A meta-analysis of reflux genome-wide association studies in 6750 Northern Europeans from the general population
Abstract
Background
Gastroesophageal reflux disease (GERD), the regurgitation of gastric acids often accompanied by heartburn, affects up to 20% of the general population. Genetic predisposition is suspected from twin and family studies but gene-hunting efforts have so far been scarce and no conclusive genome-wide study has been reported. We exploited data available from general population samples, and studied self-reported reflux symptoms in relation to genome-wide single nucleotide polymorphism (SNP) genotypes.
Methods
We performed a GWAS meta-analysis of three independent population-based cohorts from Sweden, Finland, and UK. GERD cases (n=2247) and asymptomatic controls (n=4503) were identified using questionnaire-derived symptom data. Upon stringent quality controls, genotype data for more than 2.5M markers were used for association testing. Bioinformatic characterization of genomic regions associated with GERD included gene-set enrichment analysis (GSEA), in silico prediction of genetic risk effects on gene expression, and computational analysis of drug-induced gene expression signatures using Connectivity Map (cMap).
Key results
We identified 30 GERD suggestive risk loci (P≤5×10−5), with concordant risk effects in all cohorts, and predicted functional effects on gene expression in relevant tissues. GSEA revealed involvement of GERD risk genes in biological processes associated with the regulation of ion channel and cell adhesion. From cMap analysis, omeprazole had significant effects on GERD risk gene expression, while antituberculosis and anti-inflammatory drugs scored highest among the repurposed compounds.
Conclusions
We report a large-scale genetic study of GERD, and highlight genes and pathways that contribute to further our understanding of its pathogenesis and therapeutic opportunities.
Key Points
- A genetic component of gastroesophageal reflux disease (GERD) is recognized, though gene-hunting efforts have been scarce.
- We report a meta-analysis of GERD GWA studies from three independent population-based cohorts, identifying 30 independent suggestive signals of association that show concordant risk and functional effects on gene expression.
- The detected associations establish a repository to inform follow-up efforts in independent GERD case-control studies. Our results highlight genes and pathways that contribute to further understanding of GERD pathogenesis and therapeutic opportunities.
1 Introduction
Gastroesophageal reflux disease (GERD) is a common gastrointestinal (GI) disorder, and represents a frequent reason for visits in primary care practices. GERD is currently defined as a condition that develops when the reflux of gastric content causes symptoms or complications.1 The prevalence is high, and is estimated that in Western countries up to 20% of the adult population has typical reflux-related symptoms.2 Heartburn, regurgitation, and difficulty swallowing are common in GERD, but only 50% of sufferers present with a full spectrum of symptoms, hence its diagnosis often represents a challenge.3 GERD can be classified as erosive or non-erosive esophageal reflux disease (NERD), with the former associated with increased risk of esophageal ulcer, esophageal stricture, Barrett's esophagus (BE, histologically proven esophageal columnar metaplasia), and esophageal adenocarcinoma (EA).1
The pathophysiology of GERD is complex and not well understood. Several mechanisms have been proposed over the years, and the definition of GERD has evolved from being a synonymous with esophagitis and hiatus hernia to a motility and acid-peptic disorder.4 Proton pump inhibitors (PPIs), which decrease the acid effects over the esophageal epithelium, are the most effective drugs in GERD treatment and have considerably improved patients quality of life. Histamine-2 receptors may also be effective in GERD, albeit to a much lesser extent than PPIs.5 However, a substantial proportion of cases (up to 40%, especially among NERD patients), still experience symptoms even after optimal acid-suppressive regimen,6 and alternative or add-on therapeutic strategies are therefore needed.
Genetic factors may play a role in the pathogenesis of reflux, as supported by observations of familial clustering and from Swedish and UK twin studies where higher concordance in monozygous (MZ) vs dizygous (DZ) pairs has been observed.7-10 Despite this, GERD gene mapping efforts have been rare and only one GWA study has been recently published.11 In this study, 23andMe data were used as discovery cohort, but the top-scoring loci failed to be replicated in a second cohort (BEACON) where cases were selected using a different disease definition.
Genome-wide single nucleotide polymorphism (SNP) genotype data have become available for the Swedish and UK twin cohorts where GERD inheritance was originally described,9, 10 and for an additional population-based sample, the Northern Finland Birth Cohort 1966 (NFBC1966),12 where reflux symptom scores are also available from questionnaire data. We exploited these resources to perform GWA studies and their meta-analysis in relation to GERD susceptibility, based on well-defined reflux symptom scores from questionnaire data.
2 Materials and methods
2.1 Study samples
2.1.1 Screening Across Lifespan Twin study (SALT)
The SALT study is part of the population-based Swedish Twin Registry that includes all twins born in Sweden.13 In the SALT study, extensive epidemiological information has been gathered from 45 750 Swedish twins, born in 1958 or earlier, who participated in telephone interviews conducted between 1998 and 2002. A structured questionnaire was used to elicit a history of reflux symptoms and classify GERD individuals accordingly. The same GERD definition was used as in previously reported GERD studies performed on SALT data.14, 15 During computer-assisted telephone interviews, trained interviewers first asked participants if they were regularly suffering from heartburn, pain behind the breastbone, or regurgitation of bitter or sour fluids into the mouth. If the answer to any of these three introductory questions was positive, the participants were then further asked about duration and frequency of symptoms, night waking, radiation of pain toward the neck, antacid relief, and use of antireflux medication, i.e., histamine-receptor antagonists or PPIs. GERD was defined by either at least weekly (i) occurrence of heartburn, (ii) regurgitation of bitter or sour fluids or (iii) pain behind the breastbone with at least one of the following: night waking, radiation of the discomfort toward the neck, antacid relief, use of antireflux medication. Individuals reporting no heartburn, regurgitations or pain behind the sternum were used as controls. One twin (priority to case) from each twin pair was included in the study, but GERD-discordant MZ pairs were discarded. Genotyping of this cohort was performed using Illumina OmniExpress 750K on 11326 SALT twins, mostly dizygotic twins. SALT participants provided informed consent, and the study protocol was approved by Karolinska Institutet ethics review board.
2.1.2 TwinsUK cohort
This cohort based at St. Thomas' Hospital in London is a volunteer cohort of over 10 000 twins from the general population.16 Harmonized genotype data (Illumina HumanHap300 BeadChip and Illumina HumanHap610 QuadChip) were accessible for 2362 subjects with GERD phenotypic information available. Phenotypic status was acquired by each individual filling a 25-item questionnaire asking about symptoms of heartburn and acid regurgitation. GERD was defined, as heartburn or acid regurgitation experienced at least once a week during the past year as previously suggested.10 Controls for GERD were subjects who reported no heartburn or reflux. Participants at the time of recruitment were unaware of the specific gastrointestinal interest of any investigators and gave fully informed consent under a protocol reviewed by the St. Thomas' Hospital Local Research Ethics Committee.
2.1.3 Northern Finland Birth Cohort 1966 (NFBC1966)
NFBC1966 is a population birth cohort including 96.3% of all births in 1966 in the two northernmost provinces of Finland, Oulu and Lapland (n=12 058).12 In 2012–2014, a questionnaire was mailed to those alive targeting general well-being and lifestyle factors, and included a GI module that allowed GERD case identification based on heartburn and/or acid regurgitation at least weekly. A subset of 5402 NFBC1966 participants with Illumina Infinium CNV 370-Duo SNP genome-wide genotypes17 was included in the current study. NFBC1966 participants gave their informed consent and the study was approved by the Regional Ethics Committee of the Northern Ostrobothnia Hospital District in January 25, 2012.
2.2 Genotype quality control (QC) and individual GWA studies
Genotype data from 2536 SALT participants (1087 GERD cases and 1449 controls), 2362 TwinsUK individuals (699 GERD cases and 1663 controls), and 1852 NFBC1966 subjects (461 GERD cases and 1391 controls) were included in the analyses. Stringent quality control (QC) pipeline was applied to the three datasets: SNPs with genotype call rate <98% and/or a Hardy-Weinberg equilibrium (HWE) P<1.0 × 10−7 were excluded, together with individuals with >2% missing genotypes and/or pairwise relatedness score >0.10. Imputation with IMPUTE2 and the 2.5 million HapMap (CEU rel22) SNP panel reference was performed separately on individual datasets at respective local computing facilities. Absence of population stratification was tested for each cohort through principal component analyses. These analyses took place at participating centers and individuals with any ancestry other than strictly European were excluded from the regression models. Individual case-control association tests were performed using regression methods implemented in PLINK (SALT), SNPTEST (NFBC1966), and GEMMA (TwinsUK).18-20
2.3 GWAS meta-analysis
Prior to meta-analysis, association data were harmonized by matching strand information using the QCGWAS package.21 A results summary of the premeta-analysis data harmonization is reported in Table S1. Summary GWAS results from individual cohorts were joined into a weighted z-score meta-analysis using METAL22 and GWAS statistics from 2 327 020 high-quality SNP markers (including 7% SNPs with summary info available only for two-third datasets). A quantile-quantile (Q-Q) plot of the association statistics in each cohort and the meta-analysis with the related genomic inflation factor (λ) is reported in Fig. S1. We considered risk loci genomic regions contained within the boundaries of association signals with P≤5 × 10−5 extended each side 50 kb from the outermost left/right marker with P≤1 × 10−3. LocusZoom (http://csg.sph.umich.edu/locuszoom)23 was used to plot the regional association signals in relation to linkage disequilibrium (LD) between SNPs. Quantile-quantile (Q-Q) and Manhattan plots were generated in R 3.2.0 using the packages qqman and ggplot2.24, 25
2.4 Post-GWAS analyses
2.4.1 Gene-set enrichment analysis
Transcripts mapping to the GERD loci were retrieved using risk region coordinates as input in Biomart R package.26 The resulting list, containing 47 annotated gene IDs, was analyzed for functional class scoring with Fisher exact tests and false discovery rate (FDR) correction (q≤0.05) in Enrichr against Gene Ontology (GO) terms (biological processes, cellular component, molecular function) using default settings.27
2.4.2 Expression quantitative trait loci (eQTL) analysis
Selected SNPs were examined for quantitative cis-effects over transcription of genes (eQTL effects). This was done by querying the latest release (which at the time of writing was version 6) of the Genotype-Tissue Expression (GTEx) database (http://www.gtexportal.org).28 GTEx contains precomputed eQTL data from 44 different tissues. For the purposes of this work, the analysis focused on the four tissues most relevant to GERD: gastroesophageal junction, esophagus mucosa, esophagus muscularis, and stomach. Enrichment for GERD eQTLs from these tissues was calculated using Fisher's test vs the total number of eQTLs from these tissues.
2.4.3 Connectivity Map analysis
The Connectivity Map (cMap)29 contains Affymetrix microarray genome-wide transcriptional expression profiles of 6000 independent experiments of cultured human cells treated with bioactive small molecules (mostly FDA-approved drugs). We interrogated these results by using the cMap2data R package to retrieve compound-specific gene expression rank orders,30 and created a core subset by extracting top 500 (upregulated) and bottom 500 (downregulated) differentially expressed genes from each drug experiment (n=1309). A hypergeometric test was then applied to test significant enrichment (P<.05) of GERD genes from GWAS risk loci in compound-specific core datasets, in order to identify drugs with strongest effects on their expression. Top-scoring drugs were clustered according to the Anatomical Therapeutic Chemical (ATC) index system (http://www.whocc.no/), after normalization for respective ATC level 3 frequencies in the whole cMap dataset, in order to show enrichment of specific drug categories (anatomical, therapeutic, and pharmacological properties).
3 Results
3.1 GWAS of GERD in independent cohorts
The demographics of GERD cases and asymptomatic controls identified based on questionnaire data in each cohort are reported in Table 1. Testing of genome-wide SNP markers in individual cohorts did not yield genome-wide significant results (P<5.0 × 10−8), though a number of independent signals of suggestive association (P<5.0 × 10−5) was identified in each GWAS (Table S2).
| Cohort | TOTGWASa | GERD | CTRLSb | M:F | Mean age |
|---|---|---|---|---|---|
| SALT | 11 326 | 1087 | 1449 | 1099:1437 | 54.4 |
| NFBC66 | 5402 | 461 | 1391 | 840:1012 | 46.0 |
| TwinsUK | 6675 | 699 | 1663 | 149:2213 | 56.4 |
| Meta-analysis | 23 403 | 2247 | 4503 | 2088:4662 | 52.8 |
- a Number of individuals with genome-wide SNP genotype data.
- b Asymptomatic subjects.
3.2 GWAS meta-analysis
Meta-analysis of individual GWA studies was carried out using METAL on a combined dataset including 6750 individuals and 2 327 020 SNP markers (Material and methods). In this analysis, whose results are summarized in Fig. 1 with a Manhattan plot, 109 markers from 30 independent genomic regions provided association signals of P<5.0 × 10−5, with best evidence for the SNP rs10852151 on chromosome 15 (P=2.3 × 10−7). As shown in Table 2, association signals at these loci showed concordant genetic risk effects in the three independent index GWA studies. Detailed regional plots of association, including linkage disequilibrium information, are reported in Fig. S2 for all GERD risk loci. When compared to previous GWAS results from the 23andMe/BEACON study, only one out of their five top-scoring loci (no other data available to compare) resulted nominally associated with GERD in our meta-analysis (rs520525, P=.011) and showed concordant genetic effects in all cohorts. In addition, none of the loci previously associated with BE and EA31-33 replicated in our study (Table S3).
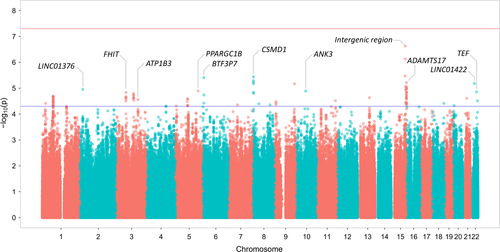
| Locus # | Lead SNP | CHR | POS | LEN | EA | EAF | Z | P | DIR (S/N/T)a | Genetic contentb |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs12128603 | 1 | 22564787 | 0.11 | T | 0.05 | 4.12 | 3.88×10−5 | +/+/+ | MIR4418, LINC00339 |
| 2 | rs782244 | 1 | 72916451 | 0.24 | A | 0.34 | −4.26 | 2.07×10−5 | −/−/− | RPL31P12 |
| 3 | rs851328 | 2 | 19407422 | 0.16 | T | 0.74 | −4.40 | 1.11×10−5 | +/+/+ | LINC01376 |
| 4 | rs11712505 | 3 | 61094353 | 0.13 | T | 0.46 | −4.34 | 1.46×10−5 | +/+/na | FHIT |
| 5 | rs7645666 | 3 | 111631591 | 0.19 | A | 0.39 | 4.31 | 1.61×10−5 | +/+/+ | PLCXD2, PHLDB2, ABHD10, TAGLN3, SLC9C1 |
| 6 | rs4234469 | 3 | 128921429 | 0.15 | A | 0.58 | 4.19 | 2.78×10−5 | −/−/− | RNF7, GRK7, HMGN2P25, ATP1B3, RP11-383G6.4 |
| 7 | rs10518512 | 4 | 127995476 | 0.19 | A | 0.22 | −4.06 | 4.99×10−5 | −/−/− | |
| 8 | rs1346576 | 5 | 4658086 | 0.11 | T | 0.66 | −4.06 | 4.84×10−5 | +/+/+ | |
| 9 | rs4704557 | 5 | 78636992 | 0.10 | T | 0.91 | 4.15 | 3.28×10−5 | −/−/− | JMY, HOMER1, BHMT |
| 10 | rs10065109 | 5 | 80358051 | 0.16 | C | 0.77 | −4.21 | 2.58×10−5 | +/+/+ | RASGRF2 |
| 11 | rs10078478 | 5 | 149082704 | 0.10 | A | 0.89 | −4.37 | 1.27×10−5 | +/+/+ | PPARGC1B, MIR378A, RN7SL868P |
| 12 | rs2326825 | 6 | 6806171 | 0.11 | T | 0.33 | −4.62 | 3.91×10−6 | −/−/na | BTF3P7, RPI1-80N2.2 |
| 13 | rs4585693 | 7 | 57577800 | 0.29 | A | 0.74 | −4.08 | 4.42×10−5 | +/+/+ | MIR3147, VN1R28P, SAPCD2P2, ZNF716, NCOR1P3 |
| 14 | rs12546471 | 8 | 4181065 | 0.12 | C | 0.61 | 4.63 | 3.64×10−6 | −/−/− | CSMD1 |
| 15 | rs7827627 | 8 | 40021859 | 0.10 | T | 0.03 | 4.11 | 4.02×10−5 | +/+/na | C8orf4 |
| 16 | rs1159046 | 8 | 113777669 | 0.25 | A | 0.91 | -4.09 | 4.40×10−5 | +/+/+ | CSMD3, MIR2053 |
| 17 | rs1755608 | 9 | 6641895 | 0.13 | A | 0.80 | 4.15 | 3.28×10−5 | −/−/− | GLDC, RPL23AP57, RN7SL123P, RPS3AP54, RNF2P1, RP11-390F4.3 |
| 18 | rs10987145 | 9 | 128853643 | 0.13 | T | 0.32 | −4.50 | 6.75×10−6 | −/−/− | |
| 19 | rs10761438 | 10 | 61780209 | 0.10 | A | 0.36 | −4.36 | 1.29×10−5 | −/−/− | ANK3, CCDC6 |
| 20 | rs11218771 | 11 | 122588584 | 0.11 | T | 0.64 | 4.17 | 3.02×10−5 | −/−/− | UBASH3B |
| 21 | rs4628837 | 13 | 78805278 | 0.20 | A | 0.35 | 4.23 | 2.32×10−5 | +/+/+ | RNF219-AS1 |
| 22 | rs10852151 | 15 | 92083295 | 0.11 | A | 0.57 | 5.17 | 2.34×10−7 | −/−/na | SLCO3A1, CRTC3, RP11-387D10.2 |
| 23 | rs7175566 | 15 | 94149785 | 0.12 | T | 0.65 | −4.45 | 8.55×10−6 | +/+/+ | |
| 24 | rs4965272 | 15 | 100584739 | 0.13 | T | 0.74 | −4.52 | 6.07×10−6 | +/+/+ | ADAMTS17, RNA5SP402, AC022819.3, CTD-3076O17.1 |
| 25 | rs591107 | 18 | 74089140 | 0.11 | A | 0.74 | 4.11 | 3.89×10−5 | −/−/− | ZNF516 |
| 26 | rs6076667 | 20 | 4367040 | 0.13 | A | 0.79 | −4.07 | 4.71×10−5 | +/+/+ | CDS2 |
| 27 | rs10451749 | 21 | 24978573 | 0.13 | A | 0.98 | −4.22 | 2.46×10−5 | +/+/na | |
| 28 | rs2213770 | 22 | 27244685 | 0.11 | A | 0.28 | 4.51 | 6.57×10−6 | +/+/+ | LINC01422, RP1-90L6.2 |
| 29 | rs2073167 | 22 | 41791536 | 0.18 | T | 0.56 | 4.35 | 1.38×10−5 | −/−/− | ZC3H7B, TEF, RNU6-495P, TOB2, PHF5A, ACO2, SGSM3, EP300, L3MBTL2, RANGAP1, POLR3H, CSDC2, PMM1, DESI1, NHP2L1, MEI1, NDUFA6, CYP2D6, RP4-756G23.5, CTA-223H9.9 |
| 30 | rs11705127 | 22 | 48023818 | 0.10 | T | 0.61 | 4.17 | 3.07×10−5 | −/−/− | LINC00898 |
- SNP, single nucleotide polymorphism; CHR, chromosome; POS, genomic coordinates according to human genome reference version 19 (hg19); LEN, locus length in Mb; EA, effect allele in the three cohorts; EAF, effect allele frequency; Z, meta-analysis z-score; P, P-value.
- a Directions of genetic effects in each cohort (S=SALT, N=NFBC1966, T=TwinsUK).
- b Transcript mapping to each GERD risk locus and transcripts associated with eQTL signal in four GERD-relevant tissues from GTEx database.
3.3 Functional genomic analyses of GERD risk loci
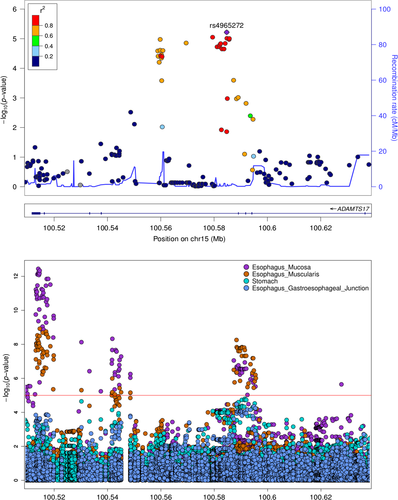
We investigated GERD loci for correlations between genotype and tissue-specific gene expression levels, as plausible regulatory mechanisms associated with regional genetic effects on disease risk. We used publicly available GTEx28 precomputed eQTL data from four selected tissues most relevant to GERD, namely gastroesophageal junction, esophagus muscularis, esophagus mucosa, and stomach. This analysis revealed that GERD risk regions are significantly enriched for eQTLs from these tissues (P<.01), seven of which correspond to genes mapping inside the actual risk loci: ABHD10, RNF7, RASGRF2, BTF3P7, C8orf4, GLDC, and ADAMTS17. The latter, in particular, maps to a GERD risk locus showing multiple significant markers (best P=6.1 × 10−6 for SNP rs4965272) and strong significant SNP-eQTLs correlations as evidence that genetic variation affects gene expression by acting on putative regulatory elements (Fig. 2).
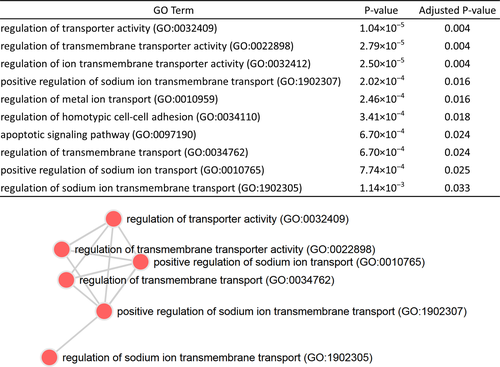
In total, 47 genes/transcripts (unique Ensembl IDs) map within the 30 GERD suggestive risk loci (Table 2). We performed GSEA in order to identify biological pathways and gain mechanistic insight for the observed associations. Of note, functional class scoring of the 47 GERD genes returned the gene ontology (GO) terms regulation of transporter activity (GO:0032409), regulation of transmembrane transporter activity (GO:0022898), regulation of ion transmembrane transporter activity (GO:0032412), regulation of homotypic cell-cell adhesion (GO:0034110), and positive regulation of sodium ion transmembrane transport (GO:1902307) as the most significantly enriched (q≤0.05 after FDR correction) “biological process” categories (Fig. 3).
3.4 Connectivity map analysis of GERD risk genes
To gain further insight into the mechanisms behind the observed associations, and the potential exploitation of this information for clinical purposes, we sought to identify known drugs or compounds that are able to affect gene expression at the reported GERD loci. We performed drug-target enrichment analyses using the Connectivity Map (cMap),29 a public repository of drug-induced genome-wide microarray transcriptional profiles from human cells treated in vitro with more than 1300 different compounds, and screened for drugs that were significantly (P≤.05) enriched for GERD genes among their top 1000 targets, irrespective of the direction of the effect on gene expression (i.e., up- or down-regulation). Among the nine cMap drugs from the ATC A02B category peptic ulcer and GERD, omeprazole showed significant effects on GERD risk gene expression (P=.032) (Table 3). Of note, several other drugs produced high scores in this analysis, with best statistical evidence obtained for the corticosteroid fludroxycortide (Table 3). Likewise, when the ATC drug classification system was taken into account, the categories J04A (treatment of tuberculosis), D11A (other dermatological preparations), J05A (direct acting antivirals), and A07E (intestinal anti-inflammatory agents) resulted most represented after normalization for their relative abundance in the cMap database (Fig. S3).
| Drug | Score | P | ATC full | ATC level 3 |
|---|---|---|---|---|
| GERD drugs | ||||
| Omeprazole | 3 | 3.23×10−2 | A02BC01 | A02B |
| Carbenoxolone | 2 | 0.127 | A02BX01 | A02B |
| Nizatidine | 2 | 0.127 | A02BA04 | A02B |
| Pirenzepine | 2 | 0.127 | A02BX03 | A02B |
| Proglumide | 2 | 0.127 | A02BX06 | A02B |
| Cimetidine | 1 | 0.365 | A02BA01 | A02B |
| Famotidine | 1 | 0.365 | A02BA03 | A02B |
| Lansoprazole | 1 | 0.365 | A02BC03 | A02B |
| Ranitidine | 1 | 0.365 | A02BA02 | A02B |
| All cMap drugs | ||||
| Fludroxycortide | 6 | 1.04×10−4 | D07AC07 | D07A |
| BAS-012416453 | 5 | 9.09×10−4 | — | — |
| Flumequine | 5 | 9.09×10−4 | J01MB07 | J01M |
| Heliotrine | 5 | 9.09×10−4 | — | — |
| Ionomycin | 5 | 9.09×10−4 | — | — |
| Levocabastine | 5 | 9.09×10−4 | S01GX02, R01AC02 | S01G, R01A |
| LM-1685 | 5 | 9.09×10−4 | — | — |
| Maprotiline | 5 | 9.09×10−4 | N06AA21 | N06A |
| Meclofemic_acid | 5 | 9.09×10−4 | — | — |
| Methylergometrine | 5 | 9.09×10−4 | G02AB01 | G02A |
| Piracetam | 5 | 9.09×10−4 | N06BX03 | N06B |
| Promazine | 5 | 9.09×10−4 | N05AA03 | N05A |
| Sisomicin | 5 | 9.09×10−4 | J01GB08 | J01G |
| Spiramycin | 5 | 9.09×10−4 | J01FA02 | J01F |
| 0316684-0000 | 4 | 6.18×10−3 | — | — |
| 5211181 | 4 | 6.18×10−3 | — | — |
| Aconitine | 4 | 6.18×10−3 | — | — |
| Adipiodone | 4 | 6.18×10−3 | V08AC04 | V08A |
| Amantadine | 4 | 6.18×10−3 | N04BB01 | N04B |
| Atractyloside | 4 | 6.18×10−3 | — | — |
| Atracurium_besilate | 4 | 6.18×10−3 | — | — |
| Baclofen | 4 | 6.18×10−3 | M03BX01 | M03B |
| Benzbromarone | 4 | 6.18×10−3 | M04AB03 | M04A |
| Bethanechol | 4 | 6.18×10−3 | N07AB02 | N07A |
| Bicuculline | 4 | 6.18×10−3 | — | — |
| Bisoprolol | 4 | 6.18×10−3 | C07AB07 | C07A |
| Bupropion | 4 | 6.18×10−3 | N07BA02 | N07B |
| Cefapirin | 4 | 6.18×10−3 | J01DB08 | J01D |
| Cefazolin | 4 | 6.18×10−3 | J01DB04 | J01D |
4 Discussion
A genetic component of GERD has been long recognized,7-10 though gene-hunting efforts have been scarce. We report here a meta-analysis of GWA studies from three independent population-based cohorts, including two original twin cohorts where reflux heritability was first described (SALT and TwinUK). Similar to previous work in other GI conditions,34 we adopted a population-based approach, exploiting available genotype data in relation to a questionnaire-based classification of reflux that has reported diagnostic performance comparable to that of family practitioners and, to a lesser extent, gastroenterologists.3 We identified 30 independent suggestive signals of association with GERD, coming from genomic regions that (i) show SNP concordant genetic risk effects in individual GWA studies and (ii) include genes associated with plausible functional pathways of potential relevance to reflux pathophysiology. Hence, although current sample size did not allow reaching genome-wide significance level (for instance, 4200 cases would be needed to achieve 80% power to detect association with P<5.0 × 10−8 at the ADAMTS17 locus), the observed associations may represent bona fide GERD risk signals, which warrant further investigation in follow-up studies. Unfortunately, we could not properly compare our data with a recently published GERD GWAS, in which the authors investigate GERD genetic predisposition in relation to BE and EA.11 As there was no complete summary statistics available, we could only investigate five reported top loci. Only locus showed borderline significance in our meta-analysis dataset (P=.011), with concordant risk effects in both studies and each individual cohort; it includes the PRRX1 gene, a putative MEF2 target involved in the development of diverse muscle types.35 MEF2 family, important regulators of muscular, neural, and immune system development.36 Of note, among other defects, subjects with MEF2C report abnormalities of GI motility, including gastroesophageal reflux disease, dysphagia, and constipation.36
The importance of using unified disease definitions is highlighted by the fact that neither the combined analysis of 23andme and BEACON led to consistent findings despite much larger sample size,11 nor could we convincingly replicate most of the reported top association signals in our GWAS meta-analysis. Because GERD is a known risk factor for BE and EA, we also tested seven known BE/EA risk loci31-33 in our GERD meta-analysis but failed to detect significant association, which leaves open the question as to whether GERD risk loci (once unequivocally identified) also affect predisposition to these conditions.
Our GWAS-downstream analyses of gene content at GERD-associated regions have provided initial insight as to the potential mechanisms affecting disease risk, which might help explain the statistical observations. GSEA returned several FDR-corrected significant results for GO categories related to the regulation of ion channel and transport functions, and cell-cell adhesion properties. Notably, these pathways are involved in the maintenance of mucosal epithelial integrity, which protects against luminal damage from acid and acid-pepsin: cell membrane and intercellular junctional complex constitute the structural barriers to H+ diffusion in the GI tract, while intercellular buffers and basolateral membrane transporter proteins constitute the functional components of the epithelium. In particular, the latter protect by neutralizing H+ within cytoplasm and intercellular space and by transporting H+ from cytoplasm to intercellular space, as in the case of the Na/H and the Cl/HCO3 exchangers.37 A dysregulated expression of cell-to-cell adhesion proteins, may be behind the impaired mucosal integrity present in GERD, similar to what has been proposed to affect other areas of the gastrointestinal tract in irritable bowel syndrome and functional dyspepsia.38-41
Although traditionally viewed as different entities, ion channels and ion pumps are increasingly recognized to share several functional and physiological features.42, 43 For this reason, dysregulation of ion channel transport as one of the potential genotype-driven mechanisms affecting GERD risk is noteworthy also in relation to its therapeutic treatment. PPIs mechanism of action involves the inhibition of the H+, K+-adenosine triphosphatase (ATPase) pumps, thereby leading to potent acid suppression. An interesting gene mapping within one of the 30 GERD loci is ATP1B3. This gene codes for the beta-3 subunit of a Na+/K+-ATPase, a ubiquitous integral membrane protein that is responsible for maintaining intracellular Na+ and K+ gradients and, hence, contributes to modulate cell membrane potential. Other channels have been previously involved in GERD pathogenesis, for example, TRPV1 expression is increased after acid exposure. This gene, which is also expressed in the esophageal mucosa,44 acts as a molecular integrator of inflammatory responses to noxious stimuli. Results using antagonists of this receptor to treat human pain and heat thresholds have already been published.45, 46
In order to provide further insight into genotype-phenotype associations, we investigated GERD loci for the presence of eQTL effects. We focused our analysis on cis-eQTL, which are mainly thought to regulate proximal genes by affecting their transcription, possibly through variations in genomic sequence that affect binding affinity of transcription factors and other regulatory elements.47 Exploiting the latest GTEx data release,28 we focused our analysis on four most selected GERD-relevant tissues, namely gastroesophageal junction, esophagus muscularis, esophagus mucosa, and stomach. We made the observation that GERD risk regions are enriched for significant eQTLs from these tissues, with esophagus muscularis showing the highest number of eQTLs mapping to GERD loci. Seven known genes were associated with eQTL effects in this tissue (ABHD10, RNF7, RASGRF2, BTF3P7, C8orf4, GLDC, ADAMTS17), representing priority candidates to be further investigated. In addition, two other genes belonging to the category of ion channels and transporters provided interesting results, namely SLC9C1 (a Na+/H+ exchanger) associated with eQTLs in the gastroesophageal junction, and SLC3A1 (an amino-acid transporter) associated to eQTLs in the esophagus mucosa.
Our findings suggest that ADAMTS17 may be the best candidate to be further investigated in future genetic and functional approaches. This gene codes for a member of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) protein family, which comprises 19 secreted proteases primarily associated with the extra-cellular matrix and involved in a wide range of human biological processes with potential roles in arthritis, cancer, angiogenesis, atherosclerosis, central nervous system disorders, and fertility.48, 49 ADAMTS17 is widely expressed in human tissues but the function of this protein has not been yet determined. Mutations in ADAMTS17 have been identified as a cause of autosomal-recessive Weill–Marchesani syndrome, a well-characterized disorder in which patients develop eye and skeletal abnormalities,50 and another member of the family (ADAM10) has been proposed as a candidate for the higher rate of e-cadherin proteolytic cleavage in patients with GERD.51 Consequence of e-cadherin cleavage is an increased junctional permeability due to loss of cell-cell adhesion, a pathway that has been involved in GERD pathogenesis.
Finally, we performed a computational cMap analysis in the attempt to link the detected genetic GERD risk effects to known and, eventually, repurposed therapeutic compounds. The adopted approach had the limitation of not allowing to predict the directions of specific genotype-driven genetic effects (i.e., we worked with gene lists irrespective of their potential down- or up-regulation), hence we could only aim at identifying drugs able to impact the expression of the highest number of GERD risk genes irrespective of the overall “therapeutic” effect. We were first interested in assessing whether any cMap drug currently used for GERD treatment (ATC A02B, Drugs for peptic ulcer, and GERD) was enriched in targets among the GERD risk loci and, of note, obtained significant results for omeprazole, a known proton pump inhibitor of the H+/K+-ATPase system. We then screened all cMap drugs and detected highest scores for fludroxycortide, a drug classified as a potent anti-inflammatory. This may not be surprising, as in last years the concept of mucosal inflammation has been integrated into the current model of GERD pathogenesis, owing to a proposed role of immune cell-mediated injury and mucosal lymphocytic infiltrate in the damaging of the esophagus.52 Clustering cMap drugs according to their ATC classification (level 3) also generated results of some interest, with the class J04A (drugs for treatment of tuberculosis) showing highest scores after normalization. We speculate this may be due to the enrichment for ion channel genes from the GERD risk loci, as these are emerging as potential antituberculosis targets.53 However, it must be emphasized that cMap screening is based on the exploitation of gene expression data, hence the identification of anti-inflammatory drugs among the top-scoring hits both for single drug screening and ATC clustering may be due at least in part to the wide impact that these have on transcription compared to drugs with more specific mode of action such as PPIs.
In conclusion, in this study, we showed that population-based cohorts with associated genetic and epidemiological data provide good opportunities for GERD-focused gene mapping efforts. The risk signals detected in individual datasets and their meta-analysis establish a repository to inform follow-up efforts in independent case-control studies, and point to the involvement of plausible biological pathways that can be prioritized for future functional investigations.
Funding
This work was supported by funds from the Swedish Research Council (Vetenskapsrådet) to MDA.
Disclosures
No conflict of interest to disclose.
Author contributions
PTS and MDA study concept and design; FeB, PHG, WE, VK, NVR, MM statistical analyses; HN, MZ, FrB, FW, HT, PM, NP, JR data acquisition; FeB, PGH, WE, VK, NVR, MM, HN, MZ, MDA data analysis and interpretation; MDA obtained funding, administrative and technical support, study supervision; FeB and MDA drafting of the manuscript, with input and critical revision from all other authors.
Abbreviations
-
- BE
-
- Barrett's esophagus
-
- cMap
-
- Connectivity Map
-
- DZ
-
- dizygous
-
- EA
-
- esophageal adenocarcinoma
-
- eQTL
-
- expression quantitative trait locus
-
- FDR
-
- false discovery rate
-
- GERD
-
- gastroesophgeal reflux disease
-
- GI
-
- gastrointestinal
-
- GO
-
- Gene Ontology
-
- GSEA
-
- Gene Set Enrichment Analysis
-
- GTEx
-
- Genotype Tissue Expression database
-
- GWAS
-
- genome-wide association study
-
- LD
-
- linkage disequilibrium
-
- MZ
-
- monozygous
-
- NERD
-
- non-erosive reflux disease
-
- NFBC1966
-
- Northern Finland Birth 1966 cohort
-
- PPIs
-
- proton pump inhibitors
-
- QC
-
- quality control
-
- Q-Q
-
- quantile-quantile
-
- SALT
-
- Screening Across Lifespan Twins study cohort
-
- SNP
-
- single nucleotide polymorphism
-
- TwinsUK
-
- Twins UK cohort




