1% NP-40 and 5 mM ATP, 1 mM PMSF, 50 μM MG132, 10 nM Ub aldehyde (Sigma-Aldrich, St Louis, MO, USA), and 10 mM N-ethylmaleimide (Coolaber, Beijng, China). After 10 min of centrifugation at 12,000 rpm at 4 °C, the supernatants were incubated with prewashed p62-agarose (Enzo Life Sciences, Farmingdale, USA) for 3 hr, and then the agarose was washed twice with BI buffer and once with BII buffer (supplemented with 200 mM NaCl in BI buffer). The proteins were separated on 10% SDS-PAGE gel and subjected to immunoblotting using anti-Myc antibody (CWBIO, Beijing, China) and anti-Ub antibody (Santa Cruz, CA, USA).
2.8 In vitro cell-free degradation assay
Ten-day-old seedlings of pub30-1 and wild type were harvested and grinded into fine powder in liquid nitrogen. Total proteins were subsequently extracted in degradation buffer containing 25 mM Tris–HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT, and 10 mM ATP (F. Wang et al., 2009). After 10-min centrifugation at 17,000 g, the supernatants were adjusted to the equal concentration with degradation buffer. Purified GST-BKI1 protein (100 ng) was incubated with 50 μl total protein extracts at 23 °C. The products were subjected to western blotting with anti-GST antibody (CWBIO, Beijing, China).
2.9 In vivo degradation assay
Ten-day-old transgenic seedlings overexpressing Myc-BKI1 were pretreated with MG132 for 16 hr and then washed 5 times before being transferred to liquid MS medium containing 200 mM cycloheximide (Sigma-Aldrich, St Louis, MO, USA) to block de novo protein synthesis. The samples were harvested after 0 min, 90 min, and 180 min, and the seedlings were grinded in liquid nitrogen and solubilized with extraction buffer (100 mM Tris–HCl, pH 7.5, 300 mM NaCl, 2 mM EDTA, pH 8.0, 1% TrionX-100, 10% glycerol, and a protease inhibitor cocktail). After 10-min centrifugation at 12,000 rpm at 4 °C, the supernatant was incubated with 5 × sodium dodecyl sulfate loading buffer and separated by 10% SDS-PAGE gel and analysed by immunoblotting with anti-Myc antibody at 1:3,000 dilution (CWBIO, Beijing, China).
3 RESULTS
3.1 The pub30 mutants are more tolerant to salinity stress during seed germination stage
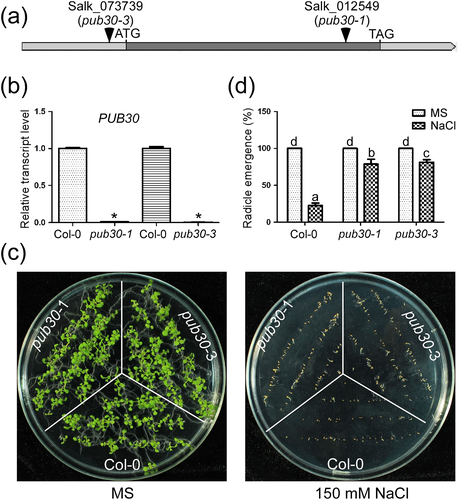
Hwang et al. (2015) reported that the Arabidopsis pub30 mutant was more tolerant to salt stress in the germination stage compared with the wild type. However, little is known about the underlying mechanism for the salt-tolerance phenotype of the pub30 mutant. Here, we aimed to elucidate the potential mechanism using two T-DNA insertion mutants, that is, pub30-1 (Salk_012549) and pub30-3 (Salk_073739; Figure 1a). Real-time PCR analysis showed that the full length transcript of PUB30 could not be detected in pub30-1 and pub30-3 mutants, suggesting that pub30-1 and pub30-3 are knockout mutants (Figure 1b). To investigate the function of PUB30 in salt stress responses, seeds of the wild type and the two pub30 knockout mutants were germinated on 1/2 strength MS medium with or without 150 mM NaCl (Figure 1c). The untreated wild-type and mutants seeds grew normally with 100% germination rate (Figure 1c and 1d). The radicle emergence rate of both wild type and mutants decreased with 150 mM NaCl treatment. The wild-type seeds were affected severely with only 22.66% emergence rate, whereas the mutants pub30-1 and pub30-3 recorded 78.75% and 81.35% radicle emergence rate, respectively (Figure 1c and 1d). A similar trend was observed when the wild-type and pub30-1 mutant seeds germinated on MS medium containing two other concentrations of NaCl (100 and 125 mM; Figure S1). These results suggest that the pub30 mutants are more tolerant to salt stress in seed germination.
3.2 Functional antagonism between two highly homologous proteins PUB30 and PUB31 in response to salt stress
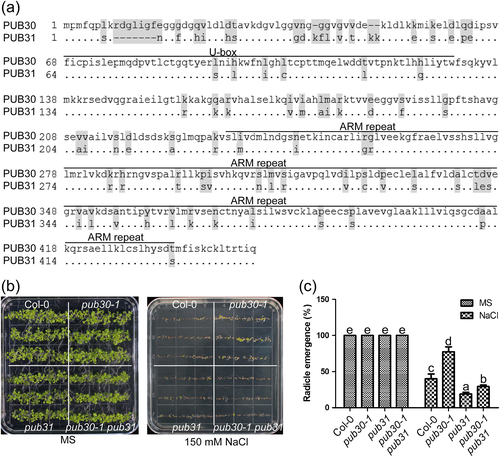
According to homologous sequence alignment, we found PUB30 shares 81% identity with PUB31 (Figure 2a). Both proteins contain a U-box domain that is essential for E3 ubiquitin ligase activity at the N-terminal region and several Armadillo (ARM)-repeat motifs that mediate protein–protein interaction at the C-terminal region (Figure 2a). In order to determine whether PUB31 has functional redundancy with PUB30, we obtained the pub31 mutant (Salk_054774) from ABRC. Quantitative RT-PCR results showed that the full length transcript of PUB31 could not be detected in the pub31 mutant (Figure S2). The pub30-1 pub31 double mutant was generated and used to observe the phenotype. There was no significant difference among the wild type, pub30-1, pub31, and pub30-1 pub31 double mutant during seed germination when they grow on normal MS medium for 12 days. However, when they grow on the MS medium containing 150 mM NaCl for 12 days, the pub31 mutant is a little sensitive to salt stress compared with the wild type, in contrast to the salt-tolerant phenotype of the pub30-1 mutant. The double mutant pub30-1 pub31 behaved in between the pub30-1 and pub31 single mutant for salt stress response and resembled the pub31 mutant phenotype in radicle emergence rate (Figure 2b and 2c). In the presence of 150 mM NaCl, the radicle emergence rate of the wild type, pub30-1, pub31, and the pub30-1 pub31 double mutant is 40.01%, 77.39%, 18.88%, and 24.80%, respectively (Figure 2c). These results suggest that PUB31 has a functional antagonism with PUB30 in salinity stress response.
3.3 Expression pattern and subcellular localization of PUB30
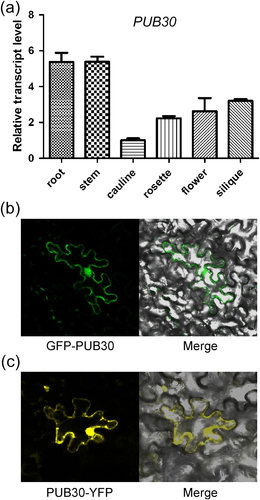
To examine the expression pattern of PUB30, real-time quantitative RT-PCR was used to detect the expression of PUB30 in different tissues, including root, stem, cauline leaf, rosette leaf, and silique. Relative higher expression level was found in root and stem, lower level in cauline and rosette leaves (Figure 3a). To explore the subcellular localization of the PUB30 protein, we constructed two plant expression vectors. In the GFP-PUB30 vector, green fluorescent protein (GFP) was fused to the N-terminus of PUB30 genomic sequence driven by the PUB30 native promoter. In the PUB30-YFP vector, yellow fluorescent protein (YFP) was fused to the C-terminus of PUB30 driven by a 35S promoter. These two vectors were transiently expressed in tobacco leaves. After incubation for 5 days, we found that these two vectors showed similar localization pattern where PUB30 was extensively localized in the cytoplasm, nucleus, and plasma membrane (Figure 3b and 3c). Additionally, the transgenic seeds harboring GFP-PUB30 in the pub30-1 mutant background were used to detect the radicle emergence on the MS medium containing 150 mM NaCl. The results showed that radicle emergence rate of two independent transgenic lines was reduced compared with that of the pub30-1 mutant, implying that the GFP-PUB30 fusion protein is functional in complementing the salt-tolerant phenotype of the pub30-1 mutant (Figure S3).
3.4 PUB30 is a functional U-box E3 ubiquitin ligase
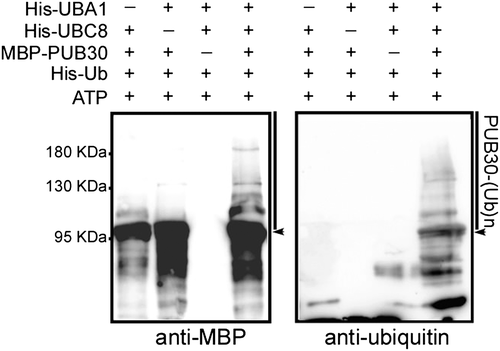
To examine whether PUB30 is a functional E3 Ub ligase, we produced a full-length Arabidopsis PUB30 protein fused with MBP in E. coli and subsequently affinity purified from the soluble fraction. In vitro self-ubiquitination assays were performed in the presence of Arabidopsis E1 (UBA1), Arabidopsis E2 (UBC8), and His-tagged Ub. Polyubiquitination was detected by anti-Ub or anti-MBP antibody only when E1, E2, and MBP-PUB30 were all present. When any of them were omitted in the assay, polyubiquitination was not detected (Figure 4). This result suggests that PUB30 possesses E3 Ub ligase activity in vitro.
3.5 PUB30 interacts with BKI1 in vitro and in vivo
To investigate whether PUB30 affects the salt stress response via ubiquitination of target protein, we performed yeast two-hybrid assay to screen its target using PUB30 as bait. Some positive clones were isolated from the cDNA library made from 12-day-old Arabidopsis seedlings. One of these clones encoded the full length BKI1 protein. The interaction between PUB30 and BKI1 was confirmed by cotransformation of AD-BKI1 and BD-PUB30 into the yeast cells (Figure 5a). Although PUB30 has highest identity (81%) with PUB31 in Arabidopsis, they have functional antagonism in response to salt stress (Figure 2). In order to investigate whether PUB31 also interacts with BKI1, AD-BKI1 and BD-PUB31 were cotransformed into the yeast cells. The result showed that PUB31 does not interacts with BKI1 although PUB30 does (Figure 5a). These results suggest that BKI1 may be the target of PUB30 in salt tolerance mechanism.
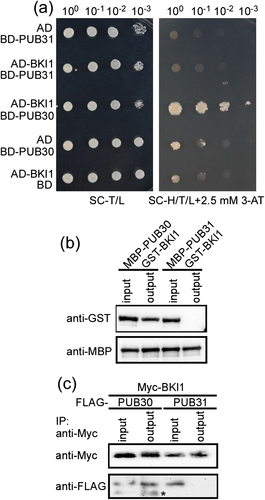
To further confirm the interaction between PUB30 and BKI1, we performed pull-down assay. PUB30 and PUB31 cDNA were cloned into pCOLD-MBP vector (Takara) containing a MBP tag, individually. MBP-PUB30 and MBP-PUB31 fusion proteins were purified from E. coli strain BL21 (DE3) with amylose resin (BioLabs). BKI1 cDNA was inserted into pCOLD-GST vector (Takara) with a GST tag. GST-BKI1 was purified with Glutathione Sepharose™4B (GE Healthcare) and eluted with reduced GST. In vitro pull-down assays showed that GST-BKI1 was pulled down by MBP-PUB30 but not by MBP-PUB31 (Figure 5b). This result further confirmed that PUB30 specifically interacts with BKI1.
To confirm the interaction between PUB30 and BKI1 in vivo, FLAG tag was translationally fused to the N-terminal region of PUB30 or PUB31 and 6 × Myc tag was translationally fused to the N-terminal region of BKI1. These gene fusions were driven by a double 35S promoter. The resulting vector Myc-BKI1 was cotransfected with FLAG-PUB30 or FLAG-PUB31 constructs into the wild-type leaf protoplasts. Total extracted protein was used to detect the expression of Myc-BKI1 and FLAG-PUB30 or Myc-BKI1 and FLAG-PUB31 in each transformation with anti-Myc antibody or anti-FLAG antibody (Figure 5c). Myc-BKI1 protein was then immunoprecipitated with anti-Myc affinity gel. The result showed that only FLAG-PUB30 is pulled-down by Myc-BKI1 and FLAG-PUB31 is not (Figure 5c). This result suggests that PUB30 specifically interacts with BKI1 in vivo.
3.6 BKI1 protein is ubiquitinated and degraded by the proteasome
U-box E3 Ub ligase specifically recognizes substrate proteins and target them for degradation by the 26S proteasome by attaching a polyubiquitin chain. Thus, we first examined whether BKI1 is degraded by the 26S proteasome. The Arabidopsis seedlings expressing Myc-BKI1 were treated with 50 μM proteasome inhibitor MG132 for different time. The total protein was extracted to detect the protein level of Myc-BKI1. The result showed that the level of Myc-BKI1 protein increased gradually as the incubation time with MG132 was prolonged (Figure 6a). Serine hydroxymethyltransferase 1, which is degraded in a 26S proteasome-dependent manner, was used as a positive control of MG132 treatment (Zhou et al., 2012). This suggests that BKI1 protein could be degraded by the 26S proteasome.
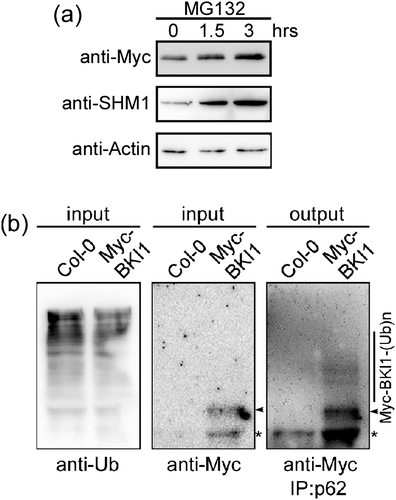
Then to check whether BKI1 protein is ubiquitinated in vivo, we used a p62-agarose matrix that is capable of binding ubiquitinated proteins (Irigoyen et al., 2014) to enrich the ubiquitinated proteins from the transgenic plants overexpressing Myc-BKI1. When the p62 resin-enriched protein fraction was detected using anti-Myc antibodies, multiple high molecular mass bands above the Myc-BKI1 band were revealed in the transgenic plants but not in the wild type, indicating that Myc-BKI1 is modified by polyubiquitin chains in planta (Figure 6b).
3.7 PUB30 mediates BKI1 protein degradation
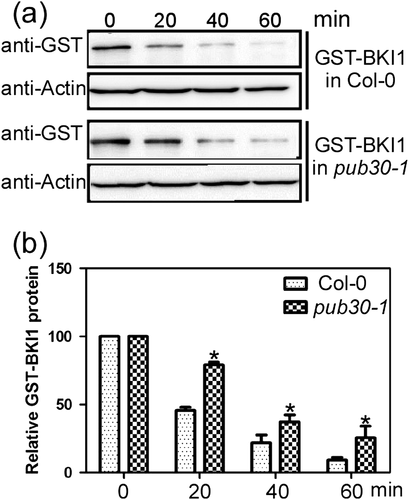
To determine whether PUB30 is required for BKI1 degradation, we used in vitro cell-free system to detect the degradation of GST-BKI1 protein expressed in E. coli. The total protein was extracted from Col-0 and the pub30-1 seedlings and incubated with GST-BKI1 protein at 23 °C for 0, 20, 40, and 60 min. Subsequently, immunoblot assay was performed with anti-GST antibody. Initially, there was no difference in the protein level of GST-BKI1 between the wild-type and the pub30-1 mutant extract. However, the protein level of GST-BKI1 was reduced more rapidly in the wild-type than in the pub30-1 mutant extract as the incubation time was increased (Figure 7a and 7b).
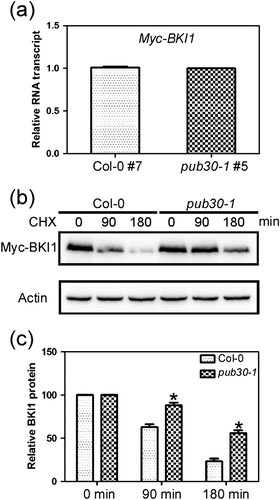
To further confirm whether BKI1 protein is targeted by PUB30 for ubiquitin-mediated degradation in vivo and to compare the BKI1 protein dynamics in the wild-type and the pub30-1 mutant, we performed an in vivo degradation assay. In this assay, the wild-type and the pub30-1 mutants were transformed with Myc-BKI1. Two transgenic lines showing comparable transcript levels of Myc-BKI1 were selected from both the wild-type and the pub30-1 mutant genotypes (Figure 8a). To increase the starting level of the Myc-BKI1 protein, these two transgenic lines were pretreated with MG132 for 16 hr as described previously (Jung et al., 2015) and then washed 5 times before being transferred to liquid MS medium containing 200 mM cycloheximide for 90 or 180 min. Total protein was isolated from the two transgenic lines and used to detect the degradation of Myc-BKI1. The immunoblot assays showed that degradation of the Myc-BKI1 protein was slowed down in the pub30-1 mutant compared with the wild type (Figure 8b and 8c). All these results suggest that BKI1 undergoes 26S proteasome-dependent degradation, and PUB30 is involved in this process.
3.8 BKI1 functions in salinity stress responses
In order to functionally characterize BKI1 in seed germination under salt stress, the transgenic seeds harboring Myc-BKI1 were sown on 1/2 MS medium with or without 150 mM NaCl for 9 days. In contrast to the wild-type, transgenic plants overexpressing BKI1 showed higher radicle emergence rate on MS medium containing 150 mM NaCl, that is, 34.42%, 87.58%, and 11.43% for transgenic line Col-0 #4, Col-0 #7, and the wild type, respectively (Figure 9a and 9c). The radicle emergence rate was positively correlated with the transcript level of BKI1 in transgenic plants Col-0 #7, Col-0 #4, and the wild type (Figure 9b).
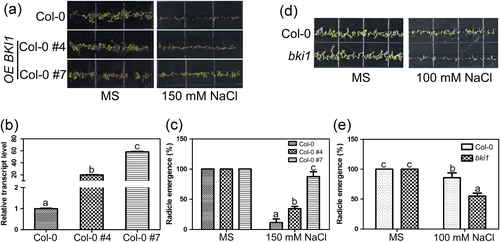
In addition, we also used the bki1 loss-of-function mutant (Jiang et al., 2015) to characterize its function in seed germination under salt stress. When the wild-type and the bki1 mutant were grown on normal MS medium, no significant difference in seed germination was observed (Figure 9d). The radicle emergence of the wild-type and the bki1 mutant grown on MS medium was almost 100% (Figure 9e). However, the bki1 mutant displayed salt-hypersensitive phenotype when germinated on MS medium containing 100 mM NaCl (Figure 9d). The fraction of seeds with radicle emergence was only 55% for the bki1 mutant, compared with 85.8% for the wild type (Figure 9e). These results suggest that BKI1 is a positive regulator of the salt-tolerance phenotype.
4 DISCUSSION
In this study, we found that the pub30-1 mutant is more tolerant to salt stress in seed germination stage compared with the wild type. The PUB30 gene encodes a U-box E3 Ub ligase that possesses E3 Ub ligase activity in vitro. PUB30 specifically interacts with BKI1 in vitro and in vivo. PUB30 regulates the stability of BKI1 protein, and the transgenic plants overexpressing Myc-BKI1 showed salt tolerance phenotype similar to the pub30-1 mutant phenotype during seed germination. Although PUB30 has highly identical sequence with its homologous protein PUB31 at the amino acid level, PUB30 and PUB31 have a reverse functionality in salinity stress tolerance in seed germination. The pub30-1 mutant is tolerant to salt stress, whereas the pub31 mutant is sensitive to salinity stress in germination stage. Phenotype of the pub30-1 pub31 double mutant resembles the pub31 mutant, suggesting the epistatic effect of PUB31 in determining sensitivity to salt stress. It was previously reported that several highly homologous proteins can have reverse functionality. In Arabidopsis, two closely related RecQ helicases AtRECQ4A and 4B share nearly 70% identity at the protein level. However, knockout of these two genes leads to antagonistic phenotypes. RECQ4A mutants show sensitivity to DNA-damaging agents and partially suppress the lethal phenotype of an AtTOP3á, a phenomenon that has previously been demonstrated for RecQ homologues of unicellular eukaryotes only. However, RECQ4B is not mutagen-sensitive and unable to suppress the induced lethality caused by loss of TOP3á (Hartung, Suer, & Puchta, 2007). Arabidopsis trithorax-like protein 1 and Arabidopsis trithorax-like protein 2 share 65% identity and 75% similarity at the protein level. However, they have functional divergence in epigenetic modification owing to their different regulatory sequences (Saleh et al., 2008).
Armadillo-repeats in U-box E3 Ub ligase are motifs that mediate protein–protein interaction, which is responsible for interaction with the substrate proteins. The ARM-repeats of PUB30 and PUB31 share 84% identity at the protein level (Figure 2a). However, only PUB30 can specifically interact with BKI1, suggesting that some amino acids in ARM-repeat are necessary for the interaction between PUB30 and BKI1. In order to elucidate the function of PUB30, identifying these amino acids in ARM-repeat would be necessary.
It is reported that the pub30-1 mutant shows salt-tolerant phenotype only in seed germination stage, whereas there is no significant difference between the pub30-1 mutant and the wild type in postgerminative growth in response to salt stress (Hwang et al., 2015). However, in our in vitro (Figure 7) and in vivo (Figure 8) degradation assay of BKI protein, 10-day-old seedlings of WT, pub30-1, and MYC-BKI1 transgenic plants in both genotypes were used because it was very difficult to harvest enough plant sample for protein degradation assay at the seed germination stage. Yet defects in BKI1 destabilization could be observed in the seedling stage of the pub30-1 mutant. These results suggest that the regulation of BKI1 stability by PUB30 still exists after the germination stage. However, the pub30 mutant has no salt-tolerant phenotype at later stages. It is possible that functions of the PUB30 and BKI1 protein or their interaction may be affected by other biochemical processes, which hinder the pub30 mutant to display salt tolerant phenotype at this stage.
The negative effect of PUB30 on salt stress tolerance seems to be limited to the seed germination stage (Hwang et al., 2015). However, BKI1 positively regulates salt tolerance response not only in the seed germination stage but also in seedling growth stage. We transferred the 5-day-old seedlings of wild type, the bki1 mutant, and transgenic plants overexpressing BKI1 to MS medium containing 0, 125, and 150 mM NaCl and kept them growing for 20 days. Without salt treatment, the primary root length of the bki1 mutant and transgenic seedlings overexpressing BKI1 were indistinguishable from that of the wild type, although the fresh weight of the bki1 mutant was less than other seedlings (Figure S5). Under salt stress treatment, the bki1 mutant was more sensitive to salt than wild type as illustrated by the retarded growth of primary root, whereas transgenic lines overexpressing Myc-BKI1 were more tolerant to salt as illustrated by the increased primary root length and fresh weight than wild type (Figure S5). These results suggest that BKI1 activity help plants to cope with high salinity at both seed germination and later stages during plant development.
It was reported that the transcript level of BKI1 is induced by 150 mM salt (Hwang et al., 2015). In our study, we found that the transcript level of BKI1 is much higher in the pub30-1 mutant compared with the wild type grown without salt treatment. In addition, BKI1 transcript level increased significantly in the pub30-1 mutant when seedlings were treated with 150 mM NaCl (Figure S6). However, the protein level of BKI1 could not be induced by salt stress (Figure S7). We speculate that the U-box E3 Ub ligase PUB30 may be the crucial player in regulating the protein level of BKI1.
All these results indicated that BKI1 is the target of PUB30 in response to salt stress in seed germination stage. The loss-of-function mutant bki1 was sensitive to salt while the transgenic plants overexpressing BKI1 showed salt-tolerant phenotype just like the pub30 mutants. However, the intrinsic molecular mechanism of BKI1 as a positive regulator in salt tolerance is unknown. BKI1 is a well-known component in BRs signal transduction. BKI1 has dual functions in BR signaling. BKI1 localized in the plasma membrane acts as a negative regulator that interacts with BRI1 and inhibits the phosphorylation activity of BRI1 (Jailais et al., 2011; X. Wang & Chory, 2006). Once BKI1 dissociates from the plasma membrane, cytosolic BKI1 interacts with a subset of 14-3-3 proteins, which can also bind the phosphorylated BES1/BZR1. Phosphorylated BKI1 antagonized 14-3-3 proteins and enhance the accumulation of BZR1 and BES1 in the nucleus to regulate the down-stream genes expression (H. Wang et al., 2011). Earlier reports have proven that BR obviously enhances the tolerance of plants to abiotic stresses (Kagale et al., 2007; Ozdemir et al., 2004). In BR signaling, BR induces BKI1 protein to dissociate from the plasma membrane to cytoplasm. We hypothesize that salt can act as a signal similar to BR to induce BKI1 dissociation from plasma membrane to activate BR signal. In order to prove this hypothesis, the transgenic seedlings harboring BKI1-YFP were treated with 50 mM NaCl, instead of 150 mM NaCl, because the high concentration of salt results in plasmolysis in a short period of time, which hinders to observe the dissociation of BKI1 from the plasma membrane. As a positive control, we employed the most active form of BR (brassinolide, BL) to treat the transgenic seedlings and found that BKI1-YFP signal transfers to the cytoplasm to form vesicles compared with the mock (Figure S8). When 50 mM NaCl was used, we found that BKI1-YPF also dissociated from the plasma membrane and formed many vesicles in the cytoplasm (Figure S8). Therefore, similar to BL, salt can also induce the dissociation of BKI1 from plasma membrane. BKI1 protein accumulates in the pub30-1 mutant in contrast to the wild type, which may enhance BR signaling in the pub30 mutants, resulting in tolerance to salinity in seed germination stage. Additionally, we found that low concentration of BL obviously promotes the seed germination. The pub30-1 mutant showed more vigorous early root growth than the wild type when exogenous BL was applied, suggesting that the intracellular BR signal maybe enhanced in the pub30-1 mutant (Figure S9). Further studies are necessary to elucidate the BR signaling in the wild type and the pub30 mutant under salt stress conditions and to understand the underlying molecular mechanism of BKI1 and PUB30 in response to salt stress.
According to a previous study (Hwang et al., 2015), the pub30 mutant showed normal response to ABA at the seed germination stage. To determine whether the bki1 mutant has altered responsiveness to ABA at the seed germination stage, we sowed the wild-type and the bki1 mutant seeds on MS with or without ABA. The results showed that there was no significant difference between the wild-type and the bki1 mutant in radicle emergence when grown on MS or MS containing different concentrations of ABA (Figure S4). These results suggest that PUB30 and BKI1 are not involved in response to ABA at seed germination stage.
ACKNOWLEDGMENTS
We thank Dr X. L. Wang at the Huazhong Agricultural University for the generous gift of the bki1 mutant. We also thank Dr Y. F. Fu at the Institute of Crop Sciences, China Academy of Agricultural Sciences to help us order the T-DNA insertion lines from ABRC. We also appreciate Dr Q. Xie at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for technical assistance with in vitro self-ubiquitination assay. This work was supported by the National Natural Science Foundation of China (Grant 31570280 to J. Z.), the Agricultural Science and Technology Innovation Program of China and a Core Research Budget of the Non-profit Governmental Research Institution (Institute of Crop Science, Chinese Academy of Agricultural Sciences), and Central Public-interest Scientific Institution Basal Research Fund. No conflict of interest declared.




