Population genomics of Corsican wildcats: Paving the way toward a new subspecies within the Felis silvestris spp. complex?
Sandrine Ruette and Sébastien Devillard share equal responsibilities.
Abstract
In the context of the current extinction crisis, identifying new conservation units is pivotal to the development of sound conservation measures, especially in highly threatened taxa such as felids. Corsican wildcats are known by Corsican people since a very long time but have been little studied. Meaningful information about their phylogenetic position is lacking. We used ddRADseq to genotype phenotypically homogenous Corsican wildcats at 3671 genome-wide SNPs and reported for the first time their genetic identity. We compared this genomic information to domestic cats Felis silvestris catus from Corsica and mainland France, European wildcats F. s. silvestris and Sardinian wildcats F. s. lybica. Our premise was that if the Corsican wildcat, as a phenotypic entity, also represents a genetic entity, it deserves conservation measures and to be recognized as a conservation unit. Corsican wildcats appeared highly genetically differentiated from European wildcats and genetically closer to Sardinian wildcats than to domestic cats. Domestic cats from Corsica and mainland France were closer to each other and Sardinian wildcats were intermediate between Corsican wildcats and domestic cats. This suggested that Corsican wildcats do not belong to the F. s. silvestris or catus lineages. The inclusion of more high-quality Sardinian samples and Near-Eastern mainland F. s. lybica would constitute the next step toward assessing the status of Corsican wildcat as a subspecies and/or evolutionarily significant unit and tracing back wildcat introduction history of in Corsica.
1 INTRODUCTION
Because of human activities, biodiversity is currently facing an extinction crisis (Brummitt et al., 2015; Ceballos et al., 2015; Cowie et al., 2017, 2022; Hallmann et al., 2017; Thomas et al., 2004). This crisis represents a threat to global biodiversity but also to humanity and its activities (Ceballos et al., 2015, 2020; Diaz et al., 2019) making biodiversity conservation, besides a moral obligation, an urgent need to ensure a viable environment for future human generations. Biodiversity is defined according to three levels (genetic, species and ecosystem diversities) which, to be adequately protected, need to be characterized and their functioning understood. For instance, among the information necessary for the design of suitable conservation and management strategies, species, subspecies or evolutionarily significant unit delineation and biogeography characterization are part of the first steps (Coates et al., 2018). Identifying populations of the same species and/or unique gene pools, indeed, helps to assign a legal status to this genetic diversity that can then be considered in conservation plans, favouring the protection of both the species and the global biodiversity (Coates et al., 2018). Genetic diversity is crucial to preserve since it is linked to the fitness of individuals, the short-term maintenance of populations, but also to the adaptive potential of populations, and thus, to the ability of species to cope with environmental changes (DeWoody et al., 2021; Frankham et al., 2004; Hoban et al., 2021 and references therein). In addition to this conservation significance, identifying new and previously ignored specific entities is also a positive and enthusiastic message sent to both the conservation biology community and governments, underlying that work performed in investigating biodiversity might be successful. Defining conservation units is nevertheless challenging, although the advance of population genomics approaches greatly improved our understanding of the species-population continuum and facilitated the identification of relevant units (Coates et al., 2018; Funk et al., 2012; Hohenlohe et al., 2021).
In the case of wild species having domestic relatives, it is even more challenging. Indeed, many of the wild ancestors of domestic species are understudied and have uncertain status due to the frequent lack of clear phenotypical demarcation and the hybridization with domestic relatives which may participate to obscure delineation signals (Smith et al., 2022). One example of such species of which the conservation is challenged partly due to the existence of domestic population are wildcats Felis silvestris. Although felids are one of the most intensively studied group and the target of large conservation efforts, wildcats do not benefit from the same level of concern as the largest and most charismatic species of this group (Albert et al., 2018; Anile & Devillard, 2020). Despite being considered as “Least Concern” by the IUCN, Felis silvestris is experiencing a population decline and is considered an endangered taxon, at least locally, in some countries, where it is protected by European laws (Yamaguchi et al., 2015). Threats to wildcats include human activities (e.g., persecution, road kills), habitat loss and fragmentation and hybridization with feral domestic cats Felis silvestris catus (Yamaguchi et al., 2015). The Felis silvestris lineage in Eurasia is actually subdivided into several subspecies, according to their geographical ranges: the African-Near Eastern wildcat (F. s. lybica), the Asian wildcat (F. s. ornata), the European wildcat (F. s. silvestris), the Southern African wildcat (F. s. cafra) and the Chinese cat (F. s. bieti) (Driscoll et al., 2007). F. s. silvestris diverged first, 230,000 years before present (BP) while F. s. cafra, F. s. ornata and F. s. lybica diverged from each other more recently (173,000 years BP, Driscoll et al., 2007). The African wildcat F. s. lybica is now recognized as the ancestor of domestic cats (Driscoll et al., 2007, 2009; Ottoni et al., 2017). The domestication process was initiated in the Near East during the Neolithic, about 10,000 years ago, and F. s. lybica then most probably followed human migration toward Europe as a commensal species (Baca et al., 2018; Faure & Kitchener, 2009; Nilson et al., 2022; Ottoni et al., 2017; Vigne et al., 2004).
During the migration from the Fertile Crescent to Europe, ancestral representatives of several mammalian species were introduced on Mediterranean islands, such as mouflons (Ovis gmelini musimon), with individuals being first introduced in Cyprus around 10,500 years BP and reaching Sardinia and Corsica 3000–4000 years later (Poplin, 1979; Vigne, 1992; Zeder, 2008) but also of red foxes (Vulpes Vulpes), house mice (Mus musculus), wild boars (Sus scrofa) or weasels (Mustela nivalis) and probably wildcats (Cucchi et al., 2020; Faure & Kitchener, 2009; Lebarbenchon et al., 2010; Ottoni et al., 2013). Some of these later species, such as sheep (Ovis aries) and pigs (Sus domesticus), were then domesticated and are thus thought to have been introduced on these islands as early domestic animals, while others, such as the house mouse and most probably cats, migrated as commensal species (Cucchi et al., 2020; Faure & Kitchener, 2009). The domestication process of animals at those times must nevertheless have remained very primitive (Rezaei, 2007; Zeder, 2008, 2012), and populations that since inhabit these islands might then represent unique gene pools needing to be recognized as such taxonomically and to benefit from conservation strategies based on scientific knowledge, as evidenced for mouflons or weasels (Guerrini et al., 2015; Lebarbenchon et al., 2010; Portanier, Chevret, et al., 2022). This is particularly relevant to Felis species. Indeed, F. s. lybica remains that dated back to around 9500 years BP were discovered in Cyprus (Vigne et al., 2004). In addition, Sardinian wildcats are genetically closer to F. s. lybica than to F. s. silvestris (Mattucci et al., 2016; Randi et al., 2001) and show some morphological specificities that led to their consideration as a specific variety F. s. lybica var. sarda (Mura et al., 2013). This highlights the singularity of the Sardinian wildcat gene pool, which might be one of the unique representatives of ancestral F. s. lybica introduced to the Western Mediterranean. Island populations, indeed, often evolved with low levels of competition due to both geographic and genetic isolation, which results in a high level of endemism and the creation of biodiversity hotspots, which are high-priority conservation areas (Kier et al., 2008; Loso & Ricklefs, 2009; Myers et al., 2000; Whittaker & Fernández-Palacios, 2007).
Through time, Sardinia and Corsica have shared a large part of their history, being one single landmass during the last glaciation but also sharing similar history of colonization by humans and genetic ancestry (Grimaldi et al., 2001; Tamm et al., 2019 and references therein). A high phylogenetic proximity has also been evidenced for several species they host (e.g., mouflon, Portanier, Chevret, et al., 2022; weasel Mustela nivalis, Lebarbenchon et al., 2010; Corsican red deer Cervus elaphus corsicanus, Doan et al., 2017). While the presence of wildcats in Corsica has been known for a long time, with Lavauden describing them for the first time in 1929 as “Felis reyi” and Arrighi and Salotti mentioning them in a scientific publication in 1988 (Arrighi & Salotti, 1988; Lavauden, 1929; see also Vigne, 1992), no further scientific studies have been performed on these animals. Based on fur characteristics, Arrighi and Salotti (1988) classified Corsican wildcats as Felis silvestris lybica, while Vigne (1988) classified other specimens as domestic cats returned to wildlife. Corsican people, nevertheless, have always distinguished “u ghjattu-volpe” (Corsican “cat-fox”), representing wildcats, and “u ghjattu insalvaticu”, representing feral cats. In accordance, the IUCN Cat Specialist Group reports Corsican wildcats as Felis lybica reyi (Kitchener et al., 2017). It is thus still unclear if these cats represent feral domestic cats or “true” wildcats and, in the latter case, if they are representatives of Felis silvestris silvestris or of Felis silvestris lybica, as in Sardinia. A major link is thus lacking in cat history in the Western Mediterranean and Europe. The main reason for the lack of recent studies on Corsican wildcats was that none have been detected in Corsica from the early 1980s to the 2000s. However, in 2008, a shepherd accidentally captured an animal that was probably a Corsican wildcat since it showed a similar phenotype to the one historically described (Arrighi & Salotti, 1988; Lavauden, 1929). This detection revived a monitoring program on the Corsican wildcat and during the 2010s, several noninvasive detections occurred through camera trapping. It allowed us to hypothesize that the Corsican wildcat was still present and to notice that these individuals were phenotypically similar across locations. Their phenotypes differed from domestic cat phenotypes and, as previously reported (Arrighi & Salotti, 1988; Lavauden, 1929), from European wildcat phenotypes. The Corsican phenotype was described as close to the Sardinian and African wildcat phenotype (see Arrighi & Salotti, 1988 and Materials and Methods below, but see Lavauden, 1929). Since then, multiple individuals that would phenotypically match the description of the Corsican wildcats have been detected and captured. It thus appeared urgent to resolve the taxonomic position of this Corsican wildcat phenotype.
Taxonomic uncertainties such as this one might indeed have deleterious effects on species conservation. On one hand, for the Corsican wildcat itself, since it would need to be officially recognized as a wild conservation unit to benefit from European conservation measures (Yamaguchi et al., 2015). On the other hand, it is necessary to confirm that Corsican wildcat are not feral cats since these may have significant detrimental effects on some vulnerable species, especially in island ecosystems (Nogales et al., 2013). If confirmed as a wild entity, Corsican wildcat conservation would also favour the preservation of numerous other species and habitat types since, like many carnivores, wildcats are an umbrella species (Jerosch et al., 2009, 2018; Noss et al., 1996; Virgos et al., 2002). It is a critical point since islands of the Mediterranean Basin are biodiversity hotspots hosting many endemic species (e.g., Escoriza & Hernandez, 2019; Grill et al., 2007; Jeanmonod et al., 2015) and particularly threatened by climate change (Courchamp et al., 2014; Ducrocq, 2016; Giorgi, 2006; Myers et al., 2000).
In the present study, the first one performed at such a scale on Corsican wildcats, we thus aimed to determine if Corsican wildcats are genetically distinct from domestic cats, Sardinian wildcats and European wildcats using population genomics approaches. We were particularly interested in evaluating if the Corsican wildcat should be considered as a management unit (MU) with a unique gene pool (Moritz, 1994; Palsbøll et al., 2007), a result that would favour a better conservation of Felis silvestris and open discussions about the definition of a new F. silvestris subspecies. To achieve this goal, we used double-digest restriction-site-associated DNA sequencing (ddRADseq) to genotype samples of wild and domestic cats from both Corsica and mainland France as well as Sardinian wildcats at genome-wide SNPs. Our premise was that if Corsican wildcats, as a phenotypic entity, also represent a distinct molecular entity, then the Corsican wildcat deserves conservation measures and conservation unit delineation.
2 MATERIALS AND METHODS
2.1 Study sites, capture protocol and phenotypic data
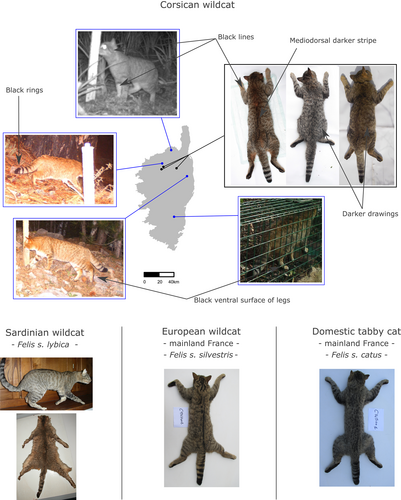
Overall, 72 samples were available for DNA sequencing, among which 10 and 12 were domestic cats from Corsica and mainland France, respectively; 20 and 23 were wildcats from Corsica and mainland France, respectively; and seven were Sardinian wildcats (Tables 1, S1). Corsican wildcat samples were obtained from 13 locations on the island (Figure 1, Table S1). Samples were collected from individuals captured in box-traps baited with fish, road-killed or old dead animals examined by Dr. M. Salotti and Dr B. Condé (Natural History Museum of Nancy, France) (see Tables 1, S1). All of these samples showed homogeneous phenotypes (Figure 2) and corresponded to the criteria described by Dr M. Salotti (Salotti, 1992). The coat of the Corsican wildcat indeed differs from that of the European wildcat and the domestic tabby cat but is close to that of the Sardinian wildcat. It is composed of two phases, a fawn grey and a fawn brown, on which are present characteristic drawings of darker tones or even black. The ventral surface of the forelegs and hind legs are black, and black lines are drawn on the forehead, the cheeks, the occipital and cervical regions, the neck, the forelegs and the hind legs. A darker mediodorsal stripe is present. The tail ends in three well-formed black rings, which are wider on their dorsal part, and in two or three sketches of rings less and less visible toward the trunk of the animal (Figure 2).
| Population | Subspecies | N | Sampling mode | Sampling years | Sample type |
|---|---|---|---|---|---|
| Domestic Corsican | F. s. catus | 1 | Road killed animal | 2014 | Ear punches in ethanol |
| 9 | Lived veterinarian animals | 2014 | Hair | ||
| Domestic mainland | F. s. catus | 12 | Road killed animals | 2010–2016 | Ear punches in ethanol |
| Wild Corsican | F. s. lybica? | 9 | Capture | 2016–2017 | Ear punches in ethanol or dried, hairs, tail tissue, dried skin |
| 2 | Dead animals | 2010–2018 | |||
| 6 | Dead animals | 1986–1992 | |||
| 3 | Museum specimen | 1964–1966 | |||
| Wild mainland | F. s. silvestris | 23 | Road killed animals | 1996–2016 | Ear punches in ethanol |
| Wild Sardinia | F. s. lybica | 7 | Museum specimen, road-killed animals | NA | Dried skin, Ear puch or liver in ethanol |
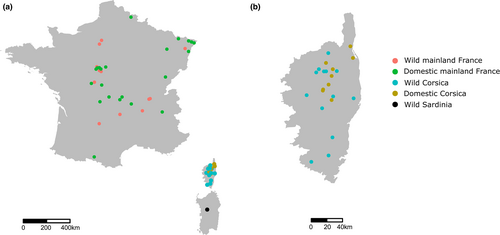
For captured animals, box-traps were placed in habitats known to be used by Corsican wildcats thanks to a monitoring that occurred in autumn 2016 using camera traps. When captured, animals were anaesthetized using Domitor (0.1 mg/kg; Orion Pharma) and revived with Antisédan (0.2 mg/kg; Orion Pharma) to ensure a rapid reversal of sedation. Before release, ear punches and hairs were sampled for molecular analyses and photographs were taken for phenotypic description. Trapping and handling were performed by trained French Biodiversity Agency (OFB) officers according to the appropriate national laws governing animal welfare, following the ethical conditions detailed in the specific accreditations delivered by the Ministry in charge of ecology in agreement with the French environmental code (Art. L. 411–1 and L 421–1 and R. 411–1). Domestic Corsican cats were sampled thanks to collaborations with veterinarians (Figure 1, Table 1). Mainland domestic and wildcat samples were obtained from road-killed animals monitored by the French Biodiversity Agency (Figure 1a). Assignment to either European wildcats Felis silvestris silvestris or domestic cats was confirmed genetically (following O'Brien et al., 2009; Devillard et al., 2014). Finally, Sardinian wildcats (n = 7, Table 1) were obtained from museum specimen and road-killed animals as a courtesy of M. Zedda.
2.2 DNA extractions, ddRADseq library preparation and sequencing
Genomic DNA was extracted using either NucleoSpin Tissue (Macherey-Nagel) or Blood and Tissue (Qiagen) kits (see Table S1), following the manufacturers' recommendations. Samples were thus lysed overnight at 56°C using proteinase K, and DNA was then purified and isolated using purification columns. The double-stranded DNA concentration of each sample was then quantified using a Qubit dsDNA HS Assay kit (Life Technologies), and purity was measured using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). ddRADseq libraries were prepared using 2–1200 ng of DNA according to sample quality (i.e., 1200 ng when possible, less otherwise, Table S1). The libraries were prepared based on the protocol of Peterson et al. (2012) but with modifications, in particular concerning the preparation of the adapters (see Henri et al., 2015). The detailed protocol can be found in Data S1. Briefly, it consisted in the double digestion of the DNA in each sample using SbfI and MseI enzymes (New England Biolabs), followed by ligation of P1-adaptors (including unique barcodes 8 and 10 bp long) and a common P2-adaptor. All samples were then pooled into a single sequencing library. Size selection of fragments (between 350 and 450 bp) was performed using Blue Pippin technology (Sage Science), and the Agilent 2100 Bioanalyser system (Agilent Technologies) was used to verify fragment sizes. The enrichment step was performed using 15 cycles of polymerase chain reaction (PCR) with the Library Amplification kit (Kapa Biosystems), and the enriched library was subjected to a final purification and quantification step (Qubit and a quantitative PCR, Library quantification kit for Illumina platform, Kapa Biosystems). The pooled library was sequenced on lanes of HiSeq 3000 and NovaSeq 6000 Illumina sequencers by the GeT-PlaGe sequencing platform (Genotoul) and allowed the procurement of 150-bp reads. As a first assessment of sequencing success, a single-end sequencing run was performed on 23 samples (five Corsican wildcats, nine mainland domestic cats, six mainland wildcats and three Sardinian wildcats). All these individuals, except the three Sardinian for which all available DNA was included in the first run (i.e., no more available DNA), were sequenced again using paired-end sequencing. This second sequencing run also included all other available samples (i.e., total of 20 + 49 samples). We thus obtained reads for the 72 previously mentioned individuals, among which 20 were sequenced twice (once single- and once paired-end). These 20 individuals served as sequencing duplicates to calculate sequencing error rate (see below). Two negative controls were also included in the paired-end run to control for contaminations.
2.3 SNP calling and filtering
The quality of the raw reads was checked using FastQC version 0.11.5 (Andrews, 2010). Reads from each sample (unique individuals and sequencing duplicates, n = 92) were demultiplexed using the process_radtags module of the Stacks version 2.52 pipeline (Catchen et al., 2011; Rochette et al., 2019) using the -c, -q and -r options to remove any read with an uncalled base, discard reads with low-quality scores and rescue barcodes and RAD-tags. The adapter sequence search was also activated with two mismatches allowed. After demultiplexing, the total numbers of retained reads were 108,368,724 and 686,123,726 reads for single-end and paired-end sequencing, respectively. Reads (paired or single-end) were then mapped using the Burrow-Wheeler Aligner version 0.7.15 (BWA, Li & Durbin, 2009) and the mem algorithm with default parameters values against the reference genome of domestic cat Felis_catus_9.0 (GenBank accession number GCA_000181335.4, Buckley et al., 2020, downloaded from Ensembl Genome Browser http://mart.ensembl.org/Felis_catus/Info/Index) previously indexed (bwa -index). Output files were then converted to BAM format and sorted using samtools version 1.9 (Li et al., 2009), which was also used to discard reads with a mapping score < 30.
The Stacks refmap.pl module was then used to build loci and identify and genotype SNPs for each sample. This first run allowed us to identify 23 samples poorly genotyped due to a low number of reads (all <1.4 M with 20 individuals having <100,000 reads). These individuals also showed a gstacks effective coverage <10 and a high proportion of missing data (around 100% for most samples). Unfortunately, all samples from Sardinia were among these low-quality samples. We thus reran refmap.pl as well as the populations module without the low-quality samples using two filtering settings: -r = 0.80, meaning that at least 80% of the individuals in a population are required to process a locus for that population, and -p = 3, meaning that a locus must be present in at least three populations to be processed. These parameters were chosen as a result of several runs of the populations module, varying -r, -p and -R (the minimum percentage of individuals across populations required to process a locus) to obtain a good compromise between the number of SNPs and the missing data quantity (per individuals and SNP). Only the first SNP per locus was kept, avoiding linkage disequilibrium issues in population genetics analyses (write-single-snp option). Using populations we also filtered the data set to exclude SNPs showing a heterozygosity higher than 70% (to avoid paralogous loci) and a minimal allelic frequency higher than 0.05 (i.e., an allele must be present at least X times to be kept, with X = 0.05 × 2 N, and N = number of individuals; with 51 individuals, an allele must be present five times). SNPs were then additionally filtered using vcftools version 0.1.14 (Danecek et al., 2011) to keep only biallelic SNPs, alleles with a minor allele frequency >0.05 (since few SNPs that were kept by Stacks were actually eliminated by the Vcftools filter for minor allele frequency), a maximal percentage of missing genotypes of 25% and genotypes with a minimal coverage of 10 and a maximal coverage of 780 (i.e., twice the mean depth). Vcftools was also used to calculate the mean depth per individual and the proportion of missing SNPs per individual (Table S1). After filtering, we used duplicates to calculate a sequencing error rate. To this end, we compared genotypes at each SNP for each pair of duplicates and calculated the error rate as the ratio of the number of SNP that had different genotypes and the total number of SNPs. We then excluded one of the two duplicates, keeping in the final data set the one showing the least missing data.
Using the same setups and filters, except for the populations module for which we parameterized p = 5, we aimed at obtaining a reduced data set in which some Sardinian individuals could be kept. We thus excluded three Sardinian individuals having <5000 reads and proceeded with the four others (between ≈ 16,000 and 85,000 reads). Using -p = 5 allowed us to keep only SNP sampled in all populations, including the Sardinian one, and we obtained a data set of 249 SNP in which all individuals had less than 15% of missing SNP (and 0% for the four Sardinian remaining individuals). This reduced data set was then processed, as the “large data set”, as follows. Using the R version 4.1.0 (R Core Team, 2021) packages vcfR version 1.12.0 (Knaus & Grünwald, 2017), pegas version 1.0–1 (Paradis, 2010) and q-value version 2.24.0 (Storey et al., 2021), we excluded SNPs showing departure from Hardy–Weinberg (HW) equilibrium (q-value <0.05) in at least one of the populations considered (i.e., tests were performed within each population since departure from Hardy–Weinberg equilibrium is expected due to possible genetic differentiation between European, Corsican, Sardinian wildcats and domestic cats). Finally, using the R package pcadapt version 4.3.3 (Luu et al., 2017; Privé et al., 2020), we filtered for outlier SNPs. The number of principal components (PCs) retained was chosen as the one before the straight line in the scree-plot (i.e., subsequent PCs only accounted for random variation, Cattell's rule, as recommended by Luu et al., 2017). To be conservative, all SNPs showing p-values < .05 after adjustment through q-values, the false discovery rate (Benjamini & Hochberg, 1995) or Bonferroni correction, were excluded from further analyses.
2.4 Population genetics analyses
Population genetic structure was investigated using both large and reduced data sets through several approaches. We first used a principal component analysis (PCA) performed using the R package adegenet version 2.1.3 (Jombart, 2008; Jombart & Ahmed, 2011). To investigate the genetic structure more in depth we then used a discriminant analysis of principal components (DAPC, adegenet), which allowed us to identify the optimal number of genetic clusters (K) using a k-means algorithm (find.clusters) and assign individuals to clusters (the DAPC itself). We ran find.clusters for K varying from 1 to 10, using 10,000 iterations and 1000 different starting points. The optimal number of clusters was identified as the one minimizing the BIC values, or, for competing solutions, as the smallest value for which there was a clear break with preceding values. A cross-validation procedure was then used to identify the optimal number of principal components to retain to perform the DAPC, which was subsequently applied to assign individuals to a cluster and calculate membership probabilities.
The Bayesian clustering algorithm implemented in the STRUCTURE software version 2.3.4 (Pritchard et al., 2000) was also used to cluster individuals without a priori information about their population of origin and determine their membership coefficients. We used the admixture and correlated allele frequency models for a varying number of clusters (K, from 1–5), with 10 independent repetitions for each K value and an MCMC length of 1,000,000 iterations (burnin: 100,000). The optimal number of clusters was determined using both the likelihood of each K (Ln Pr(X|K)) and the method described by Evanno et al. (2005) as implemented in STRUCTURE HARVESTER version 0.6.94 (Earl & vonHodt, 2012). Independent runs for the optimal K were combined using CLUMPP version 1.1.2 (Jakobsson & Rosenberg, 2007) as implemented in CLUMPAK (Kopelman et al., 2015). We also applied the maximum likelihood approach implemented in ADMIXTURE version 1.3 (Alexander et al., 2009; Zhou et al., 2011) to estimate individual ancestries. Data sets were formatted using PLINK version 1.9 (Chang et al., 2015; Purcell et al., 2007). ADMIXTURE was run 10 times for each value of K from 1 to 5 and a 10-fold cross validation (CV) was applied to then determine the optimal K value. All other parameters were default parameters. Optimal K value was identified as the one minimizing the average CV-errors value across the 10 runs. For the optimal K value, independent runs were combined using CLUMPP as implemented in CLUMPAK.
Finally, based on a priori population assignations, global and between-populations genetic differentiation indices (FST, theta estimator, Weir & Cockerham, 1984) were calculated using StAMPP R package (Pembleton et al., 2013). Confidence intervals and significance of FST values were obtained using bootstrap over loci (n = 1000 repetitions). The package hierfstat version 0.5–7 (Goudet & Jombart, 2020) was used to determine genetic diversity indices (observed and expected heterozygosity, allelic richness and FIS), while nucleotide diversity (π) was obtained by running the Stacks populations module using only the SNPs that passed all previously described filters (white-list).
3 RESULTS
3.1 Sequencing and data filtering
The Fastqc quality check revealed no sequencing issues. The per base sequence quality was >28 all along reads 1 for paired-end sequencing and all along reads for single-end sequencing. Reads 2 showed a slightly lower quality (22–26) of the first three bases of reads. After demultiplexing, the mean number of retained reads per sample was 8,833,359 for paired-end sequencing and 4,152,531 for single-end sequencing. Negative controls obtained only few reads (of a total 1144 and 56,160, only 542 and 5696 were retained after process_radtags filtering; most of the reads were discarded because no Rad tags were detected, confirming that no DNA was present in the negative controls). When the 23 low quality samples were excluded, the mean gstacks effective coverage was 60×. Most (95%) of the primary alignments produced by BWA were kept. A total of 903,417 loci of an average of 260 bp were built, but most were discarded by populations filters. A final number of 11,884 present in at least three populations and in 80% of individuals within each population were obtained, leading to 5446 SNPs genotyped before SNP filtering.
After filtering using vcftools, 4249 SNPs remained, showing a mean coverage of 330× and a mean of 10.4% missing data. The genotyping error rate per individual, calculated thanks to the 20 duplicated individuals, was low (mean of 3.7%). After removing sequencing duplicates from the data set, 228 SNPs appeared to depart from HW equilibrium in at least one of the four populations, and 350 were detected as outliers by pcadapt (keeping four PCs, see Figure S1). These SNPs were thus excluded to obtain a final data set of 3671 genotyped in 51 unique individuals (9 wild Corsican, 9 domestic Corsican, 12 domestic mainland and 21 wild mainland cats) with moderated missing data proportions (2.3 and 3.8% for mainland wild and domestic cats, respectively, and 7.1 and 34.5% for Corsican wild and domestic cats, respectively). Domestic Corsican samples were of low quality (from hairs with low DNA quantity, see Table S1), which may explain such higher missing data rate. In the reduced data set (i.e., the one including four Sardinian individuals), 15 SNPs were excluded for departing from HW equilibrium in at least one of the five populations and 32 were detected as outliers by pcadapt (keeping three PCs). The final reduced data set included 202 SNP and 60 individuals (11 Corsican wildcats, 10 Corsican domestics, 12 mainland domestics, 23 mainland wildcats and 4 Sardinian wildcats). All individuals had <15% of missing data (mean = 4.01%, 2.33%, 3.25%, 1.97% and 0% for wild Corsican, domestic Corsican, domestic mainland, wild mainland and Sardinian wildcats, respectively).
3.2 Population genetic structure including mainland European wildcats
The PCA performed to explore the population genetic structure revealed a clear genetic distinction between all four populations (Figure 3a). The first axis explained 27.2% of the variance contained in the data and separated wild mainland cats from all other individuals. The second axis explained 4.9% of the variation and showed a distinction between Corsican wildcats and domestic cats from both Corsica and mainland France. The pattern revealed by the PCA first axis was in accordance with patterns revealed by clustering approaches. Indeed, DAPC k-means and Bayesian STRUCTURE algorithms both pointed to the presence of two genetic clusters (see Figures S2 and S3; BIC was minimal for K = 2, while Ln Pr(X|K) started to plateau after K = 2). The DAPC, performed using one discriminant function and 20 PCs (cross-validation procedure), as well as the STRUCTURE approach evidenced a strong genetic distinction between European wildcats from mainland France and all other individuals. In both analyses, one cluster grouped all mainland wildcats, while the second one was composed of all other individuals (domestics from mainland France and Corsica, wildcats from Corsica, Figure 3b,c). In the DAPC, the assignation success was one (i.e., assignation of k-means algorithm and DAPC itself were exactly the same), and posterior membership probabilities were also equal to one for each individual in its genetic cluster, indicating strong support for this genetic structure. In STRUCTURE, all individuals also had high posterior probability of membership: all were equal to 1 for cluster 1 (European wildcats) and ranged from 0.99 to 1 for cluster 2 except for one individual (Figure 3c). Similar results were observed using ADMIXTURE (Figures S4, S5).
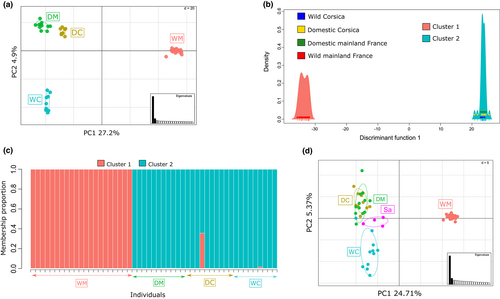
Finally, FST values also confirmed the marked genetic differentiation between European wildcats from mainland France and all other populations and highlighted a significant genetic differentiation between Corsican wild and domestic cats as well as domestic cats from mainland France (global FST = 0.40(0.39–0.41)IC95%, see Table 2 for pairwise values). The lowest FST value was obtained between domestic cats from mainland France and from Corsica. Interestingly, European wildcats from mainland France exhibited the lowest genetic diversity (Table 3). Similar results were obtained while performing the PCA and calculating FST on the reduced data set including four Sardinian individuals (Figure 3d, Table 2). Indeed, wild mainland individuals exhibited large differentiation with all individuals (including Sardinian ones) and were separated from all other individuals in the first axis of the PCA (Figure 3d). The second axis separated Corsican wildcats from domestic individuals and Sardinian individuals were intermediate between these two groups (Figure 3d). Using ADMIXTURE, wild mainland individuals were also isolated from all other individuals (Figures S4, S12).
| WC | DC | DM | Saa | |
|---|---|---|---|---|
| DC | 0.069 (0.061–0.077)*** | |||
| DM | 0.067 (0.061–0.073)*** | 0.015 (0.011–0.019)*** | ||
| Saa | 0.043 (0.018–0.069)** | 0.050 (0.016–0.92)** | 0.033 (0.007–0.061)* | |
| WM | 0.518 (0.505–0.530)*** | 0.547 (0.531–0.561)*** | 0.507 (0.494–0.519)*** | 0.693 (0.635–0.742)*** |
- Abbreviations: DC, domestics from Corsica; DM, domestics from mainland France; Sa, wildcats from Sardinia; WC, wildcats from Corsica; WM, wildcats from mainland France.
- ***p < .001, **p < .01, *p < .05 after Bonferroni correction.
- a For Sardinian individuals, FST values were calculated using the reduced data set (202 SNP).
| Population | n | H o | H e | A r | Allele number | π | F IS |
|---|---|---|---|---|---|---|---|
| WC | 9 | 0.24 ± 0.20 | 0.27 ± 0.19 | 1.77 ± 0.39 | 6282 | 0.23 ± 0.003 | 0.08 ± 0.35 |
| DC | 9 | 0.25 ± 0.19 | 0.28 ± 0.18 | 1.80 ± 0.37 | 4474 | 0.22 ± 0.003 | 0.09 ± 0.34 |
| DM | 12 | 0.25 ± 0.17 | 0.28 ± 0.17 | 1.80 ± 0.34 | 6850 | 0.24 ± 0.003 | 0.08 ± 0.29 |
| Saa | 4 | 0.11 ± 0.17 | 0.16 ± 0.21 | 1.43 ± 0.50 | 289 | 0.16 ± 0.02 | 0.23 ± 0.45 |
| WM | 21 | 0.11 ± 0.14 | 0.12 ± 0.15 | 1.43 ± 0.39 | 5956 | 0.10 ± 0.002 | 0.07 ± 0.21 |
- Abbreviations: Ar, allelic richness (rarefaction method, El Mousadik & Petit, 1996); He, expected heterozygosity; Ho, observed heterozygosity; π, nucleotide diversity calculated on variant positions, FIS, averaged inbreeding coefficient overall loci. Population: DC, domestics from Corsica; DM, domestics from mainland France; Sa, wildcats from Sardinia; WC, wildcats from Corsica; WM, wildcats from mainland France.
- a For Sardinian individuals, values were calculated using the reduced data set (202 SNP).
3.3 Population genetic structure excluding mainland European wildcats
Since we were interested in determining if Corsican wildcats were genetically close to domestic cats, Sardinian wildcats or represented a unique gene pool, we repeated all the previous analyses but excluding European wildcats from mainland France. The populations module outputted a total of 6846 loci present in all three populations and in 80% of individuals within each population, leading to 3494 SNPs. After filtering using vcftools (maximal depth = 960), 3378 SNPs remained. Among these SNPs, 116 departed from HW equilibrium in at least one population, and 149 were detected as outliers by pcadapt (keeping three PCs, see Figure S6). The final data set consisted of 3113 SNPs genotyped in 30 unique individuals (9 wild Corsican, 9 domestic Corsican, 12 domestic mainland). It is noteworthy that, following removal of mainland European wildcats, the missing data rate was drastically reduced in Corsican domestic cats (2.05%). Among the 3113 SNPs genotyped, 156 exhibited private alleles for Corsican wildcats; 129 and 222 for Corsican and mainland domestic cats, respectively. Using the reduced data set while excluding wildcats from mainland France, 258 SNP were genotyped by the populations module, among which 17 were excluded for departing from HW equilibrium and one was detected as outliers by pcadapt (keeping two PCs). A total of 240 SNPs was thus genotyped in 37 unique individuals (11 wild Corsican, 10 domestic Corsican, 12 domestic mainland and 4 Sardinian wild individuals).
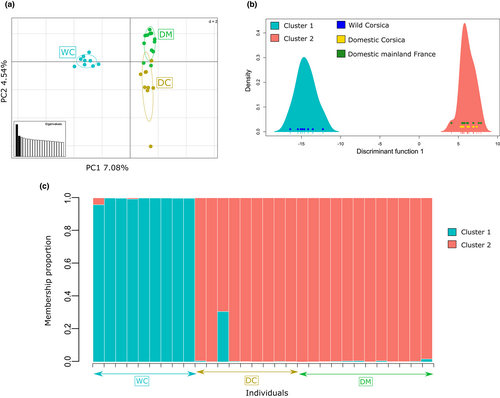
For the large data set, the first axis of the PCA explained 7.08% of the variance contained in the data and separated wild Corsican cats from all domestic individuals (Figure 4a). The second axis explained 4.54% of the variation and showed some distinction between domestic Corsican cats and mainland domestic cats. The results of the DAPC k-means algorithm were less clear. Indeed, the BIC was minimal for K = 1, although a break in slope was visible for K = 2 (Figure S7). We nevertheless looked at clustering results when choosing K = 2 to evidence, if present, genetic differentiation between Corsican wildcats and domestic cats. In doing so, the DAPC (performed using 18 PCs [cross-validation procedure] and one discriminant function) revealed patterns in accordance with the one observed on the PCA. Indeed, one cluster grouped all Corsican wildcats, while the other grouped Corsican and mainland domestic cats (Figure 4b). Although it was not the optimal clustering solution, analyses performed using K = 2 received strong support since, in DAPC, the assignation success and the posterior membership probabilities were equal to one. It is noteworthy that, when performed using a priori assignations (i.e., based on sampling and not on k-means algorithm), the DAPC led to similar results and discriminated between Corsican wildcats and domestic cats. Using the STRUCTURE Bayesian clustering approach, both the Evanno method and the Ln Pr(X|K) suggested that K = 2 was the optimal number of clusters (Figure S8). As previously, the two clusters separated Corsican wildcats and domestic cats from mainland France and Corsica, and membership proportions were very high in each cluster, ranging between 0.95 and 1 except for one domestic individual from Corsica (Figure 4c). Although using ADMIXTURE the most supported K value was K = 1, as with the DAPC, when considering K = 2 the two clusters separated Corsican wildcats and domestic cats with high membership probabilities (Figures S4, S5). Finally, FST values corresponded to previous observations, since global FST was 0.05(0.04–0.50)IC95%, and pairwise FST values were also of this order of magnitude, with the lowest FST value obtained between domestic cat populations (Table 4).
| WC | DC | DM | |
|---|---|---|---|
| DC | 0.065 (0.058–0.072)*** | ||
| DM | 0.062 (0.056–0.068)*** | 0.014 (0.010–0.018)*** | |
| Saa | 0.037 (0.013–0.059)** | 0.044 (0.015–0.081)* | 0.035 (0.009–0.062)* |
- Abbreviations: DC, domestics from Corsica; DM, domestics from mainland France; Sa, wildcats from Sardinia; WC, wildcats from Corsica.
- ***p < .001, **p < .01, *p < .05 after Bonferroni correction.
- a For Sardinian individuals, FST values were calculated using the reduced data set (240 SNP).
When considering the reduced data set including four Sardinian individuals, the PCA confirmed the intermediate position of Sardinian wildcats between Corsican wildcats and domestic individuals as well as the clear separation of Corsican wildcats (Figure 5a). As for the large data set, the DAPC results were less clear since the BIC was minimal for K = 1 but a break in slope could be detected for K = 2 (Figure S9). Looking at clustering results with K = 2, Corsican wildcats grouped in one cluster while domestic cats grouped in the second one (Figure 5b). Two of the Sardinian individuals (SAR004 and SAR005, Table S1) grouped with Corsican wildcats and two grouped with domestic cats. One Corsican wildcat (C1820001, not present in the large data set) grouped with domestic cats. STRUCTURE also detected the presence of several genetic groups since Evanno's method pointed to K = 4 as the optimal number of cluster while Ln Pr(X|K) suggested K = 3 (Figure S10).
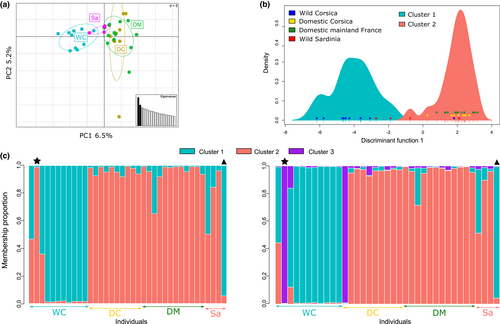
We thus examined membership proportions for both K = 2 (as supported with the large data set), K = 3 and K = 4. Both K = 2 and K = 3 clustering solutions isolated Corsican wildcats in one cluster and grouped domestic cats in another one (Figure 5c). The same Corsican wildcat as previously was assigned to the domestic cluster (C1820001, star on Figure 5c) and two additional individuals showed intermediate membership proportions. Sardinian, while all except one (SAR005, triangle on Figure 5c) assigned to the domestic cluster, also showed some degree of admixture with Corsican wildcats. When considering K = 3, similar results were observed except that the third genetic group clustered one Corsican wildcat (C1820001) and one domestic cat from Corsica (Figure 5c). Membership probabilities when considering K = 4 showed similar results except that domestic cats were shared between clusters 2 and 4 (Figure S11). ADMIXTURE analyses confirmed all these results, although K = 1 received the most support as the optimal the number of clusters (Figure S4) and considering K = 3 led to inconclusive results (Figure S12). Overall, ancestry coefficients were more intermediate using ADMIXTURE than using STRUCTURE (Figure S12). FST values agreed with an intermediate position of Sardinian wildcats between wild Corsican and domestics since the lowest FST value was observed between Sardinian and Corsican wildcats, followed by FST between Sardinian and domestics and the highest value was observed between Corsican wildcats and domestics (Table 4).
4 DISCUSSION
By investigating, for the first time, Corsican wildcats using ddRAD sequencing and population genetics approaches, we evidenced their genetic uniqueness by revealing a very strong genetic differentiation with European wildcats and a non-negligible genetic differentiation with domestic cats from both Corsica and mainland France as well as with F. s. lybica from Sardinia. Domestic cats were genetically closer to each other than to Corsican wildcats, a result that, in conjunction with phenotypic particularities (coat patterns) and genetic differentiation with Sardinian wildcats, opened the discussion about the necessity of recognizing the Corsican wildcat as a management unit in need of conservation measures and further ecological studies. Because of the low DNA quality of Sardinian samples, we only relied on a reduced data set including F. s. lybica but it suggested a higher genetic proximity of Corsican wildcats with Sardinian wildcats than with domestic cats. This called for further comprehensive studies including more Sardinian high-quality samples but also Sardinian domestic cats and mainland Near-Eastern F. s. lybica that will allow the precise repositioning of Corsican wildcats in the Felis phylogeny and to characterize the divergence between Sardinian and Corsican wildcats and original F. s. lybica.
4.1 Genetic structure of Corsican wildcats
Although they have been known by the Corsican people since ancient times, Corsican wildcats have been little studied and never using genetic approaches. It was thus unknown if these individuals were feral domestic cats or “true” wildcats, as assumed given their phenotypic particularities reminiscent of those of wild Felis silvestris spp., and if these “ghjattu-volpe” diverged from European wildcats Felis silvestris silvestris. Here, both multivariate and genetic clustering approaches, as well as genetic differentiation measures, revealed a strong genetic divergence between European and Corsican wildcats (FST = 0.50). This confirmed that Corsican wildcats do not belong to the F. s. silvestris subspecies. The absence of mainland F. s. lybica in our data set prevented us from making direct conclusions about the genetic proximity between Corsican wildcats and F. s. lybica. However, the inclusion of few Sardinian individuals on a smaller number of SNPs revealed that Corsican wildcats were genetically closer to Sardinian wildcats than to domestic cats. Since Sardinian wildcats are described as F. s. lybica, and although that would need to be ascertain in future studies, this may argue in favour of a non-negligible genetic proximity between Corsican wildcats and F. s. lybica. In addition, both Corsican and Sardinian wildcats genetic proximity with domestic cats suggested that they might share a common ancestry with domestic cats and thus be wild representatives of F.s. lybica. Indeed, since domestic cats diverged from F. s. lybica, a genomic proximity between domestic cats and F.s. lybica has been observed (Driscoll et al., 2007).
It is even more true since, in cats, the domestication process is likely to have been less intensive than in other species and to have had less strong impacts on behavioural, morphological and genomic characteristics (Faure & Kitchener, 2009; Mattucci et al., 2019; Montague et al., 2014). Preliminary analyses using mitochondrial DNA additionally confirmed the genetic proximity of F. s. lybica (including one individual from Sardinia), domestic cats and Corsican wildcats (Data S2, Figure S13, Tables S2–S3). This corresponded to the intermediate position we observed for Sardinian individuals between domestic cats and Corsican wildcats (Figure 5). Alternatively, this intermediate position, the assignment of some individuals to the domestic cluster and others to the Corsican clusters and the presence of admixed individuals (Figure 5) may suggest that Sardinian wildcats actually derived from Corsican wildcats and are nowadays victim of domestic introgression. In addition, increasing the number of genetic clusters did not separate Sardinian wildcats but instead created spurious results (Figures 5, S11, S12). However, in a previous study using microsatellite markers, Mattucci et al. (2013) detected no admixture between domestic cats and Sardinian wildcats. The intermediate position we observed in the present study may thus result from a sampling bias toward hybrid individuals. A comprehensive sampling scheme including more Sardinian wildcats and also Sardinian domestic cats as well as mainland F. s. lybica would allow to clarify the relations between Sardinian and Corsican wild and domestic cats.
The non-negligible level of genetic differentiation existing between Corsican wildcats and domestic cats from both the mainland and Corsica, and more importantly the closer proximity of domestics from the mainland and Corsica, suggested that Corsican wildcats are not domestic feral cats. Indeed, if the Corsican wildcat was feral, we would have expected a higher genetic proximity between wild and domestics from Corsica than between domestics from mainland and Corsica. This close proximity of domestics from distant geographic areas might be in line with the differing histories of domestic and wild cats. Cat domestication started in the Near East around 10,000 years ago, and these commensal organisms then probably colonized Central Europe by following Neolithic farmers (Baca et al., 2018). Phylogenetic analyses nevertheless also revealed that the genetic make-up of contemporary domestic cats actually resulted from both Near Eastern and Egyptian cats, the latter colonizing Europe much later, during the Roman Empire (around 2000 years BP, Ottoni et al., 2017; Baca et al., 2018). This genetic history of domestic cats might explain the higher genetic proximity observed between Corsican and mainland domestic cats, arguing in favour of a different origin of Corsican wild and domestic cats. The former could be a representative of Neolithic commensal F. s. lybica, and the latter a representative of contemporary domestic cats introduced much later on the island.
The degree of genetic differentiation we observed between domestic and Corsican wildcats (FST = 0.06–0.07, Table 4) was in accordance with values observed between F. s. lybica from Sardinia and North Africa and domestic cats, although differences in the genetic markers used makes the comparison challenging (Mattucci et al., 2016). It is, in addition, noteworthy that, in the SNP panels investigated here the genome representation might be biased. Indeed, by requiring that loci should be present in at least 80% of individuals in all populations (r = 0.8, p = 3 or p = 4 in Stacks analyses), while we considered probably divergent populations, we might have sampled the SNPs genotyped in the most conserved genome regions, leading to an underestimation of the genetic differentiation existing between populations. In addition, we considered SNP panels excluding SNP identified as outliers and thus showing the largest FST values (pcadapt). The loci may be outliers for different reasons (e.g., being under selection) but may also reflect the genetic divergence between populations. Along with the reduction of statistical power due to the small number of individuals involved in our analyses, such conservative genome representation might explain why the DAPC pointed to K = 1 as the optimal number of clusters. This conservative genome representation may also explain why European wildcats showed the lowest genetic diversity (Table 3). Indeed, since this population was the most divergent, sampling SNP in the most conserved regions of the genome may have led to sample less variable portions of the European wildcat genome. The relatively low genetic diversity observed here do not correspond to what was previously observed in French European wildcats (Portanier, Léger, et al., 2022).
4.2 Conservation implications
The genetic distinction of Corsican wildcats from domestic, European and Sardinian wildcats calls for conservation considerations. The Mediterranean basin is one of the most threatened biodiversity hotspots (Ducrocq, 2016; Giorgi, 2006; Myers et al., 2000), and identifying conservation units on Mediterranean islands might favour its preservation by allowing its biodiversity to be recognized at a legislative level (Coates et al., 2018). Based on our assessment of Corsican wildcat population genetic structure, we argue that this possibly unique organism should be considered as a conservation unit within the Felis silvestris species complex. Several denominations exist on the population-species continuum when speaking of conservation units, such as evolutionarily significant units (ESUs) and management units (MUs) (see Moritz, 1994; Palsbøll et al., 2007; Funk et al., 2012; Coates et al., 2018 for reviews). These concepts of infraspecific categories have definitions that may still be under debate and that evolve through time, but some consensus seems to emerge from the scientific literature. It is nowadays mostly accepted that ESUs should be defined as “populations that have substantial reproductive isolation, which has led to adaptive differences so that the population represents a significant evolutionary component of the species” (Palsbøll et al., 2007). While the ESUs concept includes notions of adaptive divergence and reproductive isolation, the MUs concept relies on the demographic independence of populations since they are defined as “populations of conspecific individuals among which the degree of connectivity is sufficiently low so that each population should be monitored and managed separately” (Palsbøll et al., 2007). To be recognized as an ESU, Corsican wildcats would need to be replaced more precisely in the Felis silvestris spp. phylogeny, and a study on adaptive genetic markers would be necessary to evidence adaptive divergence between this population and other ones, such as the Sardinian one and mainland Near-Eastern F. s. lybica. While the present study does not allow to make conclusions about ESUs, we nevertheless suggest that Corsican wildcats constitute a MU, since they inhabit an island and, to our knowledge, no translocations have occurred from Sardinia or other wildcat populations for many thousand years. It is noteworthy that the genetic differentiation detected between Sardinian and Corsican wildcat may result from this long-term isolation, which may have led to adaptive divergence but also favoured strong genetic drift if both populations have low population sizes (no data available at present).
5 CONCLUSION AND PERSPECTIVES
Investigating, for the first time, Corsican cats that have been described as wild for a very long time by the Corsican people, and using genome-wide SNPs obtained through next-generation sequencing, we evidenced that Corsican wildcats, “ghjattu-volpe”, might represent a new unique gene pool in Corsica and a Management Unit within the F. silvestris spp. complex. Although the sample size and data set composition prevented us from making conclusions about the definition of the Corsican wildcat as an ESU or a subspecies, our results open the way to further studies. Description of the Corsican wildcat as a subspecies would aid immensely in the conservation of this MU, since species and subspecies are better recognized legislatively than other infra-specific categories, especially in the European laws (Coates et al., 2018). Our study also illustrated how using relatively low-cost sequencing strategies and reduced genome representation can be useful for conservation purpose. It illustrated, for instance, how these approaches can reveal cryptic diversity previously overlooked using less powerful genetic markers and bring enthusiastic news in the current context of extinction crisis. It also emphasized the importance of focusing more on understudied Corsican fauna, and more generally on understudied insular fauna, which can be made up of numerous undiscovered MUs or ESUs.
Future works on the Corsican wildcat should focus on repositioning it more precisely in the Felis silvestris phylogeny and on evolutionary history by including more F. s. lybica samples from mainland Near East and from Sardinia and investigating adaptive divergence as well as demographic history, but also on gaining knowledge of their ecology. Implementing comprehensive biological monitoring would allow the effective conservation of the Corsican wildcat by bringing knowledge on their ecological niche, habitat suitability, spatial distribution and spatial ecology, the dynamics of their populations, their population genetic structure at finer spatial scales, including both landscape genetic connectivity and hybridization with domestic cat investigations. All these knowledges would help to protect Corsican wildcats but also bring important information about their interaction with other Corsican species, which would in turn improve discussions and debates about the conservation of anthropochorous species (Gippoliti & Amori, 2002, 2006). In addition to the investigation of demographic history, an urgent pursuit of the present study regards hybridization since hybridization between mainland wild and domestic cats has been reported (Beugin et al., 2016, 2020; Say et al., 2012) and might also occur between Corsican wild and domestic cats. Further studies involving higher sample sizes and sampling wild and domestic individuals living in close proximity would allow to determine if hybridization with domestic cats does not threaten Corsican wildcats as it does for European ones (Yamaguchi et al., 2015). For this purpose, the ddRAD sequencing data generated in the present study could allow to derive ancestry informative markers for Corsican wildcat and SNP particularly suitable to investigate introgression, as previously done for European wildcats and domestic cats (Mattucci et al., 2019; Nussberger et al., 2014). If future studies reveal an absence or a low rate of hybridization with domestic cats, Corsican wildcats could be seen as living fossils of African-Near Eastern wildcats introduced several thousands of years ago, offering astonishing opportunities to study cat domestication.
AUTHOR CONTRIBUTIONS
E.P., P.C., P.B., F.S., S.R. and S.D. conceptualized and designed the research. P.B. and F.S. supervised the fieldwork in Corsica. M.Z. provided samples from Sardinia. H.H. and C.R. conducted laboratory steps. E.P. performed data analyses with the contribution of H.H., A.E.F and P.C. during the first steps of the project. E.P. wrote the manuscript with feedback from S.D., S.R. and H.H. and all the authors. All authors contributed to interpreting the results and approving the manuscript.
ACKNOWLEDGEMENTS
We warmly thank all the professionals from the Office Français de la Biodiversité (OFB) for their technical support in sampling the different populations, especially Charles-Antoine Cecchini, Valérie Grisoni, Laurence Henry and François Léger. We also gratefully acknowledge the CC Laboratoire de Biométrie et Biologie Evolutive/Pôle Rhône-Alpes de Bioinformatique (PRABI) for providing computer resources, Fanny Dens as a student for her field and laboratory work and Jeremy Larroque for helpful discussions about SNP filtering and data analyses. We are grateful to veterinarians Dr Marc Memmi, Dr Bernard Fabrizy, Dr Anne Guiard-Marigny, Dr Nathalie Kilburg, Dr Marion Terrazzoni, Dr Claud d'Angeli and Dr Pascal Jugnet for collecting Corsican domestic cat samples and to the Museum of Natural History of Nancy, the Museum of Natural History of Strasbourg and Dr M. Salotti (Corsican University) who collected samples from furs. We also thank the sequencing GeT-PlaGe sequencing platform (Genotoul, Castanet-Tolosan, France). This research was funded by the Office de l'Environnement de la Corse (OEC), the OFB, the Direction Régionale de l'Environnement, de l'Aménagement et du Logement Corse (DREAL), the University of Lyon and the Laboratoire de Biométrie et Biologie Evolutive. We are also grateful to the two anonymous reviewers for their useful comments that helped us to improve the quality of this study.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
BENEFIT-SHARING STATEMENT
Benefits generated: A research collaboration was developed with scientists from the countries that provided genetic samples. All collaborators are included as coauthors. The results of research have been shared with the provider communities and the broader scientific community (see above) and the research addresses a priority concern, in this case, the conservation of organisms being studied. More broadly, our group is committed to international scientific partnerships, as well as institutional capacity building.
Open Research
DATA AVAILABILITY STATEMENT
Raw sequence reads and related metadata have been deposited in the SRA BioProject PRJNA811807. Sample accession numbers are reported in Table S1. The final SNP data sets are available from the FigShare repository (https://doi.org/10.6084/m9.figshare.19298864.v1). The 65 new mitochondrial sequences are available from European Nucleotide Archive with accession nos. OW373342-OW373406.




