Atypical life cycle does not lead to inbreeding or selfing in parasites despite clonemate accumulation in intermediate hosts
Abstract
Many parasites utilize asexual and sexual reproduction and multiple hosts to complete their life cycles. How these taxa avoid inbreeding is an essential question for understanding parasite evolution and ecology. Aquatic trematodes that require multiple host species may benefit from diverse genetic parasite assemblages accumulating within second intermediate hosts prior to sexual reproduction in definitive hosts. However, Cotylurus species are able to utilize the same snail species as first and second intermediate hosts, potentially resulting in the accumulation of genetically identical clones (clonemates) prior to sexual reproduction. In this study, we developed and analysed novel microsatellite loci to determine if clones are accumulating within snail hosts prior to ingestion by bird hosts and the effects this could have on parasite inbreeding. Contrary to previous studies of aquatic trematodes, significantly large numbers of clonemates were present within snails, but full-sibs were not. Genetic structure was present over a relatively small geographical scale despite the use of vagile definitive hosts. Phylogenetic analysis identified the Cotylurus sp. clones as belonging to a single species. Despite the presence of clones within snails, mating between clones/selfing was not common and heterozygosity is maintained within individuals. Potential issues with clones mating may be mitigated by the presence of snails with numerous clones, the consumption of many snails by bird hosts and parasite clone recognition/avoidance. Use of the same host species for multiple life stages may have advantages when parasites are able to avoid inbreeding and the required hosts are common.
1 INTRODUCTION
Inbreeding, or mating between close relatives (Yengo et al., 2019), can reduce gene diversity, heterozygosity and recombination rates, and therefore influence evolutionary forces such as genetic drift and natural selection (Charlesworth, 2003; Criscione et al., 2011; Detwiler et al., 2017; Prugnolle et al., 2005), and cause inbreeding depression. Inbreeding depression is a decline in fitness due to inbreeding and can occur when increased homozygosity causes the expression of deleterious recessive alleles (partial dominance hypothesis) or decreases the frequency of higher fitness heterozygous loci (overdominance hypothesis) (Charlesworth & Charlesworth, 1987; Roff, 2002). However, inbreeding can also allow natural selection to purge deleterious recessive alleles from a population, and the likelihood of this occurring can depend on the level of inbreeding and the severity of the fitness decrease the deleterious alleles cause (Charlesworth & Charlesworth, 1987; Crnokrak & Barrett, 2002; Lagrue & Poulin, 2009; Rieger et al., 2013; Roff, 2002). The impacts of inbreeding have important fitness consequences for diverse taxa and are of interest to many biological disciplines, including evolution, ecology, conservation biology/captive breeding programmes and human health (Charlesworth, 2003; Hedrick & Kalinowski, 2000; Willoughby et al., 2015; Yengo et al., 2019). Darwin himself conducted extensive experiments to better understand the effects of inbreeding in numerous plant species and, for obvious personal reasons, speculated on the negative effects of inbreeding in humans (Álvarez et al., 2015; Darwin, 1868, 1876).
Inbreeding is of particular interest in host–parasite systems because many parasites utilize life cycles that can cause the aggregation of related individuals within hosts. For example, the transmission of genetically identical parasite clones (clonemates) to definitive hosts can cause inbreeding among parasites if clonemates reproduce with each other (Criscione et al., 2011, 2022). Digenetic trematodes utilize sexual and asexual reproduction and multiple hosts to complete their life cycles. The typical pattern among digeneans consists of asexual clonal reproduction in a molluscan first intermediate host producing mobile cercariae, infection of a second intermediate host in which the trematode forms a relatively immobile larval stage, and final development to adult reproductive stage in a definitive vertebrate host (Cribb et al., 2003; Poulin & Cribb, 2002). This three-step trophic transmission process may decrease the accumulation of clonemates prior to sexual reproduction within the definitive host, allowing for outbreeding and the avoidance of inbreeding effects after asexual reproduction (Brown et al., 2001; Criscione & Blouin, 2006; Rauch et al., 2005).
The creation of numerous clonal infective stages during asexual reproduction creates the opportunity for clonemates to encounter each other during sexual reproduction within definitive hosts. Reproduction between clonemates has the same effect as self-fertilization (selfing) and can lead to increased homozygosity and cause inbreeding depression (Criscione et al., 2011; Prugnolle et al., 2005). Inbreeding depression may be more likely to occur when large numbers of clonemates are transmitted to definitive hosts together. In addition to clonemates, clumped transmission of closely related clones, such as full-siblings, could increase inbreeding (Detwiler & Criscione, 2017). The accumulation of genetically diverse clones within second intermediate hosts prior to ingestion by definitive hosts may decrease the chances of clonemates reproducing and reduce inbreeding (Criscione & Blouin, 2006; Rauch et al., 2005). Mixing of clones can occur within second intermediate hosts through the acquisition of parasites from numerous first intermediate hosts via either the host's mobility, the movement of water, the proximity of numerous infected first intermediate hosts or a combination of these (see Valdivia et al., 2014, and references therein).
Studies examining the within-host clonal diversity of aquatic trematodes have revealed high levels of clonal diversity and few clonemates within second intermediate hosts, supporting the role trophic transmission may play in maintaining trematode clonal diversity (Keeney et al., 2007a, 2007b; Lagrue et al., 2009; Leung et al., 2009; Rauch et al., 2005; Valdivia et al., 2014). When clonemates are encountered, it is probably from synchronous infections from a proximate first intermediate host (Keeney et al., 2007a, 2007b; Leung et al., 2009). Clonemates may be more likely to accumulate in species that do not utilize aquatic, three-host life cycles. Semiterrestrial species that use two animal hosts to complete their life cycles, such as Schistosoma mansoni, including those utilizing metacercaria, such as Fasciola hepatica and Fascioloides magna, potentially accumulate clonemates in definitive hosts (Beesley et al., 2021; Mulvey et al., 1991; Prugnolle et al., 2002; Prugnolle, Choisy, et al., 2004; Theron et al., 2004). This is probably facilitated by the clumped transmission of clonemates after leaving snails and aggregating in small pools (Sc. mansoni) or on vegetation (Fascioloides magna) (Criscione & Blouin, 2006). The terrestrial species Dicrocoelium dendriticum utilizes a three-host life cycle involving two hosts with relatively low dispersal (snails and ants) and snails produce discrete packages of related cercariae “slime balls” that may facilitate clumped transmission (Criscione et al., 2020).
Members of the trematode genus Cotylurus have an alternative aquatic life cycle using the same snail species as first and second intermediate hosts (Figure 1). Adult Cotylurus parasitize the intestinal tract of waterfowl and parasite eggs enter the water with host faeces. After hatching, miracidia penetrate first intermediate host snails belonging to the families Lymnaeidae, Planorbidae and Physidae, depending on the particular species (Niewiadomska, 1971; Olivier & Cort, 1941), and form cercariae-producing sporocysts. Released cercariae can utilize the same species of snails as second intermediate hosts and upon penetration form tetracotyle metacercariae (Cort et al., 1941). However, based on experimental studies with Cotylurus flabelliformis and C. lutzi, snails with active sporocysts develop temporary immunity to infections from their own cercariae (Basch, 1970; Campbell, 1973; Cort et al., 1941, 1945; Nolf & Cort, 1933; Winfield, 1932). Furthermore, cercariae do not actively attack the snails shedding them (Basch, 1970; Campbell, 1997). Cotylurus cercariae may infect other snail families as second intermediate hosts, including Planorbidae, but they are less suitable hosts (Campbell, 1997). However, if snails, either typical hosts or nonhosts, are infected with other trematode species' sporocysts or rediae, the Cotylurus cercariae can encyst as hyperparasites (Basch, 1970; Campbell, 1973; Cort et al., 1941). Mature tetracotyles encyst within snails in ~3–6 weeks (Cort et al., 1944; Olson, 1974; Ulmer, 1957). Birds, primarily waterfowl, shorebirds and wading birds, are infected when they ingest snails with mature tetracotyles (Niewiadomska, 1971). There is recent molecular evidence of some species of Cotylurus using leeches as second intermediate hosts, although these were distinct species from those used in the current study (Pyrka et al., 2021). Therefore, the basic life cycle traits of Cotylurus involve using the same species of hosts as both first and primary second intermediate hosts even if there are behavioural or immunological factors that separate the stages in different individual snails. This pattern of host use could help ensure transmission to definitive hosts since these hosts could be physically close to each other in the environment. However, this could have genetic consequences resulting in clonemates infecting the same second intermediate hosts.
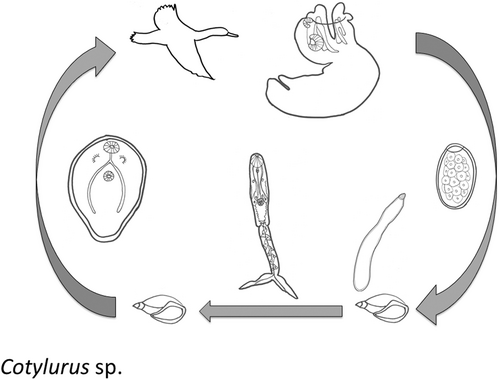
Given the evolutionary incentive for clone mixing and empirical support for clone mixing in aquatic species, are there species in which large numbers of clonemates are transmitted together from second intermediate hosts and, if so, what impact does this have on parasite inbreeding? The utilization of the same relatively sedentary species as first and second hosts makes Cotylurus species ideal candidates for accumulating clonemates and addressing these questions. This study quantifies the clonal diversity and relatedness of a Cotylurus species' tetracotyles within snail second intermediate hosts to determine if clonemate and/or related clone (full-sib) transmission is occurring. In addition, we examine the extent of inbreeding and genetic structure to shed light on the impact potential clonemate transmission has had on this species.
2 MATERIALS AND METHODS
2.1 Field sampling, snail dissection and parasite collection
As part of a broader food web study, snails were collected during two visits to three emergent cattail wetlands in May–July 2015 (Table S1). Two sampling sites were located in Barrington, IL, USA, at Crabtree Nature Center and were located 0.75 km apart (Table S1). Crabtree Entrance Pond (Crabtree Entrance) was the smallest site at 0.87 ha with ~60% open water and 100% of the shoreline occupied by cattails (Typha spp.). Crabtree 3G was the largest sampling location at 18.1 ha with 60% open water and 100% of the shoreline vegetated with 90% Typha spp. and 10% Juncus spp. The third sampling site was located along Oakwood Road (Oakwood) in Oak Creek, WI, USA, ~86 km from the other two sampling locations. This site was intermediate in size at 10.1 ha, with only 10% open water, and 100% shoreline vegetated with 75% Typha spp. and 25% invasive reed canary grass (Phalaris arundinacea). All three sites were relatively shallow, and the depth of snail sampling locations never exceeded 3 ft along the shoreline.
Standardized dipnet sweeps occurred every 10 m across a 100-m stretch of each pond's perimeter in order to collect up to 50 target snails for parasite assessment. The snail communities differed at each site. Crabtree Entrance contained Physella sp. and Stagnicola exilis, while Crabtree 3G supported populations of four snail species, Gyraulus sp., Physella sp., Planorbella sp. and St. exilis. Oakwood had populations of three snails, Physella sp., Planorbella sp. and St. exilis. Snail taxa were identified both morphologically and through DNA barcoding of the cytochrome oxidase I (COI) gene fragment.
Snails were collected in 1-L Nalgene bottles and placed in a cooler for transport to the laboratory within 2–4 h of collection. After 3–4 h of acclimation to dechlorinated laboratory water, snails were placed in individual 50-ml centrifuge tubes in 12 h of darkness followed by 12 h under fluorescent light to encourage the emergence of trematode cercariae. Snails that shed cercariae were maintained individually in the laboratory for other experiments. Snails that did not shed were crushed and examined under a stereomicroscope (10–40× magnification) for the presence of immature sporocysts or rediae, and immature or mature metacercariae, including tetracotyles. The majority of dissections occurred within 1 week of collection. It is unlikely that any tetracotyles collected were the result of infections taking place in the laboratory because snails were separated individually within 5–8 h and any snails actively shedding cercariae remained isolated after that time. Furthermore, the 2- to 3-week-long development time of tetracotyle stages would have prevented laboratory infections from biasing our data set because only mature, fully developed tetracotyle stages were counted and preserved for analysis (Basch, 1969).
Three snails included in this data set were also infected as first intermediate hosts with other species of trematodes and therefore maintained individually in the laboratory for 9–69 days after collection until dissection. Because these snails were no longer exposed to any other snails shedding cercariae, their tetracotyle infections represent the natural infection levels transmitted in the field prior to collection. Snails were dissected and all tetracotyle samples were counted and preserved in 2-ml cryovials in 80%–95% ethanol prior to DNA extraction.
At Oakwood and Crabtree Entrance we did not detect any tetracotyles in any snails other than St. exilis. At Crabtree 3G we observed tetracotyles in four Physella (number of tetracotyles per snail ranged from three to 93). However, for the purposes of this study we included only tetracotyles collected from St. exilis. The field sampling at these locations also revealed few leeches with none recovered at Oakwood, one at Crabtree Entrance and three at Crabtree 3G, although none were collected or examined for metacercariae.
2.2 Microsatellite development and amplification
A subset of mature tetracotyles from each snail was utilized for genetic analyses. Our goal was to include ~40 tetracotyles from each snail, when available. We analysed all of the tetracotyles in a snail when fewer than 40 tetracotyles were present. DNA for genotyping was extracted from individual mature tetracotyles by placing them in 200 μl 5% Chelex with 0.1 mg/ml proteinase K and incubating at 60°C for 2–12 h and 95°C for 8 min. Microsatellite primers were developed following the methods outlined for the freshwater snails Valvata tricarinata and Promenetus exacuous (Yurco & Keeney, 2018). Following this protocol, ~3 μg of DNA from sporocysts from a single St. exilis snail host was sent to the University of Wisconsin-Madison Biotechnology Center for ION Torrent PGM sequencing using ~25% of a 318 chip. Resulting DNA sequences were examined for perfect tetranucleotide repeats with a minimum length of eight repeat units using msatcommander version 1.0.8 (Faircloth, 2008). Microsatellite loci were amplified using three primer polymerase chain reactions (PCRs) (Schuelke, 2000) with the fluorescently labelled microsatellite primer tag (CAGTCGGGCGTCATCA) as a third primer. One primer from each locus-specific pair also contained this 5′ primer tag sequence (Table S2) and the second primer contained a 5′ GTTT “pigtail” (Brownstein et al., 1996). PCRs included 1× Type-it Multiplex PCR Master Mix (Qiagen), 0.2 μm standard locus primer, 0.02 μm locus primer with tag sequence, and 0.2 μm fluorescently labelled tag in a total of 10 μl. Thermal cycling conditions included an initial heat activation for 5 min at 95°C, 30 cycles of 30 s at 95°C, 90 s at 60°C, 30 s at 72°C and a final extension of 30 min at 60°C.
Individual locus PCRs were pooled post-PCR to create three sets of two to four loci labelled with the dyes NED, 6FAM, PET and VIC (Table S2). Genotyping was performed on an ABI 3730xl 96-Capillary Genetic Analyser at the DNA Analysis Facility at Yale University. Microsatellite allele peaks were scored using geneious version 8.1 (Kearse et al., 2012).
2.3 Multilocus genotype/clonemate identification and clone diversity
Identification of multilocus genotypes (MLGs) was initially conducted over all sample sites with 10 loci using genalex version 6.503 (Peakall & Smouse, 2006, 2012) and missing data were ignored. This allowed us to identify putative matching MLGs despite missing data and identify if any MLGs were recovered from different sites. The same sets of MLGs were recovered after two loci (CFL5507 and CFL19848) were removed (see Results). After removal of six sets of MLGs (31 tetracotyles) missing data at a single locus (CFL21360), genclone version 2.0 (Arnaud-Haond & Belkhir, 2007) was used to determine the probability that identical MLGs within each sample location were the product of sexual reproduction (psex) vs. asexual reproduction. psex is the probability of obtaining the observed number of identical MLGs in the sample by sexual reproduction. psex < .05 for n = 2 indicates all copies of that MLG are the products of asexual reproduction and can be considered clones (Gregorious, 2005). The removed tetracotyles were then added back into the data set and psex was calculated for them without locus CFL21360. Given the persistence of linkage disequilibrium in Crabtree Entrance after full-sibs were removed (see below), psex values were also estimated in this site using a subset of five loci not displaying linkage disequilibrium (CFL21, CFL2226, CFL4491, CFL15967 and CFL16762). genclone version 2.0 was also used with the 10- and eight-loci data sets to examine pairwise allelic differences between clones to identify clones differing at only one or two loci or alleles and examine the distribution of pairwise allelic differences in order to determine if the difference between some clones were potentially the result of scoring errors or somatic mutations (Arnaud-Haond et al., 2007).
After identification of clones, the following were calculated within each site with genepop version 4.7 (Raymond & Rousset, 1995) using one representative of each clonemate: number of alleles per locus, expected and observed heterozygosities, FIS, deviations from Hardy–Weinberg expectations, and linkage disequilibrium between loci pairs. The presence of large numbers of full-sibs can have a major effect on tests of linkage disequilibrium and cause false positive results (Mangin et al., 2012; Sánchez-Montes et al., 2017). To determine if the presence of full-sibs was causing significant linkage disequilibrium in our samples, we reran linkage disequilibrium tests in Crabtree Entrance and Crabtree 3G after (i) removing one member of each full-sib dyad that was well supported (probability full-sibs >95%) and (ii) removing one member of each full-sib dyad that was moderately well supported (probability full-sibs >80%). Sample sizes in the 80% data sets were 68 for Crabtree Entrance and 35 for Crabtree 3G, suggesting differences in significance tests were not simply the result of reduced statistical power. An adjusted critical value based on the BY false discovery rate (BY-FDR) method (Narum, 2006) was used for all significance tests with multiple comparisons. The index of clonal diversity (number of clones/number of tetracotyles genotyped) was calculated for comparison with previous studies (Arnaud-Haond et al., 2007; Criscione et al., 2020).
2.4 Genetic structure
To determine if genetic differentiation has developed among hosts and sites, analyses of molecular variance (amova) were conducted utilizing the number of different alleles to calculate genetic distances and generate hierarchical F-statistics with arlequin 3.5.2.2 (Excoffier & Lischer, 2010). Individual snails were treated as populations and grouped according to sample site. Separate analyses were done for the entire data set (n = 577) and without clonemates within snails (n = 208). For analyses with clonemates removed, one copy of an identical clone was kept in each snail when multiple snails shared clones. Overall and pairwise FST values were also calculated among the three sample sites for the entire data set and with only a single clonemate within sites (n = 158) by treating sites as the populations. Multiple clonemates were removed from sites to determine if allele frequencies among different clones, and not the presence of clones, was contributing to genetic structure. The significance of results was tested with 10,000 permutations. Identical analyses were conducted for eight loci and the five loci utilized for Crabtree Entrance psex calculations.
Genetic structure within sample sites was examined by keeping one representative clonemate in each site and using Bayesian clustering with structure version 2.4.3 (Pritchard et al., 2000). Mean ln(P(D)) values (Pritchard et al., 2000) were used as our criterion for determining the most likely number of genetic clusters (K). Analyses in each site examined K values ranging from 1 to 10 and used an admixture model with five iterations, burn-in length of 200,000 and 250,000 steps in the Markov chain Monte Carlo (MCMC) procedure. As structure assumes Hardy–Weinberg and linkage equilibrium, we looked for consistency between the eight- and five-loci data sets and interpreted the results with caution. Our goal was not to accurately determine the specific number of genetic clusters within each sample site, but to determine if there is evidence of multiple genetic clusters in any of the sites.
2.5 Sibling reconstructions and distribution of siblings and clones within hosts
If multiple clonemates are commonly present in definitive hosts, it is possible for sibling parasites to be produced, infect snails (either same or proximate) and accumulate in second intermediate snail hosts, along with clonemates. To determine if sibling parasites are accumulating in snails and potentially being cotransmitted to definitive hosts, we utilized a modified version of the methodology of Detwiler and Criscione (2017) and Criscione et al. (2020). Tetracotyles within sample sites were treated as component populations (Bush et al., 1997; Detwiler & Criscione, 2017), and pedigree reconstructions were made with colony version 2.0.6.7 (Jones & Wang, 2010). Sibling relationships were obtained from the FullSibDyad and HalfSibDyad output files. Sibling relationships supported by ≥95% probability were considered well supported and included in our analyses. No half-sib relationships were supported by ≥95% probability and we only included full-sib dyads (pairs of individuals) in our analyses. Inclusion of moderately well-supported sibling relationships does not alter our findings (data not shown). The percentage of full-sib dyads within hosts (PFS) was calculated as the weighted average over hosts based on the number of tetracotyles examined from each host vs. the total number examined in the component population, and compared to the percentage of full-sib dyads over the entire component population (PEFS) (Criscione et al., 2020; Detwiler & Criscione, 2017). Following Criscione et al. (2020), we also examined the percentage of clonemate dyads within hosts (PC) vs. the percentage of clonemate dyads over the entire component population (PEC) using the same weighted average approach. These comparisons examine the frequencies of sibling and clone dyads within hosts vs. random expectations based on frequencies within component populations to determine if siblings and/or clones are being transmitted together within second intermediate hosts. Sibship analyses were conducted with unique clones (n = 158) and clonemates were subsequently added back in to the data set (n = 577) and given the same parental assignments as their clonemates (Criscione et al., 2020). colony sibling analyses for each component population utilized the full-likelihood method of sibship reconstruction with very high precision and very long runs, female and male polygamy with inbreeding for monoecious species, updated allele frequencies, five runs, sibling scaling, no sibship prior, and allelic dropout and mutation/error rates set to 0.005. The significance of clonemate cotransmission and full-sibling cotransmission were tested using G log-likelihood tests (Goudet et al., 1996) in genepop version 4.7 with genotypes treated as a single locus. To test the significance of full-sibling cotransmission without the effect of clones, a single individual of each clone was retained within hosts, and clones belonging to full-sibling groups were coded as identical. To determine if clonemate aggregation within snails (average PC) was consistent across the three sample sites, the “alphaContrastTest3” function (Scofield et al., 2012) of the package dispersaldiversity version 0.9.9001 was employed as described for the “alphaContrastTest” in Criscione et al. (2022) using R Statistical Software version 4.2.1 (R Core Team, 2022). Pairwise comparisons between sample sites were made using the “alphaContrastTest.” Oakwood snails A6 and B1 containing a single tetracotyle were removed from average PC comparisons because the diversity tables cannot have one sample.
2.6 Self-fertilization/clonemate mating estimates and identification of selfed offspring
Rates of self-fertilization, including matings between clonemates, were estimated for unique clones with rmes (David et al., 2007) using the two-locus heterozygosity disequilibrium estimation of selfing rates (s(g2)) and its associated p-value for the null hypothesis of s = 0, and the maximum-likelihood (ML) estimation of selfing rates (s(ML)) with 95% confidence intervals (CIs). Clones that were the products of selfing (selfed offspring) were identified using colony with run settings as described for pedigree reconstructions. Selfed offspring output files were examined and tetracotyles with ≥95% probability of being the products of selfing were identified as selfed offspring. This includes individuals that are the offspring of matings between two clonemates as well as individual self-fertilizations. The proportion of unique clones that are the product of selfing/clonemate matings were calculated as the number of estimated selfed offsprings/number of unique clones within the sample site.
2.7 DNA sequencing and analyses
To examine lineage divergence and species identification, a portion of the COI gene was amplified from 42 tetracotyles using the primers Plat-diploCOX1F and Plat-diploCOX1R (Moszczynska et al., 2009). The 50-μl PCRs contained 4.0 μl of DNA extraction, 200 μm each dNTP, 2.0 mm MgCl2, 0.5 μm each primer, 1× Taq buffer and 1.25 units GoTaq DNA polymerase (Promega). PCR amplification consisted of 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 50°C and 1 min at 72°C, and a final extension for 8 min at 72°C. PCR products were purified using QIAquick PCR Purification kits (Qiagen). One to two teracotyles were amplified from each host snail and 36 different MLGs were examined (identical MLGs were amplified when snails only possessed MLGs found in other snails). COI sequences were aligned with clustal w (Thompson et al., 1994) as implemented in mega x (Kumar et al., 2018). Uncorrected p-distances were calculated between haplotype pairs with mega x.
In order to confirm species identification, Bayesian inference (BI) was implemented to reconstruct phylogenies of our samples with closely related taxa within the Strigeidae. Previously published sequences of strigeids were downloaded from GenBank and combined with new sequence data generated in this study (Table S3). Tylodelphys scheuringi was used as our outgroup taxon to root our phylogeny and other strigeid taxa were selected to match recent strigeid phylogenetic analyses (e.g., Gordy & Hanington, 2019). Our COI gene fragment contained 606 bp and this data set was partitioned by codon position. To identify appropriate models of nucleotide substitution for both analyses, we used the program mrmodeltest version 2.2 (Nylander, 2004), run in paup* version 4.0b10 (Swofford, 2002). We used the Akaike information criterion (AIC) to select the best-fit models for each partition, as estimated by mrmodeltest. Phylogenetic analyses using BI were conducted with mrbayes version 3.0b4 (Ronquist & Huelsenbeck, 2003). Two simultaneous BI runs were conducted (with the default MCMC settings) and run for a total of 4.0 × 106 generations per run, sampling trees and parameters every 100 generations. We used PSRF values (output by mrbayes), together with plots of cold chain likelihood values and parameter estimates visualized in tracer version 1.5.4 (Rambaut & Drummond, 2009), to confirm stationarity and convergence of MCMC runs. Based on this evaluation, the first 1 × 106 generations from each run were discarded as burn-in.
3 RESULTS
3.1 Presence of clonemates
Ten microsatellite loci were developed for Cotylurus sp. (Table 1). Microsatellite genotypes were determined for 577 tetracotyles from 23 snails (Crabtree Entrance 241 tetracotyles from seven snails, Crabtree 3G 274 tetracotyles from nine snails, and Oakwood 62 tetracotyles from seven snails) and 1–46 (25.09 ± 16.88) tetracotyles were analysed per snail. A single tetracotyle was analysed from two Oakwood snails. Initial MLG identification with 10 loci revealed three pairs of MLG groups differing by a single locus (CFL5507) in Crabtree 3G. In each case, one group differed from the other by nonamplification of a homozygous allele. Given that these only differed at a single locus with low amplification and potentially prevalent null alleles, these were treated as three groups of identical MLGs, producing 158 unique MLGs. Two loci (CFL5507 and CFL 19848) were excluded from final analyses due to a large proportion of missing data probably due to the presence of null alleles (Table 1). Final MLG identification with eight loci recovered the same 158 MlGs (Crabtree Entrance n = 90, Crabtree 3G n = 44 and Oakwood n = 24). Six sets of MLGs (31 tetracotyles) did not amplify for locus CFL 21360. All other MLGs amplified at the eight loci. MLGs differed by a minimum of four alleles at three loci with the 10-loci data set and two alleles at two loci with the eight-loci data set. The two pairs of MLGs differing by only two alleles were from Crabtree Entrance and neither locus involved one MLG being homozygous for an allele that was shared in the other heterozygous MLG. Treatment of these as identical MLGs would not alter our findings. Pairwise allelic difference distributions were unimodal in Crabtree Entrance and Oakwood. A second “peak” was observed in Crabtree 3G and was produced by a lack of pairs differing by four alleles as two pairs of MLGs differed by three alleles and the rest by a minimum of five alleles. Genotype calls were checked for all MLG pairs differing by two or three alleles and were accurate.
| Locus | Motif | Crabtree entrance (N = 90) | Crabtree 3G (N = 44) | Oakwood (N = 24) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | A | HO/HE | F IS | N | A | HO/HE | F IS | N | A | HO/HE | F IS | ||
| CFL21 | (AATG)19 | 90 | 14 | 0.87/0.85 | −0.012a | 44 | 15 | 0.73/0.78 | 0.082 | 24 | 17 | 0.92/0.88 | −0.020 |
| CFL2226 | (AAAT)15 | 90 | 9 | 0.80/0.79 | −0.010 | 44 | 13 | 0.89/0.88 | 0.004a | 24 | 7 | 0.92/0.78 | −0.153 |
| CFL4491 | (AAAT)15 | 90 | 19 | 0.89/0.86 | −0.025a | 44 | 17 | 0.89/0.86 | −0.014 | 24 | 16 | 0.96/0.90 | −0.041 |
| CFL5507b | (ACAT)14 | 72 | 10 | 0.29/0.79 | 0.635a | 34 | 9 | 0.06/0.80 | 0.929a | 14 | 5 | 0.00/0.55 | 1.000a |
| CFL12863 | (AATC)14 | 90 | 13 | 0.92/0.83 | −0.111 | 44 | 15 | 0.84/0.84 | 0.015 | 24 | 6 | 0.75/0.72 | −0.027 |
| CFL15967 | (AAAT)12 | 90 | 10 | 0.52/0.61 | 0.154 | 44 | 12 | 0.66/0.78 | 0.171a | 24 | 12 | 0.75/0.85 | 0.143 |
| CFL16762 | (AATT)13 | 90 | 7 | 0.60/0.60 | 0.004 | 44 | 6 | 0.54/0.55 | 0.085 | 24 | 4 | 0.42/0.52 | 0.220 |
| CFL17083 | (AGAT)14 | 90 | 14 | 0.84/0.85 | 0.006a | 44 | 12 | 0.93/0.80 | −0.156a | 24 | 17 | 0.88/0.90 | 0.045 |
| CFL19848b | (AGAT)14 | 56 | 5 | 0.13/0.64 | 0.807a | 28 | 4 | 0.14/0.64 | 0.785a | 16 | 4 | 0.25/0.69 | 0.654a |
| CFL21360 | (AAAT)13 | 88 | 13 | 0.59/0.84 | 0.298a | 41 | 11 | 0.42/0.81 | 0.497a | 23 | 11 | 0.57/0.86 | 0.359a |
| Multilocusc | −0.005a | 0.023a | 0.015 | ||||||||||
- Abbreviations: A, number of alleles; HO/HE, observed heterozygosity/expected heterozygosity; N, number of unique clones successfully amplified.
- a Significant based on BY-FDR.
- b Not used for analyses.
- c Based on seven loci without evidence of null alleles (CFL21, CFL2226, CFL4491, CFL12863, CFL15967, CFL16762, CFL17083).
The psex values with the MLGs missing data removed were all <.000001 for n = 2 and <.00001 for n = 2 with all MLGs included and CFL21360 omitted. When only five loci were used to calculate psex values in Crabtree Entrance, all psex values were <.009 for n = 2 for all groups of MLGs. The index of clonal diversity was 158/577 = 27.4%.
Excluding two snails from Oakwood with a single tetracotyle, all snails possessed multiple clones with the exception of two snails from Oakwood with three and 10 tetracotyles (Figure 2). Clonemates were recovered from all hosts with multiple tetracotyles except a single snail with seven tetracotyles in Crabtree Entrance. Snails possessed either high numbers of different clones with few clonemates (snails D8, D9), little clonal diversity with numerous copies of the same clone(s) (snails D7, E4, G6, E8) or a combination of these (snails G3, C9, E2, B8). Twenty or more clonemates were recovered from three snails (snails D7, E4, A1). Twenty-seven clones were recovered from more than one snail. In all cases, clonemate sharing between hosts only occurred within sites. In total, 42 pairs of snails shared at least one clone, up to five clones were shared between snail pairs and up to five snails shared the same clone (Table S4).
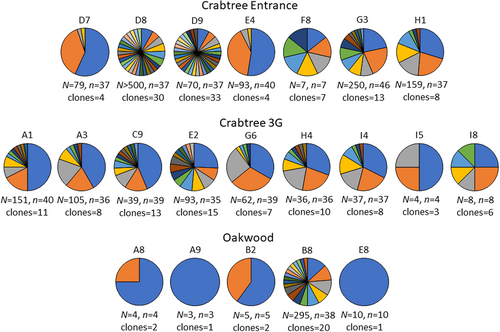
Within sample sites, the number of alleles and observed heterozygosities for the eight individual loci utilized were 4–19 and 0.42–0.96 and several loci deviated from Hardy–Weinberg expectations within sites (Table 1), based on unique clones. Linkage disequilibrium was initially detected in 13 of the 28 pairwise comparisons in Crabtree Entrance and six of the 28 pairwise comparisons in Crabtree 3G (Table S5). After removing well-supported full-sibs, linkage disequilibrium was detected in six pairwise comparisons in Crabtree Entrance and one pairwise comparison in Crabtree 3G. After removal of moderately well-supported full-sibs, two pairwise comparisons were significant in Crabtree Entrance.
3.2 Genetic structure
Hierarchical F-statistics utilizing all clonemates (n = 577) and treating snails as populations detected significant genetic differences among sample sites (FCT = 0.063; p < .0001), among all snails (FST = 0.148; p < .0001) and among snails within sample sites (FSC = 0.090; p < .0001). The same analysis with single copies of each clonemate within snails (n = 208) detected genetic structure among sample sites (FCT = 0.070; p < .0001), and among all snails (FST = 0.069; p < .0001), but not among snails within sample sites (FSC = −0.002; p = .9071). Identical analyses for both data sets utilizing five loci produced similar results (Table S6). Comparisons treating sample sites as populations using eight loci detected significant genetic structure overall (FST = 0.079; p < .0001 and FST = 0.069; p < .0001) for the full data set (n = 577) and unique clones within sites only (n = 158), respectively. Similar results were obtained using five loci (Table S6). All pairwise comparisons were significant for both data sets with FST ≥ 0.063 and p < .0001 (Tables S7 and S8). Similar results were produced using five loci (Tables S7 and S8).
structure results supported multiple genetic clusters within at least two sites. ln(P(D)) within Crabtree Entrance for the eight loci analysis supported K = 5–7, with increased variance around K = 6, while the five-loci analysis supported K = 2–3. For Crabtree 3G, eight and five loci both supported K = 2–3, with increased variance around K = 3 in the eight-loci analysis. In Oakwood, both analyses produced high variance around most estimates, with the exception of K = 1 or 2 in the eight-loci analysis (Figure S1). Longer burn-in lengths for Oakwood analyses (up to 400,000) and increased MCMC steps (up to 500,000) did not improve the results (data not shown). In all sample sites and scenarios, most tetracotyles were strongly assigned to a specific cluster with only a few individuals showing potential admixture and proportion of assignment <0.80 to a specific cluster, and snails possessed tetracotyles from multiple genetic clusters (Figure S2). The various partitions typically clustered individuals by sibling status with full-sibs grouping together, and lesser related groups of full-sibs and potential half-sibs grouping together at lower K values, indicating the influence of family-induced structure on our results (Anderson & Dunham, 2008). The lack of resolution and minimal support for genetic structure in Oakwood coincided with a lack of full-sib dyads in that site (see below).
3.3 Sibship and clone cotransmission, and self-fertilization estimates
Total numbers of dyads, clone dyads in component populations and within hosts, full-sib dyads in component populations and within hosts, and their respective percentages are presented in Table 2. No half-sib dyads were supported with ≥95% probability. Significantly higher percentages of clone dyads vs. random expectations were found within hosts in all three component populations. The “alphaContrastTest” testing average PC among sample sites was nonsignificant (p = .0905). Pairwise average PC comparisons between sample sites yielded the following results: Crabtree Entrance vs. Crabtree 3G (p = .6535), Crabtree Entrance vs. Oakwood (p = .0342) and Crabtree 3G vs. Oakwood (p = .0796). We did not detect significantly higher percentages of full-sib dyads within hosts vs. random expectations in Crabtree Entrance or Crabtree 3G. No full-sib dyads were supported with ≥95% probability in Oakwood.
| Crabtree entrance | Crabtree 3G | Oakwood | |
|---|---|---|---|
| Total dyads | 28,920 | 37,401 | 1770 |
| Total clone dyads | 1205 | 2675 | 107 |
| Clone dyads within hosts | 876 | 1000 | 87 |
| P EC a | 4.17 | 7.15 | 6.05 |
| PC and p-valueb | 19.06 p < .001 | 20.17 p < .001 | 31.22 p < .001 |
| Total full-sib dyads | 130 | 175 | 0 |
| Full-sib dyads within hosts | 25 | 31 | 0 |
| P EFS c | 0.45 | 0.47 | — |
| PFS and p-valued | 0.48 p = .510 | 0.64 p = .898 | — |
| Selfed/proportion clonese | 1/0.01 | 1/0.02 | 3/0.13 |
| s(g2) and p s(g2)f | 0.000 p = .731 | 0.000 p = .973 | 0.003 p = .389 |
| s(ML) and 95% CIsg | 0.000 0.000–0.057 | 0.000 0.000–0.045 | 0.000 0.000–0.064 |
- a Percentage of clonemate dyads over entire sample site.
- b Percentage of clonemate dyads within hosts.
- c Percentage of full-sibling dyads over entire sample site.
- d Percentage of full-siblings within hosts.
- e Estimated number of unique clones that are the products of self-fertilization or identical clone mating/proportion of unique clones that are the product of selfing (Crabtree Entrance = 1/90, Crabtree 3G = 1/44, Oakwood = 3/24).
- f Two-locus heterozygosity disequilibrium selfing rate and the probability of no selfing (p s(g2)).
- g s(ML) = selfing rate based on maximizing the log-likelihood of multilocus heterozygosity structure of the sample and associated 95% CIs.
Self-fertilization estimates were low in all sites for both the g2 and ML estimators with only Oakwood having s(g2) > 0.000 and also the lowest probability of s = 0. Upper ML 95% CIs were ≤0.064 (Table 2). Similar results were obtained with the five-loci data set (data not shown). Estimated numbers of selfed offspring from colony were also low and proportions of unique clones were consistent with selfing rate estimates, with the exception of Oakwood's being slightly elevated (Table 2).
3.4 Species identification and phylogeny of Cotylurus
Partial COI amplification from 42 tetracotyles produced six different haplotypes. Pairwise p-distances ranged from 0.00% to 0.69% among tetracotyles. Tetracotyles with identical MLGs always had identical COI haplotypes.
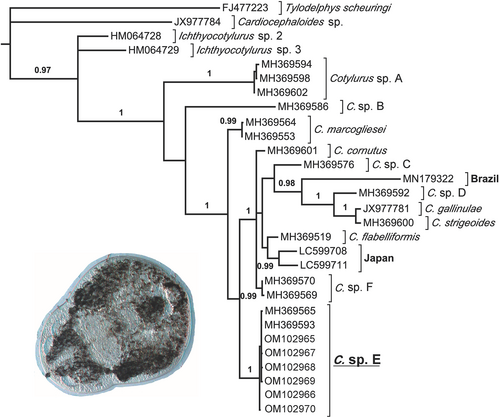
mrmodeltest 2.2 selected the following partitioned nucleotide substitution models for our data set: codon 1 (GTR+I), codon 2 (F81+I) and codon 3 (GTR+Γ). Our resulting Bayesian phylogeny showed strong support for many of the relationships among the Strigeidae and had mostly congruent hypotheses as those recovered by Gordy and Hanington (2019). More specifically, we found the genus Cotylurus to be monophyletic and sister to the genus Ichthyocotylurus (Figure 3). Within Cotylurus there appears to be a basal split between Cotylurus sp. A and C. sp. B and the other taxa. Furthermore, we found a sister relationship between the Cotylurus gallinae and C. strigeoides clade and C. sp. D, that together are sister to a species from Brazil. Although additional relationships among other Cotylurus taxa within this clade are not supported, the majority of taxa with multiple sequences were found to be monophyletic. Finally, all six haplotypes from this study matched Cotylurus sp. E of Gordy and Hanington (2019).
4 DISCUSSION
Parasite transmission dynamics are a key component of their evolution. Our study revealed that low trematode clonal diversity can develop and large numbers of clonemates can accumulate within aquatic second intermediate hosts. This is contrary to expectations as the aquatic environment facilitates clone mixing after trematode asexual reproduction within snail hosts by allowing mobile cercariae to disperse via swimming, the movements of water and host mobility (Keeney et al., 2007a, 2007b; Lagrue et al., 2009; Leung et al., 2009; Rauch et al., 2005; Valdivia et al., 2014). Our clonal diversity of 27% stands in stark contrast to the 95%–100% clonal diversities typical of aquatic trematodes with three-host life cycles (Criscione et al., 2020). Even relatively immobile aquatic intermediate hosts accumulate diverse trematode clone assemblages and few clonemates (Leung et al., 2009; Valdivia et al., 2014). Given that water movements and cercarial mobility are still occurring in our system, the ability of Cotylurus sp. to utilize the same species for two stages of its life cycle may be a major factor causing clonemate accumulation.
One potential hypothesis for clumped transmission of clonemates would be that aggregations of Stagnicola exilis snails allow large numbers of clones released from a single snail to enter a new snail prior to dispersing. If several snails release cercariae, second intermediate snail hosts will accumulate multiple copies of more than one clone, as observed in all of our sample sites. Also, less opportunity may exist for passive cercarial movement within freshwater lakes/ponds compared to studies on relatively immobile marine hosts (Leung et al., 2009; Valdivia et al., 2014), although drifting of snails and active cercarial swimming could distribute cercariae among snails (Campbell, 1973, 1997). Host aggregation may also cause multiple second intermediate hosts to become infected by the same trematode clones, which has been rare in previous aquatic studies (Keeney et al., 2007a, 2007b; Rauch et al., 2005). This could also occur by snail drifting (Campbell, 1997). Clone sharing was detected in all three sample sites, but the same clone was never recovered from different sample sites. Clonemate sharing was most common in Crabtree 3G snails (32 pairs) with five pairs sharing clones in the other sites. While we do not have data on snail density from our sites, Cotylurus tetracotyle prevalence was relatively high during most sampling visits (Table S1), supporting utilization of the same host species as a successful means of transmission to many second intermediate hosts.
While the novelty of co-occurrence of clonemates within snails is the major feature of our data, several snails, such as D8 and D9 in Crabtree Entrance (Figure 2), had the “typical” second intermediate host pattern and possessed a large number of different clones and few clonemates. Mixing of clones therefore does occur in some snails in this system. These snails were probably not infected by a “pulse” of clones from a single snail, but accumulated small numbers of different cercariae clones from multiple first intermediate hosts. Additional snails, including G3 in Crabtree Entrance, C9 and E2 in Crabtree 3G, and B8 in Oakwood (Figure 2), had a combination of large numbers of clonemates and numerous low-copy clones. While we do not have information on the clonal diversity of cercariae shed by individual snails, previous studies indicate low clonal diversity is characteristic of molluscan first intermediate hosts for many trematode species (Keeney et al., 2007a; Lagrue et al., 2009; Minchella et al., 1995; Rauch et al., 2005; Sire et al., 1999). It is likely that proximity to and the number of infected snails nearby at the time of cercarial shedding would influence clonal diversity and the extent of clonemate transmission. If a snail is not infected by large numbers of cercariae from a proximate host, the aquatic environment would still allow for the accumulation of free swimming cercariae from numerous snails. The extent to which clonemates are aggregated within hosts may vary across a species' range. While our overall comparison of average PC among sample sites was nonsignificant, the p-value was low and pairwise comparisons suggest the nonsignificant result was driven by similarities between Crabtree Entrance and Crabtree 3G, with differences existing between Oakwood and Crabtree Entrance.
We have documented clumped transmission of Cotylurus sp. clonemates from first to second intermediate hosts. An important question is how this transmission impacts the reproductive behaviours of the parasites within definitive hosts and the evolutionary consequences. The accumulation of clonemates within individual second intermediate hosts is likely to lead to the presence of clonemates reaching sexual maturity within definitive host birds, facilitating reproduction among them. Diverse mechanisms have evolved among some hermaphroditic species to facilitate outbreeding and potentially mitigate the effects of inbreeding depression, including self-incompatibility in plants (Fujii et al., 2016) and animals (Harada et al., 2008), recognition of conspecific chemical cues in polychaetes (Schleicherová et al., 2006) and chemical-mediated preference for nonself eggs in tunicate sperm (Kosman et al., 2017). Selfing by hermaphroditic trematodes is typically rare and may be a last resort when alone (Criscione & Blouin, 2006), although the frequency and success of outcrossing may be influenced by genotypic compatibility (Rieger et al., 2013). In Cotylurus, attempted outcrossings could lead to “selfing” between clonemates when they are aggregated within definitive hosts. However, despite the large number of clonemates in intermediate hosts and potential for their matings in definitive hosts, selfing remains rare in Cotylurus sp.
Two potential nonexclusive mechanisms could account for the observed low levels of selfing. First, the aggregation of clonemates could be diluted in vagile definitive hosts when they consume multiple snails containing different clones from the same or different sites. Most species of wetland dabbling ducks consume snails, particularly females during the breeding season (Drobney & Fredrickson, 1979; Kaminski & Prince, 1981; Krapu, 1979; Swanson et al., 1974). Some species such as blue-winged teals (Anas discors) consume snails in greater proportion than expected based on occurrence (Swanson et al., 1974). If definitive hosts are consuming large numbers of snails this would directly decrease the frequency with which clonemates have the opportunity to mate. The presence of snails with higher clonal diversities, such as snails D8 and D9 in Crabtree Entrance, and the overall clonal diversity of tetracotyles within sites would facilitate mixing. Criscione et al. (2022) detected high levels of clonemate aggregation of larval Dicrocoelium dendriticum in ant second intermediate hosts (PC = 44.09%–56.38%), but much less aggregation in final hosts (PC = 3.68%) and no evidence of inbreeding. We are probably observing a similar phenomenon with clone mixing in bird definitive hosts reducing the relatively high observed clonemate aggregation in snail intermediate hosts (PC = 19.06%–31.22%) and decreasing the chances of inbreeding. Second, clonemates may avoid mating with each other despite their occurrence together within the definitive host. This would require the evolution of clone/kin recognition mechanisms and, although speculative, it is possible that such inbreeding avoidance could evolve in species that are commonly transmitted with clonemates and/or siblings (Criscione et al., 2005; Prugnolle et al., 2004).
Consistent with the lack of selfing/clonemate mating, strong inbreeding effects were not detected in Cotylurus sp. Inbreeding from selfing/clonemate mating and kin-mating within bird hosts would cause heterozygote deficiencies across loci (Charlesworth, 2003; Prugnolle et al., 2005; Waples, 2015). Clones cannot persist across generations as sexual reproduction, including selfing, occurs in the definitive host, decreasing potential inbreeding effects (Criscione et al., 2011). Inbreeding could also be mitigated when different clonal lineages encounter each other since individual snails often possess clones from multiple genetic clusters. Birds moving and consuming snails from different areas would also increase reproduction between distinct clonal lineages, although survivorship time within definitive hosts may be limited (Campbell, 1973). In addition, kin-mating not involving clonemates may be uncommon since clones are frequently found together in snails, but full-sibs are not. Clonemate cercariae would be received from the same snail, but full-sibs are probably spread among first host snails from independent miracidia infections after eggs are passed with bird faeces. Related individuals infecting different snails could additionally be moved enough prior to cercarial maturation, which varies from 25 to 44 days, with an average of ~35 days post-infection depending on temperature (Basch, 1969, 1970; Nolf & Cort, 1933; Olson, 1970) to limit co-infection of the same second intermediate host snails during cercarial shedding.
Genome-wide inbreeding effects were not obvious in this study as relatively high levels of heterozygosity were observed across loci. In addition, while deviations from Hardy–Weinberg expectations were common, both negative and positive FIS values were observed (Table 2) and this variation is unlikely if inbreeding is prevalent (Criscione et al., 2011; Vilas et al., 2003). Null alleles potentially accounted for the largest deviations and positive FIS values. Deviations were also probably caused by the presence of numerous related clones (full and half-sibs) causing nonrandom sampling (Sánchez-Montes et al., 2017; Waples, 2015). Multilocus FIS values based on seven loci without null alleles were close to zero, and while statistically significant, included positive and negative values (Table 1). Negative FIS values were relatively moderate and we did not encounter individuals with more than two alleles at these loci, suggesting modest heterozygote excesses are not from duplicated loci (Detwiler & Criscione, 2011), but could result from the presence of sibling groups (Balloux, 2004).
The distribution of clonemates and closely related clones has impacted the geographical distribution of genetic variation at multiple scales. Genetic differentiation was observed among sample sites, within sample sites (potentially influenced by family groups; Anderson & Dunham, 2008) and among snail second intermediate hosts. Partitioning of genetic diversity at these levels suggests birds are probably consuming genetically distinct groups of parasites when ingesting specific snails. This is based on the presence of clonemates and not the co-infection of related, nonclonal parasites within the same individual snails. While full-sibs were present, they were not aggregated within vs. among snails and therefore do not necessarily co-infect the same bird host. While we did not include putative half-sibs, a similar pattern was observed with them. Sample sites remain genetically distinct when clonemates are removed, reflecting differences in allele frequencies from related individuals among snails in sites, even at the relatively short geographical distance (i.e., 0.75 km) between Crabtree Entrance and Crabtree 3G. Bird hosts appear to be depositing related parasite offspring in these habitats and our sampling collected aggregations of related individuals. While vagile definitive hosts can limit genetic differentiation within intermediate hosts at varying geographical scales (Blasco-Costa & Poulin, 2013; Feis et al., 2015; Jarne & Théron, 2001; Keeney et al., 2009; Louhi et al., 2010; Prugnolle et al., 2005), the clumped transmission of clonemates and dispersal of related individuals within sites is causing genetic structure to develop at this stage of the parasite's life cycle despite bird movements in our system.
In conclusion, our study demonstrates that an atypical life cycle involving the use of the same species as first and second intermediate host produces patterns that challenge many of the established norms of aquatic trematode transmission. Large numbers of clonemates accumulate within second intermediate hosts despite environmental opportunities for clone mixing prior to infection. Selfing and inbreeding effects are minimal and sufficient clonal diversity may exist in definitive hosts to promote outcrossing. Genetic structure develops over relatively small geographical distances in spite of the use of vagile definitive hosts. The ability to use the same species as first and second intermediate host may be advantageous for transmission into migratory definitive hosts when genetic diversity is maintained within parasite populations and the first intermediate host is a prey item overcoming the “trophic transmission vacuum” of Benesh et al. (2014).
AUTHOR CONTRIBUTIONS
Sarah Orlofske and Devon Keeney designed the study. Sarah Orlofske and Robert Jadin collected snails and parasites. Devon Keeney and Sarah Cobb generated molecular data. Devon Keeney, Sarah Cobb and Robert Jadin analysed molecular data. Devon Keeney, Sarah Orlofske and Robert Jadin wrote the manuscript.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGEMENTS
We are grateful to C. Criscione for his advice regarding testing the significance of trematode clone and sibling transmission data. We thank J. Block, R. Borchert, A. Carmona, O. Choi, J. Kawaguchi, J. Lujan, A. Mendoza and A. Villegas for their assistance with field sampling and snail dissections. We acknowledge the Northeastern Illinois University (NEIU) Student Center for Science Engagement for financial support of undergraduate student field and laboratory work and the NEIU Committee on Organized Research Grants in 2014 and 2015 for funding a portion of the molecular analyses. Additional financial support was provided by Le Moyne College's Research and Development Grant Program and Le Moyne College's Student Research Committee. We are also grateful for the staff of the Crabtree Nature Center in Barrington, IL, for permission to sample specific locations within the nature preserve. We thank the following institutions for the necessary permits and permissions for our field surveys and snail collection: Wisconsin Department of Natural Resources (SRL-SOD-006-2014), Forest Preserves of Cook County, and Illinois Department of Natural Resources (Scientific Research Permit NH14.5793). The manuscript was significantly improved by comments from two anonymous reviewers and the Subject Editor.
Open Research
DATA AVAILABILITY STATEMENT
Unique COI haplotype data are deposited in GenBank (accession nos. OM102965–OM102970). Individual genotype data are available on Dryad (doi: 10.5061/dryad.fbg79cnzt).




