Runs of homozygosity reveal past bottlenecks and contemporary inbreeding across diverging populations of an island-colonizing bird
Abstract
Genomes retain evidence of the demographic history and evolutionary forces that have shaped populations and drive speciation. Across island systems, contemporary patterns of genetic diversity reflect population demography, including colonization events, bottlenecks, gene flow and genetic drift. Here, we investigate genome-wide diversity and the distribution of runs of homozygosity (ROH) using whole-genome resequencing of individuals (>22× coverage) from six populations across three archipelagos of Berthelot's pipit (Anthus berthelotii)-a passerine that has recently undergone island speciation. We show the most dramatic reduction in diversity occurs between the mainland sister species (the tawny pipit) and Berthelot's pipit and is lowest in the populations that have experienced sequential bottlenecks (i.e., the Madeiran and Selvagens populations). Pairwise sequential Markovian coalescent (PSMC) analyses estimated that Berthelot's pipit diverged from its sister species ~2 million years ago, with the Madeiran archipelago founded 50,000 years ago, and the Selvagens colonized 8000 years ago. We identify many long ROH (>1 Mb) in these most recently colonized populations. Population expansion within the last 100 years may have eroded long ROH in the Madeiran archipelago, resulting in a prevalence of short ROH (<1 Mb). However, the extensive long and short ROH detected in the Selvagens suggest strong recent inbreeding and bottleneck effects, with as much as 38% of the autosomes consisting of ROH >250 kb. These findings highlight the importance of demographic history, as well as selection and genetic drift, in shaping contemporary patterns of genomic diversity across diverging populations.
1 INTRODUCTION
When populations undergo genetic bottlenecks (either due to being founded by few individuals, or as a result of a drastic population decline), it can result in the loss of genetic diversity and inbreeding (Weaver et al., 2021). In island systems, populations may be the product of multiple founding steps, for example during a stepwise range expansion (Halkka et al., 1974; Recuerda et al., 2021; Sendell-Price et al., 2021). Colonization events and associated bottlenecks can strongly shape the genetic diversity of island populations (Carson, 1971; Nei et al., 1975), which typically have reduced genetic diversity relative to their mainland ancestors (e.g., Frankham, 1997; Leroy et al., 2021; Robinson et al., 2018). The potential for a loss of genetic diversity is exaggerated by sequential bottlenecks and genetic drift as a result of long-term isolation and small population size (Gautschi et al., 2002). Limited gene flow between islands can prolong and further promote such effects on individual inbreeding (Clegg et al., 2002; Pilot et al., 2014). Over time, these forces, as well as local adaptation (Rundle & Nosil, 2005), can culminate in reproductive isolation and speciation (Comeault et al., 2015; Feder et al., 2012; Warren et al., 2012). Variation in demographic history and selective pressures across island populations can drive rapid divergence and speciation, which explains why island systems are some of the most biologically diverse habitats globally (Paulay, 1994). Thus, island populations can be key systems in which to investigate the causes and consequences of such processes.
Studying contemporary patterns of inbreeding can also provide insight into recent demography and effective population size. Inbreeding not only reflects population-level processes but can have negative consequences for individual fitness and survival, that is “inbreeding depression” (Charlesworth & Charlesworth, 1987; Charlesworth & Willis, 2009; Darwin, 1876; Doekes et al., 2021), and have implications for population persistence (Frankham, 2005; Hedrick & Garcia-Dorado, 2016; Oostermeijer et al., 1995). One powerful way to measure inbreeding is to analyse genomic runs of homozygosity (ROH), chromosome segments identical by descent (IBD), which has been used to infer population dynamics for an increasingly broad range of wild populations (Duntsch et al., 2021; Foote et al., 2021; Foster et al., 2021; Gómez-Sánchez et al., 2018; Grossen et al., 2018; Kardos, Åkesson, et al., 2017). ROH appear when individuals inherit the same chromosomal sequence from both parents (without recombination or mutation; Broman & Weber, 1999; Gibson et al., 2006; McQuillan et al., 2008). This happens more frequently with increasing parental relatedness, although ROH can also emerge when shorter identical haplotype runs are inherited from apparently unrelated individuals due to background relatedness in the population (Korf, 2013). Consequently, ROH arise due to shared ancestry in the population. Thus, long ROH segments are expected in populations which have experienced contemporary inbreeding, while shorter ROH segments indicate loss of genetic diversity from a historical founder effect or genetic bottleneck (Foote et al., 2021; Gómez-Sánchez et al., 2018; Islam et al., 2019; Kardos, Åkesson, et al., 2017; McQuillan et al., 2008; Pilot et al., 2014; Stoffel et al., 2021). In some populations ROH can arise from non-random mating even when the overall population size is large (Li et al., 2006) or from strong selection to maintain a single haplotype (Gibson et al., 2006; Kardos, Qvarnström, & Ellegren, 2017; Pemberton et al., 2012), although these are expected to occur only in a few genomic regions. The minimum length defined to identify a ROH segment has major effects on the estimation of inbreeding, and yet it is set arbitrarily. Applying different length criteria for ROH detection can reveal information about population demography across a range of time frames.
To reveal ancient dispersal or speciation events and population dynamics over long evolutionary time periods a range of modelling approaches can also be used (Beichman et al., 2018), while comparisons of shared population history can be made to infer divergence timescales between populations (Delmore et al., 2020; Excoffier et al., 2021; Patton et al., 2019; Sarabia et al., 2021; Terhorst et al., 2017). For example, pairwise sequentially Markovian coalescent (PSMC) models use patterns of heterozygosity to identify historical recombination events across a single diploid genome by inferring the time to the most recent common ancestor (TMRCA) for each independent DNA segment (Li & Durbin, 2011). These models have been used to infer the timing of dispersal or colonization events and changes to population size across a wide range of animal systems (De Jager et al., 2021; Deng et al., 2021; Escoda & Castresana, 2021; Hooper et al., 2020; Kirch et al., 2021; Nadachowska-Brzyska et al., 2016; Patton et al., 2019; Xue et al., 2015) and some plants (Izuno et al., 2016; Patil et al., 2021). Using these approaches in combination with genomic ROH distributions can enable studies to produce estimates of ancient through to contemporary demographic history.
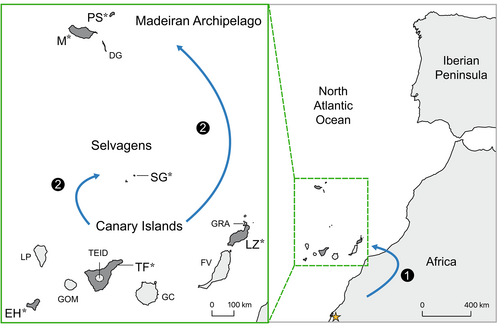
The island endemic Berthelot's pipit (Anthus berthelotii) together with its mainland sister species, the tawny pipit (Anthus campestris), offer an excellent study system to better understand the genomic and demographic underpinnings of important evolutionary processes in island species (e.g., Armstrong et al., 2018; Gonzalez-Quevedo et al., 2015; Illera et al., 2007; Spurgin et al., 2014). The ancestor of the tawny and Berthelot's pipit colonized the Canary Islands probably from mainland Africa (Voelker, 1999; see Figure 1), with Berthelot's pipit later expanding to the Madeiran and Selvagens archipelagos (Martin et al., 2021; Spurgin et al., 2014). Previous work using microsatellites (Spurgin et al., 2014) and restriction-site associated DNA sequencing (Armstrong et al., 2018; Martin et al., 2021) has revealed strong bottlenecks associated with the two independent colonization events from the Canary Islands to Madeira and Selvagens, estimated to have occurred ~8500 years ago (ka). Considerable genetic structure now exists between, but not within, Berthelot's pipit archipelago populations, with no evidence of subsequent gene flow (Armstrong et al., 2018; Illera et al., 2007; Martin et al., 2021; Spurgin et al., 2014), thus allowing us to study independent divergence histories and incipient speciation across the species range.
As yet, little detail is known about genetic divergence from the tawny pipit to Berthelot's pipit, or how population history has shaped patterns of genome-wide diversity across the Berthelot's pipit range. Here, we use whole-genome resequencing data from 11 individuals (mean 25× coverage) sampled across the range of the Berthelot's pipit, to quantify island founder events, historical fluctuations in Ne and characterize genomic signatures of inbreeding. First, we determine how genome-wide diversity and structure vary between populations and across archipelagos of Berthelot's pipit, and from its sister species. Second, we use demographic reconstruction modelling to estimate ancient (~2.5 million year ago [Ma] until 5 ka) Ne to make inferences about population divergence time frames and bottlenecks. Finally, we identify and map long (>1 Mb) and short (250 kb to 1 Mb) ROH across genomes and estimate the timing and strength of inbreeding across these populations with differing number of founding events, bottleneck severity and population isolation.
2 METHODS
2.1 Sample collection and reference genome sequencing
Berthelot's pipit is a small (~16 g), sedentary, socially monogamous, insectivorous passerine with a generation time of ~2 years (Bird et al., 2020; Garcia-del-rey & Cresswell, 2007; Illera et al., 2007). It is endemic to three Macaronesian archipelagos (the Canary, Madeiran and Selvagens archipelagos; see Figure 1) where it is relatively abundant and widespread within areas of open semi-arid coastal or alpine habitat. These islands have a relatively recent volcanic origin; 1–26 million years old (Florencio et al., 2021), and support a diversity of ecosystems (Garcia-del-rey & Cresswell, 2007). There is substantial variation in pipit pathogen exposure (specifically avian pox and malaria; Illera et al., 2008), island isolation, habitat and climate within and among archipelagos (Spurgin et al., 2012). Samples (n = 11) from six islands across the three archipelagos were selected for genome resequencing, to maximize the geological and geographical coverage (Figure 1; Table S1). Two adult individuals per population, one male and one female, were selected from El Hierro, Tenerife and Lanzarote (Canary Islands), Madeira and Porto Santo (one sample in addition to the reference sample) in the Madeiran archipelago, and Selvagem Grande (Selvagens). All individuals chosen had no pox lesions and no detected haemoprotozoa parasite infection (Illera et al., 2008). In addition, we sequenced one tawny pipit sampled during its migration in Mauritania (Latitude: 17.991703°, Longitude: −16.016672°). The birds were caught using mealworm (Tenebrio molitor) larvae-baited spring traps and sampled from different locations across each island to reduce the probability of sampling closely related individuals. Blood (~25 μL) was taken by brachial venipuncture and stored in 800 μL absolute ethanol at 4°C. DNA was extracted using the salt extraction protocol described by Richardson et al. (2001), and individuals were molecularly sexed (Griffiths et al., 1998).
A draft Berthelot's pipit reference assembly from a male bird sampled on Porto Santo in the Madeiran archipelago, generated by Armstrong et al. (2018), was used to align genome-wide sequence reads and to call genomic variants. This bird was selected due to its low genome-wide heterozygosity as identified by previous RAD-sequencing (Armstrong et al., 2018). Sequencing was performed using paired-end reads (2 × 125 bp) on an Illumina HiSeq 2500 sequencer, and assembled using discovar de novo (Weisenfeld et al., 2014). Genome completeness was assessed using cegma (Parra et al., 2007) which searched for 248 highly conserved core eukaryotic genes (61% complete; 90% partial) and busco (Simão et al., 2015), to search for 3023 vertebrate-specific single-copy orthologues (64% complete single-copy). The resulting draft assembly has a total size of 1.15 Gb (94.3% of the length of the Zebra finch [Taeniopygia guttata] genome), with contig N50 of 355 kb. For full details on the reference genome assembly see Armstrong et al. (2018).
2.2 Genome resequencing and variant calling against Berthelot's pipit genome assembly
Low Input Transposase Enabled (LITE) libraries were constructed, cleaned and sequenced using Illumina HiSeq 4000 at the Earlham Institute (Norwich, UK). High-throughput libraries were generated for each sample, pooled across four lanes of an Illumina HiSeq 4000, for paired-end sequencing (2 × 150 bp). Per-individual sequence reads with Phred quality score > Q30 (indicating per-read base call accuracy >99.9%) were merged and aligned to the indexed reference Berthelot's pipit genome using bwa-mem version 0.7.12 (Li, 2013). Once mapped, potential duplicate PCR reads were flagged using Picard tools MarkDuplicates in gatk version 4.1 (McKenna et al., 2010), and to assign read group information and validate binary alignment files (.bam) before variant discovery. Variant calling was then performed against the Berthelot's pipit reference genome using gatk HaplotypeCaller in GVCF mode, removing flagged duplicated or poor-quality reads based on default parameters. Joint genotyping was then performed across samples for each contig using gatk's GenomicsDBImport and GenotypeGVCF tools. To improve the accuracy of variant discovery and genotyping, variants were determined simultaneously across the 11 Berthelot's pipit samples and the tawny pipit sample for each data set. Contig-level VCF files were then combined using gatk SortVcf, with variants mapped to contigs <500 bp removed. Unmapped or poor-quality reads (with a root-mean-squared read mapping quality below 25) were discarded. Variants were then filtered for read strand bias (Fisher's exact test > 60 and Strand Odds Ratio > 3) and quality by depth (QD < 2) using gatk, to account for errors in read mapping.
2.3 Variant mapping to Zebra finch genome and filtering
As the draft reference genome for Berthelot's pipit is only assembled to the contig level (Armstrong et al., 2018), genomic variants were mapped to relative chromosome positions of the Zebra finch genome assembly bTaeGut1_v1.p (NCBI Assembly GCA_003957565.1; Warren et al., 2010) using the SatsumaSynteny module within satsuma (Grabherr et al., 2010), which performs well on fragmented genome assemblies (Liu et al., 2018). We used the d-genies dot-plot tool (Cabanettes & Klopp, 2018) with the default options to visually assess collinearity of the Berthelot's pipit and Zebra finch genome assemblies. This showed that high synteny exists between these genomes, although percentage identity is relatively low for most contigs (25%–75%). Despite this, only a very few short regions of the Berthelot's pipit genome are misassembled/misplaced, many of which are on sex chromosomes, which are excluded in this study (Figure S1). Output from satsuma was used to assign contigs to chromosomes, and determine their order, location and orientation. Finally, genomic variants were mapped against the satsuma output and reassigned to chromosomes using custom R scripts (RStudio Team, 2016).
Three final VCF files were generated to maximize variants that could be included in each analysis: (i) “All Pipits” with variants joint called across the 11 Berthelot's pipit individuals and one tawny pipit, (ii) “Berthelot's” data set with variants joint called across 11 Berthelot's pipit individuals, and (iii) “Tawny pipit” variants. These data were filtered for genotype quality and coverage in vcftools version 0.1.15 (Danecek et al., 2011); removing unmapped sites (--not-chr 0), sites with >2 alleles (--max-alleles 2), indels (--remove-indels) and variants with Phred-scaled quality <30, that is variant accuracy >99.9% (--minQ 30). To minimize the impact of collapsed regions in the assembly and limit the likelihood of incorrectly calling heterozygous positions, we also removed all sites at which mean read depth was <10, or more than twice the average genome-wide read depth as recommended in Li (2014) (>45 for “All Pipits,” >44 for “Berthelot's,” >55 for “Tawny pipit”; --min-meanDP 10, --max-meanDP 45/44/55). We removed sites with more than four failed genotype calls for the “All Pipits” and “Berthelot's” data sets (--max-missing-count 4) and excluded the Z chromosome from all analyses as females have systematic biases related to coverage that could affect estimates of differentiation (--not-chr Z). Individual-level data for the quality-filtered and mapped “Tawny pipit” and “Berthelot's pipit” variants are summarized in Table S1.
2.4 Genome-wide diversity and divergence across archipelagos
Genetic diversity within populations was measured as average observed heterozygosity (HO) and nucleotide diversity (π) using vcftools. To provide estimates of genome-wide nucleotide diversity, per-site nucleotide diversity was calculated and a genome-wide mean with 95% confidence intervals was generated for each individual. Individual inbreeding coefficients (FIS) were calculated across the Berthelot's pipit genomes, based on the mapped and quality filtered marker sets in both vcftools and plink version 1.9 (Chang et al., 2015). The plink method of calculating inbreeding, which uses a single-point calculation, simply reflects the proportion of heterozygous loci and is not sensitive to the presence of linkage disequilibrium (LD; Polašek et al., 2010). plink was also used to calculate individual inbreeding coefficients based on genome-wide estimates of heterozygosity; values were strongly correlated to those calculated by vcftools (Pearson correlation; r = .998), so only the values calculated by vcftools were reported.
To visualize genome-wide structure between the tawny pipit and the three archipelagos of Berthelot's pipit, a principal component analysis (PCA) was performed using plink. The strength of genetic divergence was then assessed using Wright's fixation index (FST; Willing et al., 2012). Pairwise FST values were calculated in 50-kb single nucleotide polymorphism (SNP) windows between each population using vcftools. Mantel tests were used to test for associations between PSMC colonization time estimates (see below) and mean pairwise FST values. Mantel test p-value estimates were generated from 100,000 randomized permutations, performed using the ade4 package in R (Dray & Dufour, 2007).
2.5 Ne over time and population divergence timescales
Historical fluctuations in Ne were estimated from 2.5 Ma until ~5 ka using single genomes in PSMC models (Li & Durbin, 2011), a method for studying ancient demographic history from single unphased and fragmented genomes. Population trends were mapped from before island colonization and speciation of Berthelot's pipit, through initial stages of divergence across the three North Atlantic archipelagos. Using individual-level bam files with duplicate polymerase chain reaction (PCR) reads marked, consensus genome sequences (fastq) were generated using the mpileup command (with -C50 flag to adjust the mapping quality for reads containing excessive mismatches) in samtools (Danecek et al., 2021) and the vcf2fq command from vcfutils.pl, with the Berthelot's pipit draft assembly as the reference. Each fastq file was filtered for sequencing errors by excluding sites at which the root-mean-square mapping quality of reads covering the site was <25, the inferred consensus quality was <20, and the variant read depth was either more than twice the average or <10× across the genome. All genomes had a mean read coverage >20×, variant coverage >19× and very low levels of individual missingness (<5%), enabling accurate estimation of genotype states for most sites (Han et al., 2014), which follows filtering recommendations as used to infer demographic history from PSMC modelling in other avian studies (Nadachowska-Brzyska et al., 2015, 2016).
The PSMC analyses were performed using the following fixed parameters across each individual: maximum number of iterations (N) of 30, maximum coalescent time (t) of 5, initial theta/rho ratio (r) of 1 and parameter pattern (p) of “4 + 30*2 + 4 + 6 + 10.” The above parameters were able to provide good resolution and showed more than 10 recombination events in each of the atomic time intervals within 20 iterations. These values were chosen in line with PSMC analyses conducted across other avian species (36 avian species; see Nadachowska-Brzyska et al., 2015).
Bird et al. (2020) estimate generation lengths of 2.05 and 2.20 years for the Berthelot's pipit and tawny pipit, respectively. Therefore, a generation time of 2.20 was used to scale the outputs from PSMC analyses (in the psmc_plot.pl command). A neutral mutation rate of 2.3 × 10−9 derived using a three-generation pedigree from the collared flycatcher (Ficedula albicollis, Smeds et al., 2016), which is near the average (2.28 × 10−9) reported across 38 avian species (Nadachowska-Brzyska et al., 2015), was also used.
2.6 ROH and genetic diversity across genomes
The length and distribution of ROH across individual Berthelot's pipit genomes was identified using the SNP data sets with stringent depth and quality filtering (as above). The --homozyg function in plink was implemented to identify the length and distribution of ROH. A threshold was set for the minimum length of an ROH (kb). Because strong LD typically extending up to 50 kb is common throughout the genome, especially across bottlenecked populations in the Madeiran and Selvagens archipelagos (Martin et al., 2021), short segments of homozygosity are very prevalent. As the aim was to detect and compare IBD segments to infer differing population demography across the pipit's geographical range, parameters for detecting ROH were consistent across all populations. As recommended by Meyermans et al. (2020), LD or minor allele frequency (MAF) trimming prior to ROH detection was not performed. Instead, two size categories of ROH were implemented in plink via the --homozyg function: (i) long ROH >1 Mb in length (--homozyg-kb 1000), to exclude ROH likely to be derived from ancient population processes; and (ii) shorter ROH >250 kb in length (--homozyg-kb 250), likely to reflect ancient, as well as recent, population processes.
Parameters were defined based on an assessment of sequence quality and genome SNP densities (0.1 kb/SNP), following the recommendations of Meyermans et al. (2020). The following thresholds were set: a minimum scanning window size of 50 SNPs (--homozyg-window-snp 50); a minimum density of one SNP per 50 kb on average (--homozyg-density 50; 50 kb/SNP); and a maximum gap between consecutive SNPs of 200 kb (--homozyg-gap 200). Occasional heterozygous positions within ROH resulting from sequencing errors, read mapping errors and mutations were accounted for: Specifically, it was accepted that 2% of SNPs would be heterozygous within IBD segments (--homozyg-window-het 2) and up to five missing genotype calls within a scanning window (--homozy-window-missing 5).
The TMRCA was approximated for ROH of different length classes, where the expected length of ROH L = 100/(2 t) centimorgans (cM), where t is time back to the common ancestor in generations (see, for examples, Foote et al., 2021; Peripolli et al., 2018; Stoffel et al., 2021). Recombination rate is approximated to be 1 cM/Mb, at the lower range of avian estimates, due to small Ne across Berthelot's pipit range (Backström et al., 2010; Burri et al., 2015). As Berthelot's pipits have a relatively short generation time (~2 years), the minimum ROH length threshold of >250 kb reflects the expected length when the underlying IBD haplotype has an TMRCA < 200 generations (<400 years) ago, while ROH >1 Mb correspond to an TMRCA < 50 generations (<100 years) ago.
To visualize the landscape of genetic diversity across individual genomes, nucleotide diversity was calculated across two window sizes (250 kb and 2 Mb, to assess diversity patterns at different genomic scales), each with a 20% smoothing step. The locations of long ROH (>1 Mb) and short ROH (>250 kb) were then mapped against genomic patterns of nucleotide diversity, to visually compare ROH distribution between individuals and populations.
Estimates of individual inbreeding coefficients based on ROH were derived by calculating the proportion of the autosomal genome that is covered by ROH segments above a specified length, FROH (McQuillan et al., 2008):
FROH = ∑LROH/LTOTAL.
where LROH and LTOTAL are the total length of all ROH segments and the genome, respectively. The size of the autosomal genome was considered as ~1057 Mb according to the Zebra finch reference genome assembly bTaeGut1_v1.p (NCBI Assembly GCA_003957595.1) used in this study. The correlation between FROH and FIS was measured using Pearson's correlation.
3 RESULTS
3.1 Whole-genome resequencing
Sequencing resulted in 1,030,115,042 paired-end reads (80 × 106–120 × 106 per individual), with a mean insert size of 401 bp. Genome alignment and mapping resulted in mean (±SD) read coverage of 23.6 ± 2.6× per individual, when mapped to the Zebra finch's 1.1-Gb genome (Warren et al., 2010). Reads were mapped to the contig level assembly of the Berthelot's pipit reference genome and genotypes joint called, resulting in 19,781,461 raw “All Pipits” variants of which 13,253,579 (67.6%) were mapped to the Zebra finch chromosomes. The “Berthelot's” data set resulted in 10,363,127 raw variants of which 6,953,309 (67.1%) were mapped to the Zebra finch chromosomes. The percentage of pipit genomic variants that were positioned on the Zebra finch genome is low relative to well-assembled reference genomes (e.g., Brawand et al., 2015; Scally et al., 2012) but is comparable with other studies using fragmented genome assembles (Liu et al., 2018).
Subsequent quality filtering resulted in a “All Pipits” data set with 11,575,905 autosomal mapped SNPs and a “Berthelot's” data set with 5,575,905 SNPs. Indels and SNPs with more than two genotypes were removed; the minor allele count was ≥1; >99.9% genotype variant accuracy; genotype coverage range (i.e., depth per allele) = 10–44/45; and maximum of four missing genotypes across all individuals. Individuals had low levels of missing data even prior to variant quality filtering, with no individuals having >5% missing data. The final depth of coverage for the quality-filtered SNPs was high (Jiang et al., 2019; Sims et al., 2014), with a mean 24.6× for the “Berthelot's” data set (Table S1) and 22.6× for the “All Pipits” data set.
3.2 Loss of genetic diversity during island colonization and bottlenecks
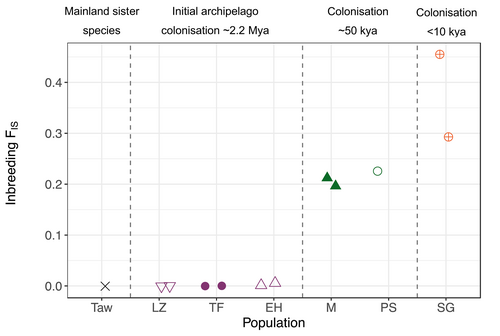
Genome-wide nucleotide diversity, heterozygosity and inbreeding for each individual are shown in Table 1. The largest reduction in all diversity measures was between the tawny pipit and Berthelot's pipit. In the tawny pipit, average heterozygosity (HO) across polymorphic SNPs was high (0.405). Across Berthelot's pipit populations in the Canary Islands, heterozygosity was 0.127–0.135 with the lowest diversity in the western island of El Hierro and the highest in Lanzarote on the eastern edge of the archipelago (Table 1). Heterozygosity was much lower in the Madeiran archipelago (0.101–0.107) and even more so in the Selvagens (0.082–0.092). Genome-wide nucleotide diversity showed a similar pattern with reduced diversity across the Madeiran (0.0011–0.0012) and the Selvagens (0.0008–0.0010) archipelagos compared to the Canaries.
| Archipelago/location | Pop. Code | Sex (individuals) | Mean π | 95% CI π | H O | FROH > 250kb/FIS |
|---|---|---|---|---|---|---|
| Mauritania, Mainland Africa | Taw | Male | 0.0047 | ±0.0010 | 0.405 | 0.002/0.000 |
| Canary Islands | LZ | Male (1) | 0.0015 | ±0.0003 | 0.135 | 0.015/0.019 |
| Female (2) | 0.0015 | ±0.0003 | 0.133 | 0.016/0.010 | ||
| Canary Islands | TF | Male (1)a | 0.0015 | ±0.0004 | 0.134 | 0.008/0.001 |
| Female (2) | 0.0014 | ±0.0001 | 0.130 | 0.039/0.044 | ||
| Canary Islands | EH | Male (1) | 0.0014 | ±0.0000 | 0.127 | 0.039/0.051 |
| Female (2) | 0.0014 | ±0.0002 | 0.132 | 0.032/0.047 | ||
| Madeira | M | Male (1)a | 0.0011 | ±0.0000 | 0.101 | 0.138/0.248 |
| Female (2) | 0.0012 | ±0.0000 | 0.107 | 0.130/0.233 | ||
| Madeira | PS | Female | 0.0011 | ±0.0000 | 0.101 | 0.136/0.261 |
| Selvagens | SG | Male (1)a | 0.0010 | ±0.0000 | 0.092 | 0.248/0.325 |
| Female (2)a | 0.0008 | ±0.0000 | 0.082 | 0.377/0.480 |
Inbreeding coefficients varied substantially between Berthelot's pipit populations and the tawny pipit (Table 1; Figure 2), with a near absence of inbreeding in the tawny pipit, increasing an order of magnitude in Canary Islands individuals (FIS = 0.001–0.051), and with high levels of inbreeding in both Madeiran populations (FIS = 0.233–0.261), and exceptionally high levels in the Selvagens (FIS = 0.325–0.480).
A PCA using the “All Pipits” SNPs showed that the strongest levels of genomic differentiation are between the tawny and Berthelot's pipit (Table 2), with the first principal component explaining 7.8% of the variation (Figure S2a). Using only the “Berthelot's” data set to perform a PCA (Figure S2b), populations separated by archipelago along both the first and second principal component, with a gradient from Selvagens to the Canary Islands to Madeira describing 2% of the genomic variation for PC1 and 1.7% for PC2. Pairwise FST results reflect those from the PCAs (Table 2). Pairwise FST between the tawny pipit and the Berthelot's pipit populations were high (FST > 0.42 with Canary Islands, and >0.36 and 0.54 for Madeira and Selvagens, respectively). Among Berthelot's pipits, populations from closely located islands within an archipelago had low FST ranging from 0.019 to 0.088, while between-archipelago populations separated by a single colonization event were marginally higher for most comparisons (FST = 0.033–0.119). Those populations having experienced two independent between-archipelago founding events (i.e., Madeira and the Selvagens) showed a significant association between genome-wide FST and log-transformed divergence time frames since colonization (r = .93, p = .044).
| Taw | TF | LZ | EH | SG | M | PS | |
|---|---|---|---|---|---|---|---|
| Taw | — | 0.424 | 0.431 | 0.438 | 0.538 | 0.516 | 0.368 |
| TF | 2.2 Ma | — | 0.019 | 0.026 | 0.106 | 0.096 | 0.033 |
| LZ | 2.2 Ma | 45 ka | — | 0.026 | 0.106 | 0.098 | 0.037 |
| EH | 2.2 Ma | 35 ka | <10 ka | — | 0.119 | 0.109 | 0.054 |
| SG | 2.2 Ma | 15—25 ka | 45 ka | 40 ka | — | 0.214 | 0.261 |
| M | 2.2 Ma | 50 ka | 50 ka | 40 ka | 40 ka | — | 0.088 |
| PS | 2.2 Ma | 50 ka | 50 ka | 40 ka | 40 ka | <10 ka | — |
- Note: Divergence times (below diagonal) are estimated from shared ancestry based on PSMC effective population sizes estimates.
- Within-archipelago island populations with potential for gene flow are coloured grey, between-archipelago populations separated by a single founding event in blue, and between-archipelago populations separated by two founding steps in orange. Populations: tawny pipit (Taw), El Hierro (EH), Tenerife lowland (TF), Lanzarote (LZ), Madeira (M), Porto Santo (PS) and Selvagem Grande (SG).
3.3 Inferring fluctuations in historical Ne and population divergence timescales
We used PSMC modelling to infer fluctuations in Ne from 2.5 Ma until 5 ka. Results show that the Ne curves of Berthelot's and tawny pipits were convergent from about 2.2 Ma, at Ne 200,000, indicating a shared ancestry and demography (Figure 3 inset). From this point in time they started to diverge to form distinct and nonoverlapping population histories (Figure 3). Our results indicate a larger ancestral Ne for the tawny pipit than across the island range of the Berthelot's pipit, with strong population growth until 150 ka (although this may be a consequence of population structure changes; Mazet et al., 2016) and more recent Ne estimates at least 10-fold higher than for Berthelot's pipit (Figure 3 inset).
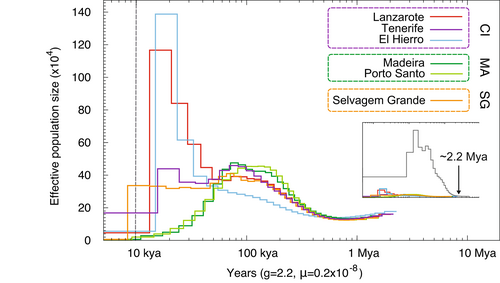
Across the contemporary Berthelot's pipit range, the PSMC results clearly indicated that all six analysed populations shared ancestry and demography for most of the investigated time period (Figure 3). The species Ne started to increase from ~200,000 ~ 1 Ma, until ~50 ka when two of three genomes from individuals from the populations of Madeira and Porto Santo reflect a gradual decline in Ne (Figure S3). Meanwhile, the Canary Island populations experienced continued population expansion or stabilization of Ne until 15 ka. A strong decline in Ne was observed in the Selvagens individuals ~7–8.5 ka. At 5000 years before the present, estimated Ne was at least four-fold greater across the Canary Island populations, compared to both Madeira and the Selvagens. Despite this strong signal of declining Ne, caution should be taken when interpreting population trends within the last 10,000 years as they are more prone to error (Li & Durbin, 2011).
3.4 Landscapes of diversity and ROH across genomes
The landscape of nucleotide diversity (π) varied significantly within individual genomes, with peaks and valleys of diversity within individual chromosomes (Figure 4). Broadly, patterns of diversity within chromosomes are reflected across the few individuals sampled (i.e., shared locations of peaks and valleys between individuals), with similar patterns in the tawny pipit and across the three Berthelot's pipit archipelagos (Figure 4; Figure S4). However, absolute diversity is three-fold higher in the tawny pipit compared to the average in Berthelot's pipit and there are only a few regions of the genome where diversity is comparable. Low levels of genomic diversity, characterized by large regions with very low genetic diversity, are observed in the more recently colonized archipelagos of Madeira and the Selvagens.

ROH genotypes were statistically quantified within two size categories indicative of the probable timing of their formation (Kirin et al., 2010; McQuillan et al., 2008): 250 kb–1 Mb and > 1 Mb. Long ROH ranged from 1 to 5.5 Mb (Figure 5), representing regions with shared ancestry 10–50 generations (~20–100 years) ago, while short ROH 250 kb–1 Mb in length originated 50–200 generations (~100–400 years) ago. No long ROH, >1 Mb, were detected in the tawny pipit genome and only a small number were detected across the Canary Island populations (Table 3; Figure S4). Signatures of IBD were very strong in the genomes of Selvagen individuals with long ROH (>1 Mb) extending across 10.8%–12.1% (130–145 Mb) of the genome, with the highest density of ROH on chromsomes 2 and 3 (Figure 4; Figure S4). The longest ROH (>4 Mb) are found in one of the Selvagens individuals, indicative of recent inbreeding (Figure 5). ROH > 250 kb are also prevelant across the genomes of individuals from the Maderian archipelago (Figure 5), with similar prevalence across the two sampled popualtions covering 11.4%–12.2% (137–146 Mb) of the genome (Table 3; Figure S3). The location of ROH varies considerably even between individuals within the same populations (Figures 4 and 6). However, some ROH locations are shared between individuals (see, for example, the Madeiran individuals; Figure S4).

| Archipelago/location | Population (individual) | No. of all ROH (>250 kb)a | Total length all ROH (kb) | No. of short ROH (250 kb–1 Mb) | Total length short ROH (kb) | No. of long ROH >1 Mb | Total length long ROH (kb) |
|---|---|---|---|---|---|---|---|
| Mauritania, Mainland Africa | Taw | 5 | 1749 | 5 | 1749 | 0 | 0 |
| Canary Islands | LZ (1) | 27 | 16,331 | 24 | 11,549 | 3 | 4781 |
| LZ (2) | 33 | 16,537 | 30 | 12,863 | 3 | 3673 | |
| Canary Islands | TF (1)a | 19 | 8808 | 19 | 8808 | 0 | 0 |
| TF (2) | 65 | 41,552 | 54 | 26,993 | 11 | 14,559 | |
| Canary Islands | EH (1) | 70 | 41,381 | 63 | 27,102 | 7 | 14,278 |
| EH (2) | 63 | 33,398 | 58 | 26,726 | 5 | 6672 | |
| Madeiran | M (1)a | 286 | 146,339 | 266 | 118,311 | 20 | 28,028 |
| M (2) | 285 | 137,354 | 264 | 110,350 | 21 | 27,004 | |
| Madeiran | PS | 296 | 143,844 | 280 | 122,945 | 16 | 20,899 |
| Selvagens | SG (1)a | 327 | 262,107 | 254 | 131,667 | 73 | 130,440 |
| SG (2)a | 594 | 398,181 | 280 | 253,352 | 94 | 144,829 |
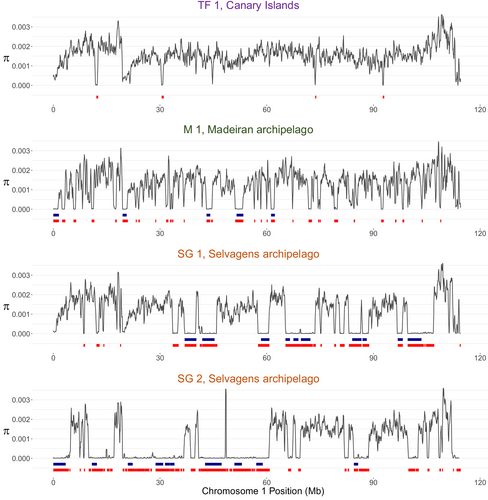
Within individuals, many ROH are clustered in chromosomal regions, suggesting these originated as larger autozygous segments, which have been eroded by mutations and recombination. As well as clustered ROH, we observe several stretches of 2–6 Mb ROH segments across genomes from both Selvagens birds, the Maderian island populations and one bird from El Hierro (a small and peripheral population in the Canary Islands).
Across all populations, measures of inbreeding based on genome-wide heterozygosity (FIS) were strongly correlated with those calculated based on the proportion of an individual's genome in ROH (FROH > 250 kb; Table 1; Pearson correlation; r = .977, Figure S5). It is important to note that substantially lower FROH are inferred when only ROH > 1 Mb are considered, as only a small proportion of autozygous segments are in contiguous loci at least 1 Mb in length (Table 3). The proportion of the genome in ROH (FROH) was very low for the tawny pipit, with only five short segments detected (FROH = 0.002), and relatively few, mostly short, ROH were detected across the Canary Island populations (FROH = 0.008–0.039). Populations that experienced strong historical founder events had a substantially greater proportion of their genome in ROH (Figure S6b, Selvagens FROH = 0.248–0.377; Porto Santo FROH = 0.136, Madeira FROH = 0.130–0.138). Short ROH were far more prevalent than ROH > 1 Mb across all populations of Berthelot's pipit, representing approximately one-third of total ROH detected across genomes (Figure S6b). While a similar number of short ROH were detected across the Madeiran and the Selvagens archipelagos, many more (>3×) long ROH were detected across the Selvagens. Finally, variance in inbreeding within populations is also apparent within Tenerife, the Selvagens and El Hierro—the total number, length and genomic location of ROH segments varied between individuals within a population (Figures S4 and S6).
4 DISCUSSION
Using whole-genome resequencing, we examined genetic diversity and demographic history of Berthelot's pipit through speciation and sequential island colonization events. We find a considerable loss of genetic diversity through colonization events, with the most significant drop during the initial mainland to island population event ~2 million years ago. We also identify genome-wide signatures of ROH resulting from a combination of ancient bottleneck effects and contemporary inbreeding.
Examining the distribution of ROH and effective population sizes (Ne) suggests three things. First, sequential colonization events are associated with strong founder effects resulting in dramatic reductions in genetic diversity across genomes. Second, ROH length correlates with the timing and severity of population bottlenecks, with high prevalence of long ROH (>1 Mb) associated with recent and severe population size reductions, probably exaggerated by limited post-colonization gene flow and high background relatedness. Third, long ROH gradually degrade to form shorter more numerous ROH, the distribution of which are in agreement with estimated fluctuations in Ne across populations. Furthermore, the data provide evidence that the initial colonization of the Canary Islands was ~2.2 Ma, which closely supports previous estimates based on mitochondrial DNA (Voelker, 1999). Our results also concur with the idea that separate secondary colonization events occurred to the Madeiran and to the Selvagens archipelagos (Spurgin et al., 2014). However, our findings suggest a much earlier colonization of Madeira 50 ka, resulting in a population bottleneck and more recent population recovery. In contrast, the Selvagens appears to have been colonized in the more recent past (<10 ka) and have experienced a further subsequent severe population reduction. Overall, we have quantified and tracked the severity of the genetic reduction since Berthelot's pipit split from its continental ancestor until the last colonization event. Such a progression is rarely tackled in genomic studies (e.g., Armstrong et al., 2018; Ibrahim et al., 2020; Recuerda et al., 2021), but it is a critical point to understanding the evolutionary history of island species.
Using whole-genome resequencing we find that genetic diversity (HO and π) shows the most dramatic reduction between the tawny pipit and Berthelot's pipit and is lowest in populations that have experienced sequential archipelago colonizations and associated population bottlenecks (Table 1). We find weak signatures of inbreeding (few short ROH, low FROH and FIS < 0.06) across the Canary Islands—the first archipelago the Berthelot's pipit colonized—but much higher levels in the Madeiran and Selvagens archipelagos, FIS > 0.2 (Figure 4, Table 1). Genetic diversity is lower than that reported among other vertebrates, including those with contracting populations (Dutoit et al., 2017; Kardos, Åkesson, et al., 2017; Yu et al., 2004), with similar nucleotide diversity to severely range restricted birds (e.g., Raso lark [Alauda razae] restricted to one islet in Cape Verde, Dierickx et al., 2020). Levels of inbreeding across the Canary Islands were comparable to various other avian island populations which have experienced strong population bottlenecks, and more severe in the Madeiran and Selvagens archipelagos: for example, historical bottleneck of 20 pink pigeon (Columba mayeri, Swinnerton et al., 2004); founder population of 33 North Island Robins (Petroica longipes, Jamieson et al., 2007); and mangrove finch (Camarhynchus heliobates) long-term bottleneck and small Ne (Lawson et al., 2017). Our comparisons of genome-wide diversity and population structure in Berthelot's and tawny pipits support the findings from previous studies of Berthelot's pipit using reduced representation RAD-sequencing (Armstrong et al., 2018; Martin et al., 2021), microsatellites and mitochondrial DNA (Illera et al., 2007; Spurgin et al., 2014). Using PCA and pairwise FST measures, we were able to further describe population structure: Berthelot's pipit diverged considerably from the tawny pipit, and moderate divergence also exists between the three Berthelot's pipit archipelagos, especially Madeira and the Selvagens (see Figure S2 and Table 2).
Using PSMC modelling, we estimate Berthelot's pipit diverged from the tawny pipit ~2.2 Ma and Ne was ~25,000 (Figure 3). These species have since become distinct, with no genomic signatures of shared ancestry since divergence and substantially lower Ne across island populations. Total Berthelot's pipit Ne steadily increased from 1 Ma until 150 ka, probably reflecting an expansion of their habitable range as volcanic activity and climate across the Canary Islands stabilized (see Figure 1). Recent population estimates suggest further ancestral splits between Berthelot's pipit populations within the last 50,000 years, which may point to earlier divergence across the three archipelagos than previously estimated (Spurgin et al., 2014). Interestingly, these results are roughly coincident with the demographic trajectories of blackcaps (Sylvia atricapilla) in Macaronesia, where resident birds on the Canary Islands, Azores and Cape Verde began to diverge within the last 30,000 years with significant reductions of their island effective population sizes (Delmore et al., 2020). It is plausible to speculate that palaeoclimatic events fuelled recurrent colonization events with similar demographic trajectories in the region (Illera et al. 2012). Small Berthelot's pipit Ne is estimated across the range at 5000 years ago (Ne < 1000 in Selvagens and Madeiran archipelago populations, and < 17,000 across Canary Island populations; Figure 3). Colonization of the Selvagens is likely to have occurred 7–8.5 ka. PSMC results do not rule out the possibility of multiple colonization events to the Madeiran archipelago, since we report differences in Ne estimated from the genomes of different individuals 10–60 ka (Figure S3). Reasons for severe population contraction across the range are unknown but may include pathogenic pressures, fitness effects of low diversity (i.e., inbreeding depression), or volcanic and climatic disturbances. PSMC models do not provide accurate information on recent Ne (present to 10 ka) and are sensitive to ancestral population structure and admixture (Li & Durbin, 2011).
We also studied patterns of genetic diversity across individual genomes to reveal signatures of demographic history. Despite differences in genome-wide levels of π, peaks and troughs of diversity were generally consistent between individuals both within the same population and across Berthelot's pipit populations and the tawny pipit (Figure 4; Figure S3), as has been reported in other avian studies (Dutoit et al., 2017). However, this is not the case for the location of ROH, for which prevalence, but not genomic location, correlates within populations (Figure S4). It is very likely that these regions represent true inbreeding instead of being consequences of shared chromosomal features (e.g., centromeres) as ROH in these regions are absent within the genomes of outbred pipits, for example across the Canary Islands (Figure 4). That the location of ROH varies strongly even between individuals within the same population also suggests that these signatures are not solely a result of strong selection within particular islands. We detected ROH across the genome of all individuals (Figure 5) and generally find (i) few short ROH in the tawny pipit and individuals from large Canary Island populations, (ii) an increased proportion of the genome in ROH (FROH) for individuals from Madeira and the Selvagens, and (iii) longer ROH in the Selvagens genomes relative to all other islands. This further supports the idea of an ancient bottleneck in the Madeiran archipelago with moderate contemporary background inbreeding, and a severe more recent bottleneck in the Selvagens, with an absence of post-colonization gene flow.
Where population contraction is very rapid it is possible there may be no signs of inbreeding (Gelabert et al., 2020), but many severely inbred species do fulfil the expectation of having numerous short ROH and a few very large ROH, such as ~17 Mb ROH in the California condor (Gymnogyps californianus, Robinson et al., 2021) and ~95 Mb ROH in a highly inbred population of grey wolves (Canis lupus, Kardos, Åkesson, et al., 2017), probably originating from the TMRCA within three generations. In the Selvagens' Berthelot's pipit population, we detect several 3- to 6-Mb ROH (Figure 5), probably originating from a common ancestor <10–20 generations (20–40 years) ago (see Figure 6). It is possible that larger autozygous regions, as reported by some other studies, do exist within Berthelot's pipit genomes but that we are unable to detect these due to unmapped or misplaced contigs within our highly fragmented genome assembly. Nevertheless, our comparisons across the Berthelot's pipit colonization range suggest the longest ROH result from contemporary inbreeding in the small, isolated population in the Selvagens.
We also detect variance in inbreeding between individuals within the same population, based on individual-level observed heterozygosity and inbreeding estimates from ROH. This is particularly clear within the Selvagens archipelago. Such variance is common in populations where there are just a handful of family groups remaining and population-wide genetic diversity is very low (Jamieson et al., 2007). Individual inbreeding is expected to fluctuate over short timescales in such populations, depending on the level of close relative mating. In a wild population it is likely that a high population average inbreeding reflects high background relatedness in the population as a result of founder effects or historical inbreeding, with individuals with exceptionally high inbreeding as a result of close parental ancestry (Brzeski et al., 2014).
High levels of inbreeding can result in inbreeding depression, which has been shown to be associated with phenotypic variation, survival and reproductive success in many natural populations (Brzeski et al., 2014; Jamieson et al., 2007; Richardson et al., 2004; Sin et al., 2021). Despite this, inbreeding does occur in nature (Kirch et al., 2021; Tian et al., 2022), and fitness may not always be severely impacted, especially in an island setting where intraspecific competition may be reduced. Furthermore, extreme and prolonged bottlenecks are thought to result in the purging of deleterious alleles (Crnokrak & Barrett, 2002; Pérez-Pereira et al., 2022; Stoffel et al., 2021), but this is not always the case (Kennedy et al., 2014). In some situations, extreme genetic bottlenecks can instead result in the fixation of deleterious alleles, and thus it can be impossible to assess relative inbreeding depression within a population (see Van Oosterhout, 2020). We cannot link inbreeding directly to fitness in Berthelot's pipit populations as we have not monitored individuals throughout their lifetime. However, it is plausible to think that high levels of inbreeding in the Selvagens archipelago may have led to inbreeding depression (Szpiech et al., 2013). Furthermore, census field population monitoring (2010–2021) suggests a further recent population crash from 315 individuals in 2010 to fewer than 73 individuals in 2021 (D. Menezes, unpublished data; Table S2). Inbreeding depression and a lack of adaptive potential may threaten the long-term viability of the Madeiran and (especially) the Selvagens populations. This may be exacerbated if they are exposed to new environmental pressures, such as introduced infectious diseases (Jarvi et al., 2001) and climate change (Wood et al., 2017). The results presented here are therefore of importance to conservation management of Berthelot's pipit, particularly the populations endemic to the Selvagens archipelago.
Despite the strong patterns of increasing inbreeding and ROH length we observed through island colonization events, our results rely on data from a small number of individuals and populations. Studying population demography using whole-genome resequencing from many more individuals across the Berthelot's pipit range would further develop understanding of the processes shaping genetic variation within and between populations, allowing investigation of shared ROH locations, for example. In addition, further research is required to understand lost or altered gene function in regions with exceptionally low diversity to uncover potential traits where variation has been lost.
5 CONCLUSIONS
Genomic tools can be used to study contemporary and historical population demography, providing an opportunity to understand how genetic diversity is shaped across populations. We assessed patterns of genetic diversity across the Berthelot's pipit contemporary range, revealing that island colonization and sequential founder events result in cumulative reductions in genetic diversity, inbreeding within populations and rapid divergence among populations. It is likely that post-colonization population expansion across the Madeiran archipelago has resulted in genetic recovery which can be observed via many short ROH segments, while the Selvagens has experienced a more recent severe bottleneck and high background inbreeding, with ROH covering as much as 37.7% of autosomes. With ongoing decline of animal populations, climate-driven range shifts and habitat fragmentation, understanding the evolutionary processes behind the loss of genetic diversity across small and isolated populations may aid conservation efforts. Taken together, our study shows how whole-genome resequencing data can be used to deepen our understanding of how past and present population history shape contemporary genetic diversity and its role in speciation.
AUTHOR CONTRIBUTIONS
CAM, DSR and LGS conceived and found funding for the project. CAM performed DNA extractions, bioinformatics and genomic analyses with contributions from LGS. Fieldwork was undertaken by LGS, DSR and JCI. Guidance on PSMC analysis was provided by KNB. The first draft of the paper was written by CAM with input from DSR and all authors. All authors contributed to discussing the results and approved the submission of the final manuscript.
ACKNOWLEDGEMENTS
We thank Matthew Clark and Lawrence Percival-Alwyn for assistance in generating the pipit reference genome. We also thank Martin Taylor, Mike Ritchie, Josephine Pemberton and three anonymous reviewers whose comments improved the manuscript. We are grateful to the Regional Governments of the Canary Islands (Ref.: 2019/5555), the Cabildo of Lanzarote (Ref.: 101/2019), the Cabildo of Tenerife (Ref.: 2019-01740) and Madeira (Ref.: 06/IFCN/2020) for their permission to sample Berthelot's pipits, the Governments of the Canary Islands and Madeira for providing accommodation, and the Portuguese Navy for transport to Selvagem Grande. The bioinformatics analyses were carried out on the High-Performance Computing Cluster supported by the Research and Specialist Computing Support service at UEA. This work was supported by Natural Environment Research Council (NERC) studentships awarded to CAM through the EnvEAST DTP (NE/L002582/1) and ECS through the ARIES DTP (NE/S007334/1). Genome sequencing was funded through a Norwich Research Park Science Links Seed Fund to DSR. JCI was funded by a research grant from the Spanish Ministry of Science, Innovation and Universities, and the European Regional Development Fund (PGC2018-097575-B-I00) and by a regional GRUPIN grant from the Regional Government of Asturias (AYUD/2021/51261).
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The genomic data supporting this study and code used to perform the data analyses within this article are openly available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.ksn02v75k (Martin et al., 2022).




