Development of an autonomously replicating viral expression system tailored for Catharanthus roseus
Catharanthus roseus is an important medicinal plant with a capacity to synthesize >130 monoterpinoid indole alkaloids (MIA). Many of these compounds, including the chemotherapeutics vinblastine, vincristine and their chemical derivatives have high economic value with diverse applications. Vincristine is the only effective antileukaemic drug that can drastically reduce white blood cell counts. Since the 1950s, it has increased the survival rate of children with leukaemia from 20% to 80% (Nejat et al., 2015). However, due to its scarcity, vincristine is one of the most expensive plant-derived compounds on the market and supply cannot meet demand (Kumar et al., 2013). Total chemical synthesis of vinblastine and vincristine is inefficient due to their structural complexity and stereochemistry and industrial production relies on the extraction of precursors from C. roseus in which the MIAs accumulate in trace amounts; for example 5.0 and 0.5 ppm for vinblastine and vincristine, respectively. Accordingly, recent attention has shifted to the development of new approaches to elevate MIA levels in C. roseus. The MIA biosynthetic pathway, however, is intricate and composed of around 30 to 50 enzymatic steps with multifaceted metabolic regulation involving intracellular compartmentalization, different cell types and tissue differentiation, making it a complex target for metabolic engineering (Dugé de Bernonville et al., 2015, Nejat et al., 2015). Furthermore, unravelling the MIA biosynthetic pathway and its regulation in C. roseus has been hindered by a paucity of technologies to express candidate genes in planta. With the exception of a virus-induced gene silencing (VIGS) system (Liscombe and O'Connor, 2011), genomic approaches that enable gene functions to be explored in vivo have been most effective in (and often limited to) hairy root and cell suspension cultures. These systems lack the required level of cyto- and tissue-differentiation essential for the expression of MIA pathway genes and, therefore, do not truly reflect MIA biosynthesis and its regulation in planta (Liscombe and O'Connor, 2011). To overcome this, we have utilized the replication machinery of a C. roseus-infecting geminivirus to develop an autonomously replicating viral expression system that provides consistent and high gene expression in C. roseus leaves.
Geminiviruses are small, circular, single-stranded DNA viruses that infect monocotyledonous and dicotyledonous plants in tropical and subtropical regions. The family Geminiviridae is divided into nine genera, of which the Begomovirus genus is the largest consisting of both monopartite and bipartite genome types (Hanley-Bowdoin et al., 2013). Geminiviruses replicate their genomes exclusively in the nucleus of the infected host cell via a rolling circle replication process (Figure 1b) initiated at a consensus stem loop structure located in the intergenic region (IR). The IR also serves to direct divergent transcription of both virion and complementary sense genes. A coat protein (CP) forms the viral capsid and mediates vector transmission, V2 and AV2 function as anti-defence proteins to inhibit post-transcriptional gene silencing (PTGS) and the Replication initiator protein (Rep) initiates viral replication. Begomoviruses encode three additional gene products, the transcriptional activator protein (TrAP; and the related C2 protein) interfere with transcriptional gene silencing (TGS) and PTGS, the replication enhancer protein (REn) is involved in viral replication and the C4 protein counteracts PTGS (Hanley-Bowdoin et al., 2013). In 2012, the complete nucleotide sequence of a bipartite begomovirus was isolated from C. roseus, the virus was named Catharanthus yellow mosaic virus (CaYMV) (Ilyas et al., 2013).
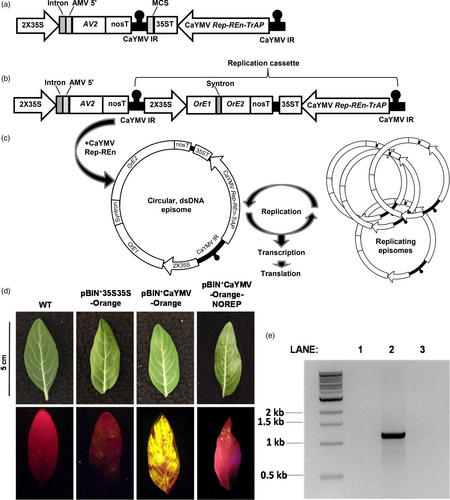
We designed a CaYMV-based replicating vector system for transient gene expression in C. roseus; illustrated in Figure 1a. The system contains the CaYMV Rep, Ren and TrAP genes under the transcriptional control of their native promoter sequences and located between repeats of the CaYMV IR. A cauliflower mosaic virus (CaMV) 35S 3′ UTR was inserted downstream of REn to effectively terminate transcription. The CP gene was not included in order to render the system non-infectious. Outside the replicative unit, but within the T-DNA, was located an expression cassette containing the CaYMV AV2 gene under the transcriptional control of an enhanced CaMV 35S promoter (2X35S), potato gbbs gene intron IV and alfalfa mosaic virus translational enhancer. The AV2 gene was included in the expression system based on its anti-defence role during virus infection. AV2 was placed outside of the replication unit to moderate its expression as AV2 proteins can trigger a hypersensitive response (Mubin et al., 2010). A multiple cloning site (MCS) was included in the replication unit in order to insert the transgene of interest.
The CaYMV-based cassette was chemically synthesized and mobilized into the binary plasmid pBIN+ to generate vector pBIN+CaYMV (Figure 1a). An expression cassette containing an Orange florescent protein reporter gene sequence and synthetic intron (syntron) under the transcriptional control of a 2X35S promoter was inserted into pBIN+CaYMV as an AsiSI-AvrII restriction fragment to create vector pBIN+CaYMV-Orange (Figure 1b). The syntron was included to prevent Orange expression in Agrobacteria. Following Rep-initiated replicative release, the single-stranded, circular episomal molecule is converted to a double-stranded DNA form via the origin of second strand synthesis and by host polymerases (Figure 1c). This form is both transcriptionally active and serves as a template for further rolling circle amplification resulting in high episomal copy number and high transgene expression levels. The vector pBIN+CaYMV-Orange was mobilized into Agrobacterium tumefaciens (strain AGL1) by electroporation and agroinfiltrated into C. roseus leaves via the lower epidermis with a needle-less syringe, at an OD600 of 1.5.
Transient expression of Orange in C. roseus leaves agroinfiltrated with pBIN+CaYMV-Orange was assessed by visualizing florescence with a Dual Florescent Protein (DFP) flashlight and a Royal Blue LED and Yellow filter. Six days after agroinfiltration, fluorescence was compared to wild-type (WT) tissue and leaves infiltrated with a non-replicating vector (pBIN+2X35S-ORANGE). Expression of Orange was significantly higher and more uniform in C. roseus leaves agroinfiltrated with pBIN+CaYMV-Orange (Figure 1d). To prove that high level Orange expression was the result of Rep-mediated replication of the expression unit, the Rep gene sequence in pBIN+CaYMV-Orange was disrupted and the REn coding sequence removed, creating vector pBIN+CaYMV-Orange-NOREP. Minimal Orange florescence was observed in leaf tissue agroinfiltrated with this vector (Figure 1d). To demonstrate replicative release from the vector and the production of CaYMV-Orange episomal DNA forms, outwardly extending primers were designed to amplify a 1100 bp sequence spanning the CaYMV IR. These primers were used in a PCR with DNA extracted from leaves agroinfiltrated with pBIN+CaYMV-Orange and pBIN+CaYMV-Orange-NOREP, 6 days after agroinfiltration (Figure 1e). No PCR product was obtained from WT leaves (Figure 1e, Lane 1). A strong PCR product of ~1100 bp was amplified from leaves agroinfiltrated with pBIN+CaYMV-Orange suggesting extra-chromosomal episome formation and Rep-mediated amplification, whereas a very weak ~1100 bp product was amplified from leaves agroinfiltrated with pBIN+CaYMV-Orange-NOREP, indicating host-mediated recombination and low-level episome formation, independent of Rep (Figure 1e, Lanes 2 and 3, respectively).
We have designed an autonomously replicating viral expression platform based on the genome of CaYMV and tailored for high and consistent gene expression in the leaves of C. roseus. Up to this point, the lack of a reliable transgene expression system in this host has been a limitation to better understanding and manipulating MIA biosynthesis and regulation. The CaYMV system could, for example, be used to express candidate genes involved in the transport of MIA intermediates between plant cells and organs, such as a recently proposed but as yet uncharacterized secologanin transporter (Kidd et al., 2019). The platform described here opens up new pathways in MIA research and product development. Our approach also highlights how viruses can be exploited to develop tailored gene expression systems for specific species that are recalcitrant to established gene expression technologies.
Acknowledgements
This work was supported by a discovery early career research award granted to C.M. by the Australian Research Council.
Competing interests
The authors have no competing interests.
Contributions
C.M., B.D. and P.W. designed the experiments, C.M. performed the experiments and analysed the results. CM and BD wrote the manuscript. All authors read and approved the final manuscript.




