Genomic basis of Y-linked dwarfism in cichlids pursuing alternative reproductive tactics
Abstract
Sexually antagonistic selection, which favours different optima in males and females, is predicted to play an important role in the evolution of sex chromosomes. Body size is a sexually antagonistic trait in the shell-brooding cichlid fish Lamprologous callipterus, as “bourgeois” males must be large enough to carry empty snail shells to build nests whereas females must be small enough to fit into shells for breeding. In this species, there is also a second male morph: smaller “dwarf” males employ an alternative reproductive strategy by wriggling past spawning females into shells to fertilize eggs. L. callipterus male morphology is passed strictly from father to son, suggesting Y-linkage. However, sex chromosomes had not been previously identified in this species, and the genomic basis of size dimorphism was unknown. Here we used whole-genome sequencing to identify a 2.4-Mb sex-linked region on scaffold_23 with reduced coverage and single nucleotide polymorphism density in both male morphs compared to females. Within this sex region, distinct Y-haplotypes delineate the two male morphs, and candidate genes for body size (GHRHR, a known dwarfism gene) and sex determination (ADCYAP1R1) are in high linkage disequilibrium. Because differences in body size between females and males are under strong selection in L. callipterus, we hypothesize that sexual antagonism over body size initiated early events in sex chromosome evolution, followed by Y divergence to give rise to bourgeois and dwarf male reproductive strategies. Our results are consistent with the hypothesis that sexually antagonistic traits should be linked to young sex chromosomes.
1 INTRODUCTION
Sex determination is extremely diverse across eukaryotes and can be controlled environmentally or genetically (Bachtrog et al., 2014). In most organisms with genetic control, the sex-determining locus is carried on a sex chromosome. Highly differentiated (i.e., heteromorphic) sex chromosome pairs, such as the XY chromosome pair in mammals and ZW chromosome pair in birds, have repeatedly evolved across the tree of life (Bachtrog et al., 2014). The first step in the evolution of sex chromosomes from autosomes in species with separate sexes is believed to be the acquisition of a sex-determining gene on the Y or W chromosome (Charlesworth & Charlesworth, 2000; Brian Charlesworth, 1996). When this is followed by recombination suppression between the new sex chromosome pair, possibly via an inversion, degeneration of the Y or W chromosome occurs, leading to heteromorphic sex chromosome pairs (Bachtrog, 2013; Charlesworth et al., 2005).
Several models to explain the repeated loss of recombination on sex chromosomes have been proposed (reviewed by Ponnikas et al., 2018). The prevailing model stipulates that sexually antagonistic selection can drive recombination suppression because it favours linkage between the sex-determining gene and a locus with sexually antagonistic effects (i.e., beneficial in one sex and detrimental in the other; Charlesworth & Charlesworth, 1980; Rice, 1987). However, the broad applicability of the sexual antagonism hypothesis of sex chromosome evolution is still debated as it has been challenging to demonstrate empirically (Ironside, 2010; Ponnikas et al., 2018). One challenge is that in old and/or heteromorphic sex chromosome systems, it is difficult to disentangle whether recombination suppression was initiated due to the presence of sexually antagonistic loci or whether sexually antagonistic loci accumulated after recombination was suppressed. Younger and/or less differentiated (i.e., homomorphic) sex chromosomes are more promising systems to test whether sexually antagonistic selection has played a role in the evolution of sex chromosomes (Bachtrog, 2013; Ponnikas et al., 2018). Indeed, sexually antagonistic traits are linked to young sex chromosomes in guppies (Almeida et al., 2020; Sandkam et al., 2021; Wright et al., 2017), stickleback (Kitano et al., 2009) and cichlids (Roberts et al., 2009). These studies highlight the value of studying traits that are known to be sexually antagonistic and then determining whether they are associated with young sex chromosome systems.
Adaptive radiations of cichlid fish harbour hundreds of recently diverged species that have young and homomorphic sex chromosomes (Ozouf-Costaz et al., 2017; Poletto et al., 2010), with a high turnover of sex determination mechanisms even among closely related species (El Taher et al., 2021; Gammerdinger & Kocher, 2018). We hypothesize that sexually antagonistic selection may play an important role in sex chromosome evolution in cichlid fishes, as many sexually dimorphic traits related to body colour, body size and behaviour are found in this lineage (Lande et al., 2001; Taborsky, 2001). Indeed, in several sexually dimorphic cichlid species from Lake Malawi, the sexually antagonistic “orange blotch” body colour patterning is associated with the evolution of a novel sex determination locus (Roberts et al., 2009). Furthermore, as cichlid sex chromosomes are young and homomorphic, studies will not be confounded by the effects of recombination suppression that happened a long time in the past. Thus, cichlids are an excellent system to investigate the sexual antagonism theory of sex chromosome evolution.
Here, we focused on the cichlid Lamprologous callipterus from Lake Tanganyika in East Africa (Figure 1a) that exhibits the most extreme sexual size dimorphism so far known among animals in which males exceed females in size. Nest-building “bourgeois” males have a 12-fold difference in body mass compared to females (Schütz & Taborsky, 2000; see Taborsky, 1998 for terminology). These males must exceed a minimum size threshold in order to carry empty gastropod shells to build a nest, and females must be small enough to fit into these shells for spawning and brood care (Schütz et al., 2006; Schütz & Taborsky, 2005). Consequently, body size is a sexually antagonistic trait in this species due to differential fitness effects of body size for shell-brooders. In addition to the bourgeois males, so-called “dwarf males” that average only 2.4% of the mass of bourgeois males also exist in this species (Figure 1a). These males compete with bourgeois nest owners for egg fertilization by applying an alternative reproductive strategy that involves wriggling past spawning females into the shell to fertilize eggs from this advantageous position (Figure 1a; Wirtz-Ocana et al., 2014). Thus, dwarf males need to be even smaller than females (Sato et al., 2004). Bourgeois males acquire more mates than dwarf males, but dwarf males are more successful during direct sperm competition in laboratory trials and in the field (Schütz et al., 2010; Taborsky et al., 2018; Wirtz-Ocana et al., 2014). Dwarf males are much rarer than bourgeois males in natural populations (von Kuerthy & Taborsky, 2016; Wirtz-Ocana et al., 2014). Negative frequency-dependent selection is thought to be responsible for the relative numbers of dwarf and bourgeois males in nature, as these tactics are fixed for life and hence should yield similar fitness returns (Brockmann & Taborsky, 2008; Parker, 1984; Taborsky & Brockmann, 2010).
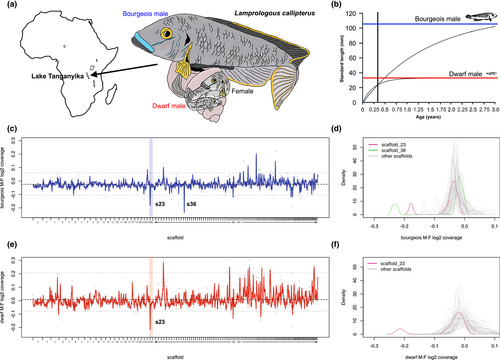
Experimental pedigrees have shown that reproductive tactic and body size in L. callipterus males adhered to the expectation of a Y-linked Mendelian trait controlled by a single locus (Wirtz-Ocana et al., 2014). Bourgeois males only sire bourgeois sons and dwarf males only sire dwarf sons, while daughters do not differ in any respect between these two types of males (Wirtz-Ocana et al., 2014). These results suggest that Y-linkage has resolved sexual antagonism over body size between males and females. However, neither the sex chromosomes nor the genes controlling male body size have yet been identified in this species. It is also unclear which of the two male types is the ancestral state.
In this study, we investigate the genomic basis of male body size in L. callipterus using whole-genome resequencing data from dwarf and bourgeois males, and their daughters (Table 1). Dwarf males initially grow at a faster rate than bourgeois males, but growth halts early in ontogeny (Wirtz-Ocana et al., 2013), and bourgeois males continue to grow indefinitely (Taborsky, 2001; Figure 1b). Thus, we hypothesized that the dwarf-determining gene may encode a growth factor. We tested whether such a factor was linked to the sex-determining region, as the sexual antagonism hypothesis outlined above would predict. To investigate this hypothesis, we first needed to characterize the sex-determining region. The karyotype of L. callipterus has not been studied, but karyotypes of other Lamprologini and related cichlids show no sex chromosome differentiation (Ozouf-Costaz et al., 2017). However, genomic data provide higher resolution into the subtle molecular differences between sexes in species with homomorphic sex chromosomes (Fontaine et al., 2017; Gammerdinger & Kocher, 2018). We compared whole genomes of males and females to identify regions of differentiation between the sexes and the morphs. In addition to L. callipterus samples, we included males and females of six related species from the Lamprologini tribe (see Table 1) for a comparative phylogenomic perspective on sex determination and body size evolution.
| Species (breeding mode) | No. of samples sequenced | ~Body size (cm) |
|---|---|---|
| Lamprologous callipterus (obligate shell-brooder) |
2 bourgeois males 2 dwarf males 4 females (1 daughter per male) |
12.0 2.0 6.0 |
| Lamprologous lemairii (facultative shell-brooder) |
1 male 1 female |
22.0 Not known |
| Lamprologous ocellatus “gold” (obligate shell-brooder) |
1 male 1 female |
5.0 3.5 |
| Neolamprologous brevis (obligate shell-brooder) |
1 male 1 female |
6.0 4.0 |
| Neolamprologous multifasciatus (facultative shell-brooder) |
1 male 1 female |
4.0 2.5 |
| Altolamprologus sp. “Sumbu Shell” (obligate shell-brooder) | 1 male | 5.5 |
| Altolamprologus calvus “Congo” (not a shell-brooder) | 1 male | 10 |
2 MATERIALS AND METHODS
2.1 Sampling and sequencing
Sampling was conducted at the breeding facility at the Ethologische Station Hasli, University of Bern, Switzerland. Genomic DNA was extracted from fin clips of males and females of five species and males only of two species for which females were not available (Table 1). The stock population of the target species Lamprologous callipterus originated from the southern end of Lake Tanganyika near Mpulungu. Two L. callipterus dwarf males and two bourgeois males, as well as one daughter per male were sampled (see Data S1 for genetic and biological relatedness) because it is easier to identify sex chromosomes in related individuals (Palmer et al., 2019). The TrueSeq DNA Library Preparation Kit was used for library preparation, and samples were sequenced at the Vienna BioCentre Sequencing Facility in Austria. The libraries were sequenced on eight lanes of an Illumina HiSeq 2500 machine generating ~125 million paired-end reads (120 bp in length) per sample.
2.2 Reference-based assembly and characterization of sex-determining region coverage
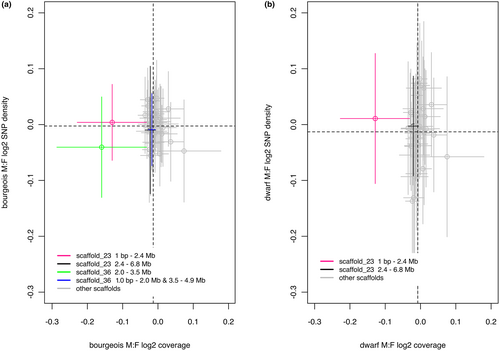
Raw Illumina reads were trimmed using fastx-trimmer (http://hannonlab.cshl.edu/fastx_toolkit v0.13). Reads were mapped to the Neolamprologous brichardi genome (Brawand et al., 2014), which was chosen because it is from the most closely related cichlid species to our focal species (Darolti & Mank, 2022). However, to verify our results, we also mapped our data to the high-quality genome of Oreochromis niloticus (Conte et al., 2017). See Section 3.1 for details. While N. brichardi is the most closely related reference genome, it is not the best assembled reference cichlid genome and there are trade-offs that must be considered when picking the best reference genome for sex chromosome mapping. As we are attempting to identify a young and homomorphic sex chromosome, we chose to use a more closely related reference. Read mapping was conducted using bowtie2 version 2.2.7 (Langmead et al., 2009) in the “very-sensitive” mode with no mixed alignments and retaining only concordantly mapped reads. We obtained an average coverage of ~25× across the genome (Appendix S1). Coverage of windows across the genome was calculated using bedtools version 2.17 (Quinlan & Hall, 2010). The genome-wide coverage analysis was performed using 1-Mb moving windows with a 0.5-Mb step-size for all scaffolds with more than three windows. The scaffold_23 and scaffold_36 coverage analysis was performed using 30-kb moving windows with a 15-kb step-size. For each window, coverage was defined as the total number of times each site was sequenced divided by the number of sites that were sequenced. The data were normalized by dividing the coverage per window with average coverage per sample. To identify candidate sex chromosomes, the following calculation was used: log2 (average male coverage/average female coverage). Negative values would indicate reduced coverage in males (i.e., a differentiated Y region). To identify regions of differentiation between bourgeois and dwarf males the following calculation was used: log2 (average dwarf male coverage/average bourgeois male coverage). Regions in the genome with significantly reduced coverage (i.e., outliers) were defined as windows in the bottom 1% of the male:female coverage distribution. A permutation test using 20,000 permutations implemented in R (version 4.0.2) was used to calculate if the coverage on scaffold_23 and scaffold_36 was significantly lower than the other scaffolds (i.e., autosomes). The permutation test was repeated after dividing scaffold_23 into two regions that represented the two peaks identified in the coverage density plots shown in Figure 1(d,f): (i) the region from 1 bp to 2.4 Mb and (ii) the region from 2.4 to 6.8 Mb. The permutation test was also repeated after dividing scaffold_36 into two regions that represented the two peaks identified in the coverage density plots shown in Figure 1d: (1) the region from 2.0 to 3.5 Mb and (ii) the region from 1.0 to 2.0 Mb and 3.5 to 4.9 Mb.
2.3 Single nucleotide polymorphism calling, FST, LD and inversion analysis
samtools version 1.3 and bcftools version 1.10.2 (Li et al., 2009) were used for calling and genotyping single nucleotide polymorphisms (SNPs) across all L. callipterus samples against the N. brichardi reference genome with default settings. SNPs passing the following filters were retained: sample and site DP ≥ 5, genotype quality ≥ 30, mapping quality ≥ 30, and missingness < 0.25. vcftools version 0.1.13 was used to calculate SNP density in dwarf males, bourgeois males and females. SNP density was normalized by the genome-wide average SNP density per sample. Permutation tests with 20,000 permutations implemented in R version 4.0.2 were used to calculate if SNP density on scaffold_23 and scaffold_36 was significantly different from the rest of the genome. FST was calculated using the popgenome R package (Pfeifer et al., 2014) in nonoverlapping windows of 30 kb. Significant differences in FST were estimated using the Wilcoxon rank sum test implemented in R version 4.0.2. Outliers were defined as having values greater than the genome-wide 95th percentile or values lower than the genome-wide 5th percentile, depending on the analysis conducted. Sample relatedness was calculated using vcftools version 0.1.13—relatedness2 option (Data S1). Linkage disequilibrium (LD) across all L. callipterus samples was calculated as r2 using the gpart package in R version 4.0.2 with an MAF filter of <0.05. LD heatmaps were plotted using the gpart LDBlockHeatmap function (Kim et al., 2019). Inversion analysis was conducted using two independent tools: lumpy version 2.13 (Layer et al., 2014) and delly version 0.7.2 (Rausch et al., 2012) with default settings on the mapped BAM files for all L. callipterus samples.
2.4 Phylogenomics
We applied a gene tree vs. species tree approach to investigate the evolution of sex chromosomes in the lamprologini clade. A tailored FASTA genome file was built for each individual using angsd version 0.930 -dofasta function (Korneliussen et al., 2014), which assembles FASTA-format genome sequences by picking the most common base per position in the mapped reads for each individual. Gene sequences for every second gene in the reference genome were extracted using bedtools version 2.17 (Quinlan & Hall, 2010) and aligned using mafft version 7.273 (Yamada et al., 2016). Using every second gene provided a genome-wide overview with reduced computation time. Individual gene trees were constructed using the raxml version 8.2.9 (Stamatakis, 2014) GTRCAT method with 100 bootstrap replicates. A genome phylogeny was estimated using a coalescent approach using astral-ii (Mirarab & Warnow, 2015) with the 11,855 gene trees and N. brichardi as the outgroup. Alignments of genes in the most diverged sex region of scaffold_23 from 1.18 to 1.28 Mb were built as outlined above but with 1000 bootstrap replicates.
2.5 Signatures of selection
As body size is an important trait across the whole Lamprologini clade, we tested for signatures of selection on the candidate dwarfism gene GHRHR. Exon sequences of the gene were extracted from the reconstructed fasta genomes generated for the phylogenies using bedtools version 2.17 (Quinlan & Hall, 2010). The exons were concatenated, and aligned using mafft version 7.273 (Yamada et al., 2016). End stop codons were removed, and pairwise dN/dS was calculated using random models in paml (Yang, 2007).
3 RESULTS
3.1 Identification of sex-linked scaffolds in Lamprologous callipterus
To identify the sex-determining region in Lamprologous callipterus, we analysed the genomic coverage of mapped reads from eight individuals (two bourgeois males, two dwarf males and four females; Table 1) to the reference congener Neolamprologous brichardi (female) genome (Brawand et al., 2014). Approximately 80% of the ~120 million paired-end reads per sample mapped to the reference genome with average genome-wide coverage of 20–30× across samples (Table S1). We compared normalized genomic coverage between males and females to identify regions of reduced coverage in males, as diverged Y reads will no longer map to the X chromosome. Coverage analysis using a 1.0-Mb moving window and 0.5-Mb step-size revealed a small region on scaffold_23 with significantly reduced coverage outlier windows (below the bottom 1% of the genome-wide average), compared to the rest of the genome, in the two bourgeois males vs. the four females (Figure 1c), as well as in the two dwarf males vs. the four females (Figure 1e). Another small region of reduced coverage was identified on scaffold_36 in the two bourgeois males vs. the four females (Figure 1c). The coverage in the outlier windows on scaffold_36 was the most extreme outlier across the genome of the bourgeois males (Figure 1d, permutation test p = 0). The coverage in the outlier windows on scaffold_23 was the second most extreme outlier across the genome of bourgeois males (Figure 1d, permutation test p < 5.0 e−5) and the most extreme outlier in the dwarf males (Figure 1f, permutation test p = 0). A comparison of coverage between dwarf and bourgeois males suggested that the small region on scaffold_23 had lower coverage in the dwarf males compared to the bourgeois males, but this was not significant (Grubbs's outlier test p = .431; Figure S1). The density profile of log2 M:F coverage of all windows on scaffold_36 was bimodal for bourgeois males: the right peak contained windows from 1.0 bp to 2 Mb and 3.5–4.9 Mb with coverage similar to autosomal scaffolds, and the left peak contained windows from 2.0 to 3.5 Mb with lower coverage (Figure 1d). The density profile of log2 M:F coverage of all windows on scaffold_23 was bimodal for both male morphs (Figure 1d,f). The left peak contained windows from 1.0 bp to 2.4 Mb with lower log2 M:F coverage, and the right peak contained windows from 2.4 to 6.8 Mb with coverage similar to autosomal scaffolds. Thus, the region from 1.0 bp to 2.4 Mb on scaffold_23, which is shared by both male morphs, is hereon considered the putative sex-linked region in L. callipterus. The region from 2.0 to 3.5 Mb on scaffold_36 is a candidate sex-linked region specific to the bourgeois males.
Combining genomic coverage and SNP density can be useful for characterizing sex-linked regions (Palmer et al., 2019; Wright et al., 2017). In regions where there is some differentiation (i.e., sequence divergence) between the X and the Y but not yet degeneration to the point where sequence reads from the Y chromosome no longer map to the reference genome, there will be more SNPs in males than in females. However, if the Y has degenerated, SNP density will be lower in males. We identified 9,135,392 SNPs across the genome of the L. callipterus males and females, of which 97,746 SNPs were on scaffold_23. For the coverage and SNP density analysis, we divided the 1-Mb windows on scaffold_23 into two subsets based on the two coverage peaks (Figure 1d,f): (i) the region from 1.0 bp to 2.4 Mb; and (ii) the region from 2.4 to 6.8 Mb. We found that windows from the 1.0 bp to 2.4 Mb region had significantly lower bourgeois M:F coverage relative to the rest of the genome (permutation test p < 5.0 e−5) but no significant increase in bourgeois M:F SNP density (permutation test p = .15; Figure 2a). The region from 1.0 bp to 2.4 Mb had significantly lower dwarf M:F coverage (permutation test p = 0) and also significantly higher dwarf M:F SNP density (permutation test p < .03) relative to the rest of the genome (Figure 2b). The windows from the region 2.4–6.8 Mb did not have significantly lower M:F coverage compared to the rest of the genome (permutation test p > .05). However, this region had significantly elevated M:F SNP density in the dwarf males (permutation test p = 4.5 e−4) but not in the bourgeois males (permutation test p = .76). For the bourgeois male-specific candidate region from 2.0 to 3.5 Mb on scaffold_36, M:F coverage was significantly reduced (permutation test p = 0), but there was no increase but rather a decrease in M:F SNP density compared to the rest of the genome. When we repeated these analyses with the Oreochromis niloticus reference genome (NB: N. brichardi scaffold_23 corresponds to O. niloticus LG18:19.3–26.1 Mbp, see Figure S2) we discovered that the N. brichardi sex region on scaffold_23 may represent a subset of a putatively larger sex region on LG18 (Figures S3 and S4). However, the genome-wide coverage results were noisier with many coverage outliers (Figure S5). This is not surprising given the large divergence times (~24 million years ago, Irisarri et al., 2018) between O. niloticus and L. callipterus, but also because the sex region in L. callipterus is young and not strongly differentiated. O. niloticus LG18:19.3–26.1 Mb was a significant outlier in the dwarf males:females comparison (Figure S5).
3.2 Refining patterns of sex linkage and candidate genes on scaffold_36
To further examine the pattern of divergence across scaffold_36 in L. callipterus bourgeois males vs. females, we calculated SNP density and FST using 30-kb windows (Figure S6). There was a decrease in M:F SNP density beyond the 5th percentile of genome-wide SNP density at 2.0–2.4 Mb and 3.6–3.9 Mb (Figure S6A). There was, however, no clear increase in FST beyond the genome-wide 95th percentile across scaffold_36 (Figure S6B). Of the 28 genes annotated in the region of reduced coverage on scaffold_36, none had a function that could putatively be associated with a phenotype that delineates the bourgeois male reproductive tactic (Data S2). However, one gene in this region, endothelin-2 (EDN2) is associated with ovary development in mice (Ko et al., 2006). Because this region only had reduced coverage in bourgeois males vs. females and did not have strong evidence of genes with sex-associated phenotypes, the region of reduced coverage on scaffold_23 was the best candidate for the L. callipterus sex region, and we therefore concentrated our further analyses on this scaffold.
3.3 Refining patterns of sex linkage on scaffold_23
To further examine the spatial pattern of divergence across scaffold_23 in L. callipterus bourgeois and dwarf males vs. females, we calculated M:F coverage, SNP density and FST in 30-kb windows (Figure 3). The most extreme coverage reduction in both males was between 1.18 and 1.28 Mb, which represented the lowest 0.001% of the genome-wide M:F coverage (Figure 3a,b). This decrease in coverage was recapitulated when both male morphs were collectively compared to the females (Figure S7A). The reduction in coverage in dwarf males vs. females (log2 M:F = −0.6) was two-fold lower than that of bourgeois males vs. females (log2 M:F = −0.3) within the 1.18–1.28 Mb region, such that the dwarf males had reduced coverage when compared to bourgeois males (log2 dwarf males:bourgeois males = −0.3; Figure S7B). This suggests that the sex region on scaffold_23 is distinct between the dwarf and bourgeois males.
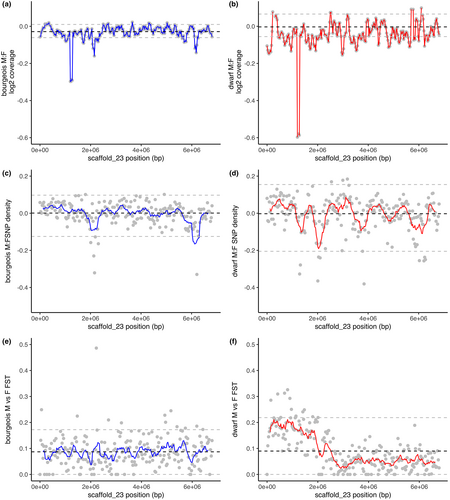
When SNP density was analysed together for all windows within the two regions of scaffold_23, there was either an increase in M:F SNP density (consistent with differentiation between the X and the Y) or no change in M:F SNP density compared to the rest of the genome (Figure 2). However, when the smaller 30-kb windows were analysed individually, windows with a decrease in M:F SNP density (consistent with degeneration of the Y) beyond the 5th percentile of genome-wide SNP density can also be seen around 2.0 and 6.2 Mb in the bourgeois M:F comparison (Figure 3c) and around 1.2, 2.0, 3.9 and 6.2 Mb in the dwarf M:F comparison (Figure 3d).
Male–female FST quantifies differences in allele frequency between the sexes, and thus can be used to detect differentiation in very young sex chromosomes (Gammerdinger et al., 2014; Vicoso, 2019). FST will identify regions where males are heterozygous and females are homozygous, consistent with expectations for a sex-linked region. We analysed FST between L. callipterus males and females and found significantly higher FST from 1 bp to 2.4 Mb of scaffold_23 compared to the remainder of scaffold_23 (Wilcoxon rank sum test p < 2.2 e−16) between dwarf males vs. females (Figure 3f). Several windows from 1.0 to 2.0 Mb exceeded the genome-wide 95th percentile of dwarf male vs. female FST with FST values between 0.21 and 0.32. FST values of 0.3 are expected for fully sex-linked regions (i.e., male XY and female XX). No such distinct pattern was observed for bourgeois males vs. females (Wilcoxon rank sum test p = .83; Figure 3e) but one window with FST of 0.49 was found around 2.3 Mb that was higher between bourgeois males and females and exceeded the genome-wide 95th percentile. We also compared FST between bourgeois males and dwarf males on scaffold_23 to identify intrasexual differentiation in the sex-determining region (Figure S8). The most significant FST between the two male-types was in windows spanning 1.08–1.11 Mb with FST = 0.49 (Grubbs's outlier test p < 6.3 e−11), which also exceeded the 95th percentile of genome-wide FST. There were also 20 other FST outliers (>genome-wide 95th percentile) spread across scaffold_23.
3.4 Recombination suppression on scaffold_23
The sexual antagonism model of sex chromosome evolution posits that recombination suppression between the sex-determining loci and sexually antagonistic loci ensures that they cosegregate (Charlesworth & Charlesworth, 1980; Rice, 1987). To determine whether there is recombination suppression on scaffold_23, we calculated r2 in order to quantify LD using all the SNPs from all L. callipterus samples across scaffold_23. It is important to note that high LD can result from either recombination suppression or strong selection. We identified eight LD blocks on scaffold_23, of which seven blocks were <0.03 Mb in size (Figure S9). The largest LD block, denoted LD-Block 5, was found between 1.0 and 1.4 Mb (Figure 4a). Thus, LD-Block-5 was perfectly centred around the above-mentioned regions of reduced M:F coverage and elevated FST. The pattern of LD was V-shaped, with the centre point of the “V” in high LD with upstream and downstream SNPs, extending from 1 bp to 1.4 Mb. However, the upstream and downstream SNPs were not in LD with each other. Inversions can produce high LD by restricting recombination during sex chromosome evolution. However, we did not find evidence of inversions on scaffold_23 using two different bioinformatics approaches (see Section 2: Methods). Our negative results could just reflect the limitation of the short-read data for identifying inversions. However, the LD pattern of inversions is expected to be more block-like and not “V” shaped as we observe in our data.
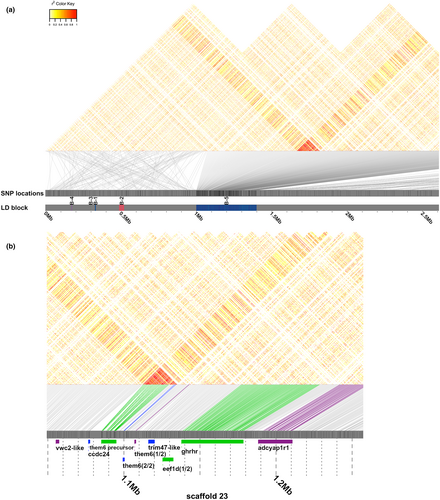
3.5 Identification of candidate sex determination and body size genes on scaffold_23
By investigating the function of genes in the sex-linked regions of scaffold_23 using www.genecards.org and a thorough literature review, we identified several genes that have known functions associated with sexual development, growth and alternative reproductive behaviours (Table 2; Data S2). The region 1.18–1.28 Mb with the lowest M:F coverage in L. callipterus had two annotated genes (Table 2; Data S2). One was pituitary adenylate cyclase-activating polypeptide type I receptor (ADCYAP1R1), a receptor of the well-characterized adenylate cyclase-activating polypeptide gene (ADCYAP, alias PACAP). ADCYAP1 was located just downstream at 1.8 Mb, within the LD-Block 5 (Data S2). Both this gene and its receptors are expressed in a wide variety of tissues, including the testes and ovaries (Vaudry et al., 2009) and are involved in oogenesis (Apa et al., 1997) and spermatogenesis in rats (Romanelli et al., 1997).
| Gene position on scaffold_23 | Gene name | Function or phenotype | Reference |
|---|---|---|---|
| 993,878–1,034,646 | B4GALT1 | Ehlers-Danlos progeroid syndrome (premature ageing), homologue B4GALT7 causes dwarfism in horses | Leegwater et al. (2016) |
| 1,085,286–1,094,884 | THEM6 precursor | Regulates THEM6 | |
| 1,099,381–1,100,314 | THEM6 (2 of 2) | Metabolic regulation of brown fat and energy expenditure | Zhang et al. (2012) |
| 1,107,073–1,107,814 | THEM6 (1 of 2) | Metabolic regulation of brown fat and energy expenditure | Zhang et al. (2012) |
| 1,125,564–1,132,363 | EEF1D (1 of 2) | Intellectual and adaptive functioning | Iseri et al. 2019 |
| 1,137,957–1,178,397 | GHRHR | Growth/Dwarfism of Sindh, little mice | Godfrey et al. (1993), Baumann and Maheshwari (1997), Perry et al. (2014) |
| 1,188,396–1,210,632 | ADCYAP1R1 | Gametogenesis | Romanelli et al. (1997); Daniel & Habener (2000) |
| 1,290,830–1,352,831 | XCR1b.3 (2 tandem duplicates) | GPCR signalling pathway member that includes ghrhra and adcyap1r1 | Fox et al. (2019) |
| 1,395,330–1,484,131 | PDE1C | Infertility, male mating-behaviour in Drosophila | Morton et al. (2010) |
| 1,842,461–1,845,858 | ADCYAP1 | Sexual behaviour in nest-building vs. non-nest-building males and female oocyte maturation in gourami fish | Levy and Degani (2012) |
- Note: For a full list of genes in the sex-linked region see Data S2.
Located 9.9 kb upstream of ADCYAP1R1 between 1.14 and 1.18 Mb was the growth hormone releasing hormone receptor (GHRHR), which is a G-protein coupled receptor (GPCR) and an important player in stimulating growth hormone synthesis and secretion from the pituitary gland (Lee et al., 2007; Figure 4, Table 2; Data S2). GHRHR was found in the elevated FST region of >0.2 between dwarf males and females (Figure 3f), and it had an FST of 0.31 between dwarf and bourgeois males (Figure S8). Mutations of GHRHR cause pituitary Dwarfism of Sindh in a remote population of Pakistanis belonging to the Sindhi ethnic group and the little phenotype in laboratory mice (Baumann & Maheshwari, 1997; Godfrey et al., 1993). Thus, GHRHR is a strong candidate for body size determination in L. callipterus. Both ADCYAP1R1 and GHRHR are in high LD with the genes at the centre of LD-Block 5 (Figure 4), and they also known to interact directly in protein–protein interaction networks in humans (Figure S10).
There were 15 genes in LD-Block 5, with two genes (THEM6 precursor and THEM6 [2 of 2]) at the centre of perfect LD at ~1.1 Mb (Table 2; Data S2). Just upstream of THEM6 (2 of 2) lies THEM6 (1 of 2). All three of these genes are members of the thioesterase superfamily, which has been associated with regulating energy expenditure in animals (Zhang et al., 2012). A detailed discussion of other interesting genes in the LD-block can be found in the Supporting Results and Discussion (Appendix S2).
3.6 Turnover of sex chromosomes in the Lamprologini
Lamprologous callipterus belongs to a large, substrate-breeding clade of cichlids in Lake Tanganyika constituting the tribe Lamprologini, where shell-brooding and small body size, particularly in females, have co-evolved multiple, independent times (Koblmüller et al., 2007; Sato & Gashagaza, 1997; Schütz et al., 2006). To resolve the evolutionary history of the sex-linked region, we compared the phylogeny of the scaffold_23 sex-linked region to the whole-genome phylogeny for L. callipterus and six other Lamprologini species sequenced in this study (Table 1). The whole-genome phylogeny was constructed using 11,855 genes across the genome, and it confirmed the strongly supported (bootstrap values = 100) monophyly of all included species (Figure 5). The tree suggests that obligate shell-brooding evolved three times in these species, or that it evolved in the common ancestor of these species after it split from N. brichardi and was subsequently lost three times.
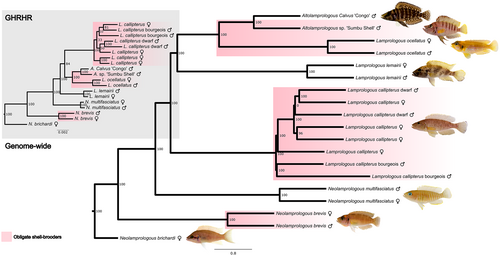
In the phylogeny of the candidate body size gene, GHRHR (Figure 5, inset), the species monophyly is maintained but the position of L. lemairii (facultative shell-brooder) and L. callipterus is switched compared to the whole-genome phylogeny. This results in a cluster of all obligate-shell brooders, except for N. brevis. The Altolamprologus calvus “Congo,” which is not a shell-brooder, is also in this cluster. The switch in topology between the whole-genome phylogeny and the sex-region phylogeny could reflect either the coalescent history of this region or introgression. In fact, F1 hybrids between L. callipterus and other snail shell dwellers have been observed in nature (Koblmüller et al., 2007).
To determine whether the sex determination system in L. callipterus is shared with other Lamprologini, we placed the L. callipterus sex-linked region in a phylogenomic context by analysing the coverage of scaffold_23 in males and females of four related species (Figure S11). The facultative shell-brooders N. multifasciatus and L. lemarii did not have outlier coverage windows (below the genome-wide 5th percentile) on scaffold_23. The obligate shell-brooders L. ocellatus had reduced coverage (below the genome-wide 5th percentile) on scaffold_23 from 1.29 to 1.34 Mb (Figure S11), but this does not overlap with the region of reduced coverage in L. callipterus at 1.18–1.28 Mb. However, it may belong to the putatively larger sex region of L. callipterus that was identified by remapping the data to the O. niloticus reference genome (Figures S2 and S3). Two copies of the chemokine receptors XCR1B.3 are found on scaffold_23: 1.29–1.34 Mb. XCR1B.3 is a member of the GPCR signalling pathway that also includes ADCYAP1R1 and GHRHR. N. brevis, an ancestral obligate shell-brooder, had outlier windows of coverage at scaffold_23: 2.14–2.16 Mb, a region devoid of genes. Overall, these observations point towards a possible shared sex-determining region on scaffold_23 in the shell-brooding Lamprologini tribe, which needs to be further investigated in a follow-up study.
3.7 No evidence for rapid evolution of the GHRHR gene across the Lamprologini
We screened the GHRHR gene for signatures of selection and found evidence for purifying selection (dN/dS <1) in all the ingroup species (L. callipterus, A. calvus “Congo,” A. sp. “Sumbu shell,” L. ocellatus) compared to the L. lemairii outgroup (Table S2). All ingroup species were obligate shell-brooders, with the exception of A. sp. “Calvus Congo,” a sister species of A. sp. “Sumbu shell.” The strongest purifying selection was in the L. callipterus lineage (dN/dS = 0.069). This was significantly lower than the other species (p = 4.0 e−8, 1.19 e−10 and 7.0 e−6 Welch's t tests; see Table S2), which had dN/dS values >0.12. We found no male-specific nonsynonymous mutations in the GHRHR gene, but we did identify two synonymous mutations (Figure S12). It may be that gene regulation and not sequence variation in GHRHR is controlling body size determination in L. callipterus. In the future it would be valuable to study the gene expression networks and alternative splicing patterns of candidate sex and body size genes of L. callipterus (Singh & Ahi, 2022; Tian et al., 2019).
4 DISCUSSION
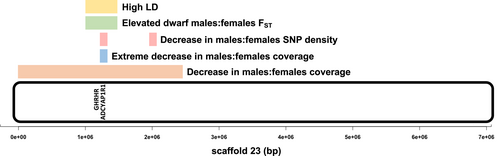
The shell-brooding cichlid species Lamprologous callipterus is a unique system to test the sexual antagonism hypothesis of sex chromosome evolution. The presence of bourgeois and dwarf male morphs that are strictly paternally inherited also offers the opportunity to study origins of Y haplotype diversity. Here we present first insights into the genomic basis of sex-linked body size determination in L. callipterus, which is under sexually antagonistic selection and the foundation of two very different alternative male reproductive tactics. We identify a small sex region in L. callipterus, with candidate genes for sex determination and sexual antagonism (Figure 6) and report the presence of two distinct Y haplotypes. We hypothesize that the L. callipterus sex chromosome may have evolved to resolve sexual antagonism over body size in males and females, followed by the evolution of Y-haplotype diversity that gave rise to two male reproductive tactics.
4.1 Lamprologous callipterus has a young sex chromosome
Using multiple lines of evidence (coverage, SNP density, LD, FST) we found scaffold_23 to be the most promising sex-linked region of L. callipterus (Figure 6). Consistent with expectations of recombination suppression and Y differentiation, we found a decrease in M:F coverage at scaffold_23: 1.18–1.28 Mb and elevated LD across scaffold_23: 1.0–1.4 Mb. This decrease in coverage and increase in LD was found in both bourgeois and dwarf males, supporting a shared Y region in L. callipterus male morphs. We also observed locally restricted decreases in male-specific SNPs in dwarf and bourgeois males on scaffold_23 between 1 bp and 2.4 Mb, which is consistent with deletions on the Y chromosome (Darolti et al., 2019). In addition, we found elevated FST between dwarf males and females from 1 bp to 2.4 Mb. Higher genetic differentiation is predicted around sexually antagonistic loci that are linked to sex-determining loci (Kasimatis et al., 2017; Kirkpatrick & Guerrero, 2014), but high FST can arise as a result of several other factors (Bissegger et al., 2020; Rowe et al., 2018). Overall, the differentiation in the sex-linked region of L. callipterus was not as extreme as that found in older therian sex chromosomes (Vicoso, 2019), probably because it is a comparatively younger sex chromosome that evolved <3 million years ago (Irisarri et al., 2018). In the context of East African cichlid radiation, however, the L. callipterus sex region would be considered old and thus shows more differentiation than that observed in other studied cichlid sex chromosomes (Gammerdinger & Kocher, 2018). Interestingly, scaffold_23 of L. callipterus maps to linkage group 18, which is one of the candidate sex chromosomes of the riverine cichlid Astatotilapia burtoni (Böhne et al., 2016; Roberts et al., 2016). Since scaffold_23 is a smaller scaffold that has been anchored to a larger linkage group in the high-quality cichlid reference genome of Oreochromis niloticus, by remapping our data to O. niloticus we found that scaffold_23 may be a subset of a putatively larger sex region. In future studies it would be important to generate long-read data for L. callipterus to enhance the resolution of its sex region.
As no orthologues of known sex-determining genes such as SRY or Dmrt1 (Doris Bachtrog, 2013) were found in the L. callipterus sex-linked region, we determined ADCYAP1R1 as the best candidate for the master sex-determining gene due to its role in gametogenesis (Apa et al., 1997; Romanelli et al., 1997; Vaudry et al., 2009). The location of ADCYAP1R1 in the most degenerated region of scaffold_23 (1.18–1.28 Mb) is consistent with predictions that a novel sex-determining gene can initiate sex-chromosome evolution and differentiation. This would point to a dose-dependent mechanism, whereby fish with two copies become female and fish with one copy become male (unless there are actually more copies of this gene). This is very different from most sex-determination loci identified so far in fish, which are either genes that have divergent alleles on the X and Y chromosomes or Y-specific gene duplications (Pan et al., 2021). However, Y-specific gene duplications cannot be identified by our analytical approach, which mapped reads to the reference genome. The ADCYAP1R1 gene is a receptor of the ADCYAP gene, which has sex-specific expression associated with sexual behaviour in nest-building vs. non-nest-building males and female oocyte maturation in gourami fish (Levy & Degani, 2012). ADCYAP1 was located just downstream at 1.8 Mb on scaffold_23 (Table 2; Data S2). Given how remarkably conserved this gene family is across vertebrates (Vaudry et al., 2009), it is likely that ADCYAP1R1 and ADCYAP1 play a functionally conserved role in L. callipterus sexual development. However, more detailed work is needed to determine whether and how these genes contribute to sex determination in L. callipterus.
4.2 Linkage between the sex-determining locus and a candidate sexually antagonistic locus
The L. callipterus candidate sex-determining gene, ADCYAP1R1, was in linkage with the candidate gene for body size determination, GHRHR (Figures 4 and 6). The sexual antagonism model of sex chromosome evolution postulates that novel sex-determination genes may arise in response to conflict between sexes over traits such as body size (van Doorn & Kirkpatrick, 2007). Since differences in body size between males and females affect reproductive fitness in the shell-brooding L. callipterus (Schütz et al., 2006; Schütz & Taborsky, 2000, 2005) and higher dwarf male–female FST (a possible indicator of antagonism) was found in the sex-determining region with the candidate body size gene (Figure 6), we propose that sexual antagonism over body size may have driven the evolution of a novel sex chromosome in L. callipterus. It is possible that this sex chromosome on scaffold_23 is ancestral to the Lamprologini cichlid tribe, but our results were unclear (Figure S11). We also cannot rule out that it may have arisen in the lamprologini ancestor and was subsequently lost in some species.
Although inversions are often found on sex chromosomes and are proposed to mediate recombination suppression (Lahn & Page, 1999; Peichel et al., 2020; Ponnikas et al., 2018), we did not find evidence of an inversion. It is possible that strong selection for low recombination facilitated the establishment of sex-specific and morph-specific polymorphisms as observed with guppy alternative male morphs (Bergero et al., 2019; Sandkam et al., 2021). The region of high LD on scaffold_23 in L. callipterus extended beyond the sex-determining gene and the sexually antagonistic body size gene to include several other candidate genes for traits, such as behaviour, physiology and morphology, that differentiate the L. callipterus male morphs (see Supporting Results and Discussion in Appendix S2). These behavioural, physiological and morphological loci are probably cosegregating with the sex-determining locus. Thus, the sex-linked region of L. callipterus may govern multiple sex and male morph-related phenotypes, consistent with “supergene” architectures found in other species with alternative male reproductive tactics (Sandkam et al., 2021; Schwander et al., 2014; Thompson & Jiggins, 2014).
4.3 Male morphs are associated with two Y-haplotypes
While the sex region on scaffold_23 was shared by both L. callipterus male morphs, there were clear differences in this region between the two males, suggesting that there are two distinct Y haplotypes in L. callipterus (Figure 3). The bourgeois males have an additional region of low coverage on scaffold_36 that is not found in dwarf males, which further supports that there are two distinct Y haplotypes. Within the shared sex region on scaffold_23, the dwarf male Y haplotype has lower coverage and may therefore be more degenerated than the bourgeois male Y haplotype, which can have important gene dosage effects (Raznahan et al., 2018). FST patterns across the sex region also suggested that the region of recombination suppression is larger in the dwarf male Y haplotype compared to bourgeois males. This would be the case if selection for linkage was stronger between the sex-determining locus and a sexually antagonistic locus in dwarf males, perhaps because the dwarf male body size was under stronger selection pressure than the bourgeois male body size. This is conceivable as the dwarf male mating tactic is strictly applicable for males small enough to enter the shell and wriggle past the female (Sato et al., 2004), whereas the bourgeois male mating tactic may be applied by males of a range of body sizes, even if passing a size threshold to carry shells greatly improves the efficiency of this mating tactic (Schütz & Taborsky, 2005). It is also possible that the much smaller population size of dwarf males compared to bourgeois males (Wirtz-Ocana et al., 2014) resulted in greater Y degeneration. Indeed, differences in the rate and extent of Y degeneration have been observed in two closely related stickleback species that share the same ancestral Y chromosome, with more rapid and extensive degeneration in the species with the smaller effective population size (Sardell et al., 2021).
But how did the second Y haplotype evolve and how is it maintained in the population? Given that the reproductive potential and evolutionary fitness of the dwarf males is critically dependent on the fitness of the bourgeois males, we hypothesize that the bourgeois male type is ancestral. It is possible that a mutation in the GHRHR candidate body size gene gave rise to dwarf males that had higher fitness due to their efficient parasitic reproductive tactic (Taborsky et al., 2018; Wirtz-Ocana et al., 2014). This high fitness allowed the new dwarfism mutation to establish and be maintained by negative frequency-dependent selection in the population (Clark, 1987; Gadgil, 1972), as the fitness of each type of males is dependent on their relative frequency in the population (Taborsky & Brockmann, 2010; Taborsky, 2008; von Kuerthy et al., 2016). Such Y haplotype diversity has also been observed in guppies (Poecilia reticulata) that experience negative frequency-dependent selection on Y-linked colour patterns, as well as in the closely related species Poecilia parae in which there are five behaviourally and morphologically distinct reproductive male morphs that each have a distinct Y-haplotype (Almeida et al., 2020; Sandkam et al., 2021).
5 CONCLUSION
Our study reveals a young sex chromosome in the Lake Tanganyika cichlid adaptive radiation that supports the sexual antagonism model of sex chromosome evolution. Our results suggest that the suite of complex traits that differ between the bourgeois and dwarf male morphs in body size, growth rate, testis/sperm size, behaviour and reproductive strategy are delineated by two distinct Y haplotypes. Future functional studies of candidate genes and long-read exploration of the L. callipterus sex-linked region provide exciting avenues to further investigate the sexual antagonism hypothesis and the evolution of male alternative reproductive tactics.
AUTHOR CONTRIBUTIONS
MT and CS conceived the study. CLP contributed to interpretation of the results. PS designed and conducted all analyses, made the figures, and drafted the manuscript, which was edited by MT, CS and CLP. All authors have read and approved the final manuscript.
ACKNOWLEDGEMENTS
We thank the ZMF facility at Med-Uni Graz for library preparation, W. Gessl for fish photographs and M. Koller for figure editing.
FUNDING INFORMATION
This work was supported by the Swiss National Foundation grant (SNF Project 310030B-138660) to MT and the Austrian Science Fund grant (FWF P29838) to CS. PS was funded by a PhD scholarship from the Austrian Centre of Limnology, University of Graz.
CONFLICT OF INTEREST
The authors declare no competing interests.
Open Research
DATA AVAILABILITY STATEMENT
WGS data have been deposited to NCBI SRA (https://dataview.ncbi.nlm.nih.gov/object/PRJNA593252?reviewer=gi3t337sqe3vmb1beknpdb9a31). Analysis code is available on https://github.com/poojasingh09/sex_chromosome_cichlid.




