Long-wavelength-sensitive (lws) opsin gene expression, foraging and visual communication in coral reef fishes
Abstract
Coral reef fishes are diverse in ecology and behaviour and show remarkable colour variability. Investigating the visual pigment gene (opsin) expression in these fishes makes it possible to associate their visual genotype and phenotype (spectral sensitivities) to visual tasks, such as feeding strategy or conspecific detection. By studying all major damselfish clades (Pomacentridae) and representatives from five other coral reef fish families, we show that the long-wavelength-sensitive (lws) opsin is highly expressed in algivorous and less or not expressed in zooplanktivorous species. Lws is also upregulated in species with orange/red colours (reflectance >520 nm) and expression is highest in orange/red-coloured algivores. Visual models from the perspective of a typical damselfish indicate that sensitivity to longer wavelengths does enhance the ability to detect the red to far-red component of algae and orange/red-coloured conspecifics, possibly enabling social signalling. Character state reconstructions indicate that in the early evolutionary history of damselfishes, there was no lws expression and no orange/red coloration. Omnivory was most often the dominant state. Although herbivory was sometimes dominant, zooplanktivory was never dominant. Sensitivity to long wavelength (increased lws expression) only emerged in association with algivory but never with zooplanktivory. Higher lws expression is also exploited by social signalling in orange/red, which emerged after the transition to algivory. Although the relative timing of traits may deviate by different reconstructions and alternative explanations are possible, our results are consistent with sensory bias whereby social signals evolve as a correlated response to natural selection on sensory system properties in other contexts.
1 INTRODUCTION
A central objective in biology is to understand changes in biological diversity through time and lineages, especially the processes of speciation and the emergence of species-rich clades. Ecological niches are multidimensional and ecological diversification may be positively correlated with niche dimensionality (Nosil & Sandoval, 2008). In this study we focus on the extremely species-rich group of tropical coral reef fishes that display extraordinarily high diversity in ecology, behaviour and (colour) phenotype (Randall et al., 1997). Reef fishes, especially those inhabiting shallow water coral reefs in the tropics, live in a light-flooded and spectrally diverse environment (Cox et al., 2021). Many are themselves, often conspicuously, colourful (Lorenz, 1962; Marshall, 2000a), and importantly, have evolved diverse visual sensitivities and tuning mechanisms (Carleton et al., 2020; Cortesi et al., 2020). The study of colourful coral reef fish living in a multidimensional adaptive landscape allows us to gain valuable insights into the evolutionary interactions between vision, visual signalling traits, behaviour and ecology (Cortesi et al., 2020).
In teleosts, a multitude of tuning mechanisms, including opsin gene evolution via duplications and deletions, sequence variability and (co)expression, is used to presumably optimize vision (i.e., spectral sensitivities) for the prevailing light environment and/or visual tasks (reviewed in Carleton et al., 2020). In the photoreceptor cells of the retina, opsins, together with a vitamin A-derived chromophore, form the functional unit of visual pigments that absorb light and constitute the first step in visual processing (Wald, 1968; Yokoyama, 2008). Vertebrate opsins can be classified based on their genealogy, photoreceptor specificity, and the spectral sensitivity they confer. Rod photoreceptors express a single rod opsin type (rhodopsin, rh1) used for scotopic vision. Cone photoreceptors, on the other hand, express four basic types of cone opsins, which mediate photopic (colour) vision: two short-wavelength (UV-blue)-sensitive genes (sws1 and sws2), a mid-wavelength (green)-sensitive gene (rh2), and a long-wavelength (red)-sensitive gene (lws) (Yokoyama, 2008).
Coral reef fishes of the superorder Acanthopterygii have evolved a number of different visual tuning mechanisms and a set of spectral sensitivities peaking anywhere between the ultraviolet (UV) and the red spectrum of light (350–600 nm) (Carleton et al., 2020; Cortesi et al., 2020; Losey et al., 2003; Luehrmann et al., 2019; Marshall et al., 2019; Phillips et al., 2016; Siebeck & Marshall, 2001; Stieb et al., 2017). While medium-wavelength-sensitivity (i.e., from blue to green) is well matched to the most prevalent light on coral reefs (Losey et al., 2003), spectral sensitivities at either end of the spectrum (i.e., UV and red) seem more likely to represent adaptations to one or more specific visual tasks. For example, damselfishes (Pomacentridae) use their UV vision to detect UV patterns of con- or heterospecifics (Siebeck et al., 2010). Herbivorous algae-feeding damselfishes also display enhanced red sensitivity (Stieb et al., 2017) as it increases algal contrast (due to the red and far-red-reflecting part of chlorophyll) when seen against the reef background (Marshall et al., 2003b). Long-wavelength-sensitivity may furthermore play a role in inter- or intraspecific communication in species that use red colour signals, such as many cichlids (Seehausen et al., 2008), guppies (Sandkam et al., 2018), medaka (Kamijo et al., 2018) or indeed other reef fishes (Marshall et al., 2003b). Once evolved, sensitivity to a specific spectral range is likely to be exploited by other visual tasks, as has been shown for guppies (Endler, 1983; Grether et al., 2005; Kodric-Brown, 1989; Rodd et al., 2002) or Old World monkeys (Fernandez & Morris, 2017) that feed on reddish fruit and also use red for social signals.
In this paper, we focused on the emergence of long-wavelength-(red)-sensitivity and its function in coral reef fishes by extending our previous work on opsin gene evolution. We mainly focused on damselfishes. This speciose family of small to medium-sized reef fishes mirrors the high behavioural, ecological and colour diversity found among coral reef fishes more broadly (Allen, 1991). Importantly for this work, damselfishes are also one of the most studied reef fish families in terms of opsin evolution. Of the 21 species analysed thus far, only benthic herbivorous but not zooplanktivorous damselfishes expressed increased levels of lws producing red-sensitive visual pigments (Stieb et al., 2017). We now aimed to reveal whether a sensory bias towards red sensitivity in algal feeding damselfishes is also exploited in yellow-red colour signalling. Our predictions were that: (1) benthic herbivory and red colours would both correlate with enhanced red sensitivity as shown by higher lws expression, (2) red sensitivity has a functional benefit in detecting algae and red colour signals, (3) specialization for algae feeding is followed by the evolution of red signalling colours and (4) following on from prediction 3, only algal feeding species have evolved red coloration. To test for a relationship of long-wavelength-sensitivity (lws expression) with feeding ecology (benthic herbivory), social signalling (long-wavelength-reflecting colours; yellow, orange, and red), or both, we combined our previous work with newly sequenced retinal transcriptomes to generate a more extensive damselfish data set. To go beyond the damselfish radiation, we also included pairs of benthic herbivorous and zooplanktivorous species from five other typical tropical reef fish families, including butterflyfishes (Chaetodontidae), angelfishes (Pomacanthidae), blennies (Blennidae), surgeonfishes (Acanthuridae), and labrids/wrasses (Labridae). To test for a functional benefit of seeing red, we next computed a damselfish visual system and modelled whether red sensitivity may improve the detection of benthic algae and the detection of yellow, orange, or red coloured conspecific signals. Lastly, by reconstructing ancestral character states, we assessed the sequence of emergence of lws expression, trophic groups and orange/red colour signals and their possible evolutionary interactions across the damselfish phylogeny.
2 MATERIALS AND METHODS
2.1 Specimen collection
Specimens were either collected from reefs surrounding Lizard Island (14° 40 S, 145° 27 E), Australia, using SCUBA and hand nets under the Great Barrier Reef Marine Park Permit (G12/35005.1) and the Queensland General Fisheries Permit (140763), or obtained from an aquarium supplier (Cairns Marine Pty Ltd, Cairns, Australia), collecting fishes from the Northern Great Barrier Reef. Fish used for molecular analysis were anaesthetized with an overdose of clove oil and killed by decapitation within 24 h after capture. Retinas were dissected out and preserved in RNAlater (Ambion) until further processing. Dissection took place during the daytime, at least 1 h after dawn and before dusk, respectively. The relative cone opsin expression has previously been shown not to be affected by time of day in several damselfish species (Stieb et al., 2016). Further, we normalized opsin expression by proportion of cone type, which was empirically determined to be the best method for removing time-of-day variation (Yourick et al., 2019). All experimental procedures were approved by The University of Queensland Animal Ethics Committee (QBI/223/10/ARC/US AIRFORCE [NF] and QBI/192/13/ARC).
2.2 Opsin gene studies
To investigate and quantitate opsin gene expression among damselfishes, we compiled a data set for 39 species [based on our previous results], n = 21 (Luehrmann et al., 2018; Stieb et al., 2016, 2017, 2019), and newly sequenced transcriptomes, n = 18, (Bioproject PRJNA747115: SAMN21876388-SAMN21876434; Table S1). We were further interested in comparing the relative opsin gene expression between herbivorous and zooplanktivorous species-pairs from various other reef fish families (n = 5 coral reef fish families). For this, we compiled a data set based on previous results (Phillips et al., 2016; Tettamanti et al., 2019) and generated new transcriptomes for the remaining species following our previously established protocols (Luehrmann et al., 2019; Musilova et al., 2019). To confirm the assignment of newly obtained opsin sequences to the correct opsin gene type/family, we used a fish-opsin reference data set to reconstruct maximum-likelihood amino acid trees using PHYML (100 bootstrap iterations) (Dereeper et al., 2008).
2.2.1 Transcriptomic sequencing and processing
Retinas were homogenized using a TissueLyser LT (Qiagen) and total RNA was extracted with the RNeasy Mini Kit (Qiagen) including an optional DNAse digestion step. RNA was quality checked with an Agilent 2100 BioAnalyser 6000 NanoChip (Agilent Technologies). RNAseq libraries were made using the TruSeq RNA Sample Preparation Kit version 2 (Illumina), and transcriptomes were sequenced as 125 bp paired reads on the Illumina platform (HiSeq2000 version 4). Samples were multiplexed at 12 samples per lane, obtaining 4–51 million sequenced reads per sample.
Transcriptomes were processed following previously published methods (Cortesi, Musilová, et al., 2015; de Busserolles et al., 2017) using the online Bioinformatics platform Galaxy version 1.0.4 (Research Computing Centre, The University of Queensland, Australia) (Afgan et al., 2015). In short, data were converted using FASTQ Groomer, quality checked using FastQC, and trimmed using customized settings in Trimmomatic. Trinity was used for de novo assembly of transcripts, with a group pair distance of 250 bp, and minimum inchworm kmer coverage of 2. Further bioinformatic analyses were performed using Geneious software (version 9.0.4). Assembled transcripts were then mapped to known and publicly available opsin genes of reference species (see Figure S1). To manually check for gene duplications, we followed previously described methods (Cortesi, Musilová, et al., 2015; de Busserolles et al., 2017). Briefly, after identification of candidate gene coding sequences, unassembled reads were mapped to the opsin gene repertoire of the species using medium-sensitivity settings (70% identity threshold). Deviating reads were then extracted by working from single polynucleotide polymorphism (SNP) to SNP by exploiting paired-end matching to cover gaps, and their consensus sequence was used as species-specific reference for repeated high-specificity (100% identity) mapping of unassembled reads until maximum obtainable sequence length was reached. To analyse differences in relative cone opsin gene expression, we mapped the unassembled filtered PE reads against the CDSs of genes extracted from the transcriptomes (as per Cortesi, Musilová, et al., 2015 and de Busserolles et al., 2017).
2.2.2 Relative opsin gene expression given as proportional single and double cone expression
Because cone opsin expression is given as a fraction of the total single (sws1s and sws2b) and total double cone expression (rh2s and lws), respectively, we reanalysed expression data gained from Stieb et al. (2016, 2017) for damselfish and from Phillips et al. (2016) for labrids to calculate the proportional expression of single and double cone opsin genes, respectively.
2.3 Spectral reflectance
We compiled spectral reflectance data on live specimens (as per Marshall et al., 2003b) for 35 species (newly generated, n = 10; from the literature, n = 25 [Cheney & Marshall, 2009; Marshall, 2000b; Siebeck, 2002; Stieb et al., 2017]), following the colour categorization in Marshall (2000b).
The spectral reflectance of different areas of the fish was measured at a 45° angle using a 200 nm bifurcated UV/visible optic fibre connected to a PX-2 pulse xenon light source (Ocean Optics) and an Ocean Optics (Dunedin) USB2000 spectrophotometer attached to a laptop computer running OOIBASE32 (Ocean Optics). A Spectralon 99% white reflectance standard was used to calibrate the percentage of light reflected at each wavelength from 300 to 800 nm. Spectral reflectance was measured for two to three individuals per species by measuring distinct colour patches (from a human point of view) as well as common areas that may reflect in UV (Marshall, 2000a), such as the surroundings of the eyes and mouth, the operculum, fins and the caudal peduncle. At least 10 measurements per area and individual were taken and subsequently averaged.
2.4 Relationship of proportional opsin gene expression with diet and coloration
To identify possible evolutionary correlations between damselfish cone opsin expression and either trophic groups or long wavelength (yellow, orange and red) coloration, we computed phylogenetic generalized least squares regressions (PGLS) using the caper package (Orme et al., 2013) in R (R Core Team, 2011). The PGLS regression estimates a maximum likelihood (ML) value of the phylogenetic scaling factor lambda (Pagel's λ), with λ = 1 indicating complete phylogenetic dependence and λ = 0 indicating no phylogenetic effect. To compare opsin expression to feeding ecology, we placed species into three different trophic groups: herbivores, zooplanktivores, or omnivores (Table S2), with omnivores known to forage on both zooplankton and algae. To test for relationships of opsin expression to different patterns of fish coloration, we grouped species first based on having yellow coloration (reflectance starting beyond 500 nm) and second, based on having orange/red (reflectance starting beyond 520 nm) coloration (including coloration of fins or bodies, or patches thereof). We first determined the historical evolutionary dependence of opsin gene expression on trophic groups, yellow, and orange/red coloration independently. If more than one of these variables had a significant effect on expression, they were subsequently tested together. Finally, we were interested in how long-wavelength sensitivity is affected when orange/red coloration is attributed to a trophic group. For this, we tested proportional double cone lws expression in species showing no orange/red coloration versus species showing orange/red coloration within the different trophic groups. This test was only possible for omnivores and herbivores as none of the zooplanktivores reflected in orange/red.
To further explore the relationship between proportional opsin gene expression and feeding strategy, we compared herbivorous and zooplanktivorous species pairs from various other reef fish families (herbivore vs. zooplanktivore): butterflyfishes (Chaetodon ulietensis vs. Hemitaurichthys polyepis), angelfishes (Centropyge bicolor vs. Genicanthus watanabei), blennies (Escenius bicolor vs. Meiacanthus atrodorsalis), surgeonfishes (Acanthurus blochii vs. Naso brevirostris), and labrids (Chlorurus sordidus vs. Bodianus mesothorax). Those species were chosen based both on their occurrence at the sampling location, reefs surrounding Lizard Island, and on their trophic group (herbivore or zooplanktivore) without respect to coloration.
2.5 Visual modelling
We performed visual modelling as we were interested in whether an additional long-wavelength-sensitive visual pigment (LWS) might enhance the fish's capability to detect benthic algae and/or conspecifics. For this, modelling was performed using a typical UV-transmitting damselfish lens (we used the lens of Dacyllus aruanus [Stieb et al., 2017]). We constructed a trichromatic damselfish visual system with known peak spectral sensitivities (λmax) (gained from microspectrophotometry measurements in Pomacentrus amboinensis [Siebeck et al., 2010]) of 370 nm (SWS1), 480 (RH2B), and 523 (RH2A) (for matching visual pigments and opsin genes, see Stieb et al. [2016]), and then added the fourth visual pigment (LWS) with a range of 525–565 nm λmax (5 nm increments) (a λmax of 560 nm was measured in Pomacentrus melanochir [Loew & Lythgoe, 1978]). While the different members of double cones can express two distinct visual pigments (as for example LWS and RH2B), one member of double cones can also coexpress two different opsin pigments (as for example LWS and RH2A), resulting in intermediate sensitivities as shown for the African cichlid fish, Metriaclima zebra (Dalton et al., 2015). These different scenarios are represented in our models by adding the fourth visual pigment (LWS) with a range of 525–565 nm λmax. Evidence that individual members of double cones are used in colour vision as independent spectral channels comes from behavioural studies in the reef fish Rhinecanthus aculeatus (Pignatelli et al., 2010).
We first calculated the quantum catch of different photoreceptors as they viewed the light reflected off different targets. For visual differentiation, we used the receptor noise limited model to quantify the relative differentiation of two colour targets (e.g., Vorobyev et al., 2001) providing the results in terms of just noticeable differences (JNDs), that is, the threshold at which two objects should be distinguishable from one another under bright illumination. Algae detection was calculated as the ability to distinguish benthic algae against different backgrounds (sand, rock, coral). Conspecific detection included comparing colours of selected damselfish species (Chromis viridis was chosen to represent a species with no yellow and no orange/red reflectance; Pomacentrus amboinensis and Pomacentrus coelestis were chosen to represent species with yellow reflectance but no orange/red; and Pomacentrus moluccensis, Chrysiptera cyanea, and Amphiprion biaculeatus represented species with orange/red reflectance) against each other or against the ambient illuminant, and also against the host anemone in the case of the anemonefish Amphiprion biaculeatus.
2.5.1 Quantum catch equations
Reflectance spectra from different targets used for quantum catch calculations were gathered as follows: fishes or the anemone were illuminated by sidewelling irradiance and algae and substrates by downwelling irradiance, both obtained from previous light measurements around Lizard Island (Stieb et al., 2016); reflectance data from the host anemone (Entacmaea quadricolor) of the anemonefish Amphiprion biaculeatus was measured anew, reflectance data from algae, the average reef, and rubble background was taken from Marshall et al. (2003b), average sand background was taken from Cortesi and Cheney (2010) and damselfish spectral reflectance was measured anew or taken from Marshall (2000a) and Stieb et al. (2017); for ambient illuminant, as a target, we used horizontal radiance obtained from previous measurements in reefs around Lizard Island (Stieb et al., 2016).
Here, we included this normalization for consistency with previous studies and for plotting the quantum catches in a trichromatic visual space. However, this correction did not impact the final differentiation calculations for either colour or luminance, as comparisons between targets involved ratios of quantum catches, where the von Kries factor ki cancels out.
2.5.2 Visual differentiation equations
As per Stieb et al. (2019), damselfishes have one single cone for each pair of double cones so that nS:nM:nL = 1:2:2. We further set the Weber fraction νI, to 0.1, based on colour experiments in other fishes (Champ et al., 2016; Cheney et al., 2019; Escobar-Camacho et al., 2017).
2.6 Ancestral character state reconstruction
To reconstruct ancestral states for proportional double cone lws expression, trophic groups and orange/red coloration, we used a damselfish phylogeny modified from The Fish Tree of Life (Rabosky et al., 2018). For this, we concatenated sequence data and constructed maximum-likelihood trees (100 bootstrap iterations) using PHYML (Guindon & Gascuel, 2003) in Geneious version 9.0.5. Nile tilapia (Oreochromis niloticus) and the black surfperch (Embiotoca jacksoni) were used as outgroups to root the tree. As no genetic markers were available for Parma unifasciata, we used markers for Parma oligolepis as a surrogate to place Parma unifasciata in the phylogeny.
Ancestral reconstruction analyses of lws expression, trophic groups and orange/red coloration were performed using the package corHMM (Beaulieu et al., 2021) in R (R Core Team, 2021). corHMM can model ancestral reconstructions that assume a correlation between two or more traits in a way that the traits coevolve with each other. This allowed us to model the transition of one trait being dependent on the transitions of the other traits. First, we reconstructed ancestral states for each trait independently. For trophic groups, we coded extant species in three possible states (herbivores, omnivores or zooplanktivores). For orange/red coloration, we used binary states (absence or presence). Expression level of lws was originally obtained as a continuous trait (proportional double cone expression in %). Since corHMM can only handle discrete data, lws expression was coded into four different categories: 0%–0.9%, 1%–9.9%, 10%–19.9% and 20%–31%. The last three categories were chosen in 10% interval; as 31% (highest observed value) was the sole value above 30%, it was included in the range 20%–31%. The first category below 1% expression represents species that have either no or a minor lws expression. Then, we performed the ancestral state reconstructions of these traits assuming their evolution to be dependent on each other. We also allowed all possible combinations between traits. For each ancestral reconstruction, we ran three different analyses, each based on a different transition rate configuration: (1) allowing different transition rates among states (all rates differ [ARD]), (2) constraining forward and reverse transitions to have the same rates (SYMmetrical rates [SYM]), and (3) constraining all transitions to have the same rate (equal transition rates [ER]). For orange/red coloration, SYM and ER become identical models because this trait is binary. To find which of these transition rate models is the best fit for our data, we used the corrected Akaike information criterion (AICc). Since some of the tips of our internal groups had missing information, we also set corHMM to reconstruct their most likely state.
We also used BayesTraits version 3.0.1 (Pagel & Meade, 2006) to reconstruct lws expression in its original form, that is, as a continuous trait. We followed the advice in the BayesTrait's manual, and we first ran a random walk maximum likelihood (ML) analysis of lws expression on the damselfish tree to obtain a general estimate of the most likely gene expression level at the root of the tree (parameter α), which resulted in a value of c. 1.5%. Then we ran a new analysis based on a random walk Markov chain Monte Carlo (MCMC) approach in order to estimate ancestral states of all nodes and tips with missing data. Given that our preliminary ML analysis pointed to a low lws expression at the root of the tree, we set our MCMC with a strong prior (exponential prior with a mean of 1%). The observation that long-wavelength sensitivity (assuming lws expression) is rarely found among coral reef fishes (Losey et al., 2003; Luehrmann et al., 2019; Phillips et al., 2016; Stieb et al., 2017), and seemingly is associated with specific behaviours, supports the usage of a strong prior on low lws expression at the root of the damselfish tree. For comparative purposes, we also set BayesTraits to reconstruct lws expression assuming an uninformative prior (uniform distribution, varying from 0 to 31%). For both settings, we ran three independent analyses with 100,000,000 generations each, with estimated parameters sampled every 10,000th generation. Mixing and convergence of the runs were confirmed.
In all ancestral state reconstructions aforementioned, we assigned the outgroups (Nile tilapia and the black surfperch) with no data on lws expression, coloration or trophic group to not impact ancestral trait states (both outgroup taxa belong to clades with considerable variation in all of our traits).
3 RESULTS
Opsin expression profiles of damselfish species are listed in Table S2, and opsin expression profiles of the other reef fish families (labrids, butterflyfishes, angelfishes, blennies, and surgeonfishes) are listed in Table 1 and Table S3. Protein-based maximum likelihood trees revealed that the newly extracted reef fish opsins (Genbank # OK350470–OK350614; Table S4) grouped with well-described opsin classes from other fish species (Figure S1a,b). Colour categories, including the presence or absence of yellow and orange/red, for damselfish and species from the other reef fish families are summarized in Table S5. The presence of orange/red is found in 10 damselfishes (Table S2) and five of the other reef fish species (Table 1). Reflectance data from various damselfish species with orange/red coloration are shown in Figure 1b(ii) and Figure S2a(ii). Reflectance data from various damselfish species with yellow or other colours are shown in Figure S2a(iii and iv). Spectral reflectance of the remaining species measured in this study are presented in Figure S3. Trophic groups of damselfish species are listed in Table S2; trophic groups of the other reef fish families are listed in Table 1.
| n | Proportional single cone expression | Proportional double cone expression | Overall expression | Trophic group | Yellow (>500 nm) | Orange/red (>520 nm) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sws1 | sws2b | sws2aα | sws2aβ | rh2b | rh2a | lws | Cone | Rod | |||||
| Chaetodontidae—Butterflyfishes | |||||||||||||
| Chaetodon ulietensis | 3 | — | — | 24.4 ± 4.1 | 75.6 ± 4.1 | 4.8 ± 2.7 | 79.8 ± 11.0 | 15.5 ± 11.9 | 15.9 ± 7.3 | 84.1 ± 7.3 | H, (BI)3, 4 | Yes13 | Yes13 |
| Hemitaurichthys polylepis | 3 | — | 86.9 ± 1.5 | 13.1 ± 1.5 | — | 56.9 ± 5.6 | 43.1 ± 5.6 | — | 14.5 ± 4.6 | 85.6 ± 4.6 | P3, 4, 5 | Yes | No |
| Pomacanthidae—Angelfishes | |||||||||||||
| Centropyge bicolor | 3 | — | — | — | 100 | 1.4 ± 1.4 | 94.4 ± 3.5 | 4.1 ± 2.0 | 15.3 ± 7.4 | 84.7 ± 7.4 | H3, 6, BI 4 | Yes13 | Yes13 |
| Genicanthus watanabei | 2 | — | — | 34.3 ± 20.5 | 65.7 ± 20.5 | 52.8 ± 4.1 | 45.9 ± 4.6 | — | 30.8 ± 4.8 | 69.2 ± 4.8 | P3, 6 | Yes | No |
| Blennidae—Blennies | |||||||||||||
| Ecsenius bicolor | 3 | (<0.01) | — | 16.4 ± 7.0 | 83.5 ± 7.0 | 0.6 ± 0.1 | 57.5 ± 10.2 | 41.9 ± 10.2 | 37.1 ± 22.4 | 62.9 ± 22.4 | H: D7, 8 | No14 | Yes14 |
| Meiacanthus atrodorsalis | 3 | 0.9 ± 1.4 | — | 98.3 ± 1.2 | 0.8 ± 1.1 | 60.2 ± 10.5 | 22.4 ± 3.7 | 17.4 ± 7.0 | 53.3 ± 15.6 | 46.7 ± 15.6 | P, (BI)3, 4 | Yes14 | No14 |
| Acanthuridae—Surgeonfishes | |||||||||||||
| Acanthurus blochii | 4 | — | — | 100 | — | 2 ± 0.5 | 56.9 ± 1.3 | 41.1 ± 1.3 | 16.1 ± 4.5 | 84 ± 4.5 | H3, 5, 7 | Yes | Yes |
| Naso brevirostris | 31 | — | 100 | — | — | 56.1 ± 1.9 | 38.1 ± 1.6 | 5.8 ± 0.4 | 17.3 ± 1.8 | 82.7 ± 1.8 | P3, 9 | Yes | No |
| Labridae—Labrids/Wrasses | |||||||||||||
| Chlorurus sordidus | 12 | — | 100 | — | — | — | 53.4 | 46.6 | 22.4 | 77.6 | H3, 10 H: D10 | Yes | Yes |
| Bodianus mesothorax | 12 | 7.6 | 92.4 | — | — | 41.3 | 43.4 | 15.3 | 42.7 | 57.3 | P11, G12 | N/A | N/A |
-
In accordance with previous studies (Luehrmann et al., 2018; Stieb et al., 2016, 2017, 2019), our transcriptomic analysis revealed that all damselfish species expressed sws1, rh2a, and rh2b, with only a subset of species additionally expressing sws2b and/or lws. Here, gene expression of longer wavelength (as indicated by increased lws expression) opsins is associated both with feeding ecology and coloration (Table S6 summarizes PGLS results):
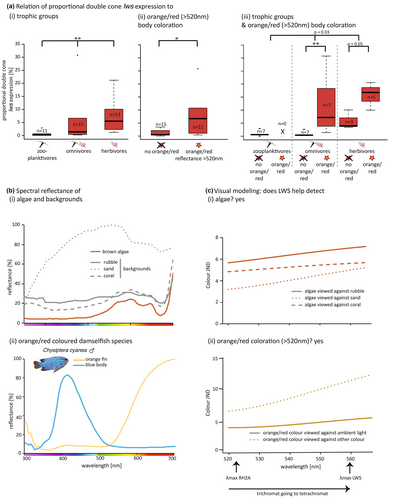
- Proportional double cone expression of lws (simplified as lws expression from now on) is associated with benthic herbivory. Among damselfishes, expression of lws was significantly associated with feeding ecology after correction for multiple testing, though it had a strong phylogenetic dependence (λ = 0.795, F1,35 = 11.43, p = .001787). Zooplanktivores showed little to none, omnivores some, and herbivores substantial levels of lws expression (Figure 1a[i], Figure S4a).
- Proportional double cone expression of rh2a (simplified as rh2a expression from now on) and lws covary with orange/red coloration. We further found a significant positive correlation between the expression of lws and orange/red, but not yellow coloration (orange/red: λ = 0, F1,23 = 7.456, p = .01192; yellow: λ = 0, F1,23 = 1.13, p = .2987) and a very strong negative correlation between the rh2a expression and orange/red, but not yellow coloration (orange/red: λ = 0, F1,23 = 15.12, p = .0007411; yellow: λ = 0, F1,23 = 0.5968, p = .4477) (Figure 1a[ii], Figure S4b).
- Lws opsin gene expression is highest in orange/red omni- and herbivores. When food and orange/red coloration were tested together, only a trend (λ = 0, F2,22 = 3.836, p = .03297) for a correlation between orange/red coloration and lws expression was observed (Table S6; Figure 1a[iii]). When species with and without orange/red coloration were compared within their respective trophic groups, orange/red omnivores showed a significant lws upregulation relative to other omnivores (λ = 0, F1,10 = 16.36, p = .002346) (Figure 1a[iii]), while a trend towards higher expression was also noticeable for orange/red herbivores (λ = 0, F1,4 = 8.083, p = .04672) (Figure 1a[iii]).
While most species within pairs from the other reef fish families (labrids, butterflyfishes, angelfishes, blennies, and surgeonfishes) expressed a core set of the same three opsin genes, they did differ in the expression of additional genes resulting in one species having a short- and the other species having a long-shifted expression profile (Table 1 and Table S3). Here, short- versus long-wavelength shifted opsin combinations covary with feeding ecology and fish coloration as they do for damselfishes. Across families, benthic herbivores had long-wavelength-shifted visual systems with pronounced lws expression: in butterfly- and angelfishes, only herbivores expressed lws; in blennies, surgeonfishes, and labrids, the herbivores expressed higher levels of lws compared to the zooplanktivores. In contrast, for all reef fish families, zooplanktivores expressed a shorter shifted single cone opsin combination compared to herbivores. Importantly, for most within-family species contrasts, the herbivorous species showed an orange/red coloration, while the zooplanktivorous species had no orange/red coloration.
- By computing a damselfish visual system, our visual models (results listed in Table S7) show that adding a LWS pigment enhances the differentiation of algae and orange/red coloration. Most algae colours against different backgrounds (for algae and background reflectance, see Figure 1b[i] and Figure S2a[i]) increased in contrast (in terms of JND) when going from potentially tri- to tetrachromatic vision (Figure 1c[i] and Figure S2b[i]). When comparing fish colours (for orange/red-coloured damselfish species, see Figure 1b[ii] and Figure S2a[ii]; for non-orange/red species, see Figure S2a[iii and iv]), adding the fourth spectral channel mostly improved the contrast of red (Figure 1c[ii] and Figure S2b[ii]), but not of yellow or other colours (Figure S2b[iii and iv]), against the ambient illuminant and against other fish colours. In the case of the red anemonefish, Amphiprion biaculeatus, it also increased its contrast against the host anemone (Figure S2b[ii]; for anemone reflectance, see Figure S2a[i]).
-
Reconstructions of character states across the damselfish phylogeny using the package corHMM revealed ancestral states and possible evolutionary relationships among lws expression, trophic ecology and orange/red colour signalling (Figure 2, Figures S5–S7). AICc results for competitive models based on distinct transition rate arrangements (ER—all rates are equal, ARD—all rates differ, and SYM –rates for forward and reverse transitions are same) are given in Table S8. ER was the best fit for our traits in all ancestral reconstructions, except for the reconstruction with trait-dependence between trophic ecology and lws expression, which was better described by ARD (Figure 2 and Figure S5). Reconstructions with less support are shown in Figures S6 and S7.
Ancestral reconstructions of each individual trait (lws expression, trophic groups, and orange/red colour) are shown in Figure S5 and display similar patterns as the trait-dependent analyses. Ancestral state reconstruction of proportional double cone lws expression as a continuous trait (using BaysesTrait) did differ depending on the prior used (Figure S8). When assuming an uninformative prior (uniform distribution with lower and upper bounds of 0% and 31%, respectively), the deepest nodes suggest very high upregulation of lws expression, which tends to decrease towards the tips (Figure S8a). A more constrained prior (exponential distribution with mean equals to 1), produced more similar results to the corHMM reconstructions, with most ancestral nodes showing very low levels of lws expression (Figure S8b).
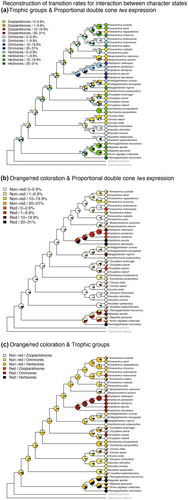
When looking at the evolutionary interactions between trophic groups and lws expression, basal nodes showed the lowest lws expression (0%–0.9%), and this low lws expression was either combined with omnivory or herbivory (Figure 2a). Among basal nodes, omnivory was often, herbivory sometimes and zooplanktivory never the dominant state. As omni- and herbivory both include algal feeding, algal feeding with highest probability precedes the emergence or increase of lws expression. Importantly, when reconstructions of internal branches become less ambiguous, we note that zooplanktivory always retained zero to the lowest lws expression levels (e.g., zooplanktivorous species of the genus Chromis or the genus Neopomacentrus). Herbivory, on the other hand, tends to correlate with higher lws expression as seen, for example, in the herbivorous Stegastinae, and in several species within Pomacentrinae like Dischistodus prosopotaenia and Dischistodus perpicillatus as well as Pomacentrus wardi, Pomacentrus australis, Pomacentrus adelus and Pomacentrus chrysurus, Chrysiptera brownrigii and Neoglyphididon nigroris. However, internal branches being dominated by omnivory can either retain a low lws expression (e.g., within the genus Dascyllus) or be associated with an increased lws expression (e.g., within Amphiprionini).
Important for the association of orange/red colour signals with lws expression is the fact that basal nodes and most internal branches have no orange/red colour signals and no or very low lws expression (Figure 2b). Transitions to orange/red coloration are accompanied with an increase in lws expression at the tips of several species within Pomacentrinae (e.g., in Pomacentrus moluccensis, Chrysiptera cyanea and Neoglyphidodon nigroris) and in basal nodes of Stegastinae and Amphiprionini that include the species with the highest lws expression among all damselfishes. Prominently, orange/red colour signalling is always associated with a rise in lws expression, but a rise in lws expression is also seen in algal feeding species that lack orange/red colour signalling (like the herbivorous Dischistodus prosopotaenia as well as Pomacentrus wardi, Pomacentrus australis, and Pomacentrus chrysurus). However, no clear pattern can be observed as to whether increased lws expression precedes or follows the emergence of orang/red colour signals.
Finally, for the interaction of trophic groups and orange/red colour signals, it is most notable that orange/red colour signals are missing at basal nodes and evolve exclusively on branches reconstructed as subtending algal feeding clades (like the herbivorous Stegastinae and omnivorous Amphiprionini) or first emerge on the tips of the tree, that is., in present-day algal feeding species (Figure 2c). In contrast, omni- and herbivory evolved independently from and before orange/red coloration across the damselfish phylogeny. This is also true for the clades Amphiprionini and Stegastinae in which orange/red colour signals occur early in internal branches but an omnivorous or herbivorous state, respectively, lacking orange/red signals still precedes the origin of the colour signals.
4 DISCUSSION
The mechanisms for shifting spectral sensitivities to long wavelengths in fishes are diverse. Long-wavelength shifts can be achieved by changes in LWS sequence structure (Carleton et al., 2005), a chromophore shift (A1–A2), or may include yellow/orange carotenoid-based optical filters (de Busserolles et al., 2015; Douglas et al., 1998; Kondrashev, 2008; Saarinen et al., 2012; Siebeck et al., 2003; Terai et al., 2017). This study highlights that in major reef fish families, long-wavelength shifts in visual sensitivity have been achieved by turning on or increasing proportional double cone lws opsin gene expression (simplified as lws expression from now on). An increase in LWS opsin protein implies that more photoreceptors across the retina increase their photon catch at long (red) wavelengths, making them overall more sensitive to red. While visual models (this study) provide theoretical support that adding LWS increases the fish's ability to detect red signals, only behavioural experiments can directly provide evidence for colour vision and response to red signals.
Here we show that in damselfishes and likely in several other reef fish families, lws gene expression and by virtue long-wavelength-(red)-sensitivity is related to benthic herbivory and orange/red coloration. Moreover, orange/red colour signals only evolved in association with algal feeding. For a summarizing figure with all tested coral reef fish species and traits, see Figure 3.
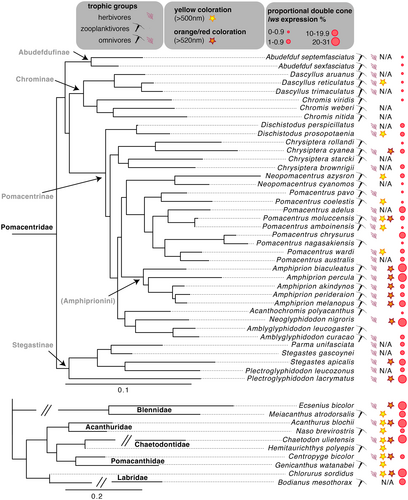
4.1 Opsin gene expression tuned to feeding ecology: Lws associated with benthic herbivory
Studying 39 damselfish species with representatives from four of the five subfamilies, we found that lws expression was highest in herbivores feeding on benthic algae followed by omnivores. In contrast, almost no expression was found in zooplanktivores (Figure 1a[i]). Corresponding patterns were also found across phylogenetically diverse reef fish families (Figure 3). Moreover, across these families, either parts or the entire visual palette/repertoire was shifted towards longer wavelengths in benthic herbivorous species. Examples are a shift from sws1 (347–383 nm) to sws2 (397–482 nm), sws2b (395–425 nm) to sws2a (439–475 nm), or sws2aα (448 nm) to sws2aβ (457 nm) in single cones, and/or a shift from rh2b (472–484 nm) to rh2a (518–528 nm) or rh2 (452–537 nm) to lws (501–573 nm) in double cones (Table 1 and S3) (λmax gained from Bowmaker, 2008; Cortesi, Musilová, et al., 2015; Hofmann & Carleton, 2009; Musilova et al., 2021; Spady et al., 2006; Yokoyama, 2008).
Sensitivity to either end of the visible spectrum of the light has previously been associated with foraging in various vertebrates. For example, UV-sensitivity in fishes is generally thought to enhance the efficiency of predating on UV-absorbing or scattering zooplankton (Browman et al., 1994; Loew et al., 1993; Rick et al., 2012). Although not all zooplanktivorous reef fishes expressed sws1, they always expressed shorter-shifted single cone opsins (i.e., sws2a instead of sws2b or sws2βa instead of sws2aα) compared to herbivorous species (Table 1 and S3).
In terrestrial forest species, long-wavelength-sensitivity improves distinguishing between items of brown forest litter (Lythgoe & Partridge, 1989). Similarly, long-wavelength-sensitivity in primates helps increasing the contrast of yellow and orange fruit against green foliage (Osorio & Vorobyev, 1996; Regan et al., 1998). Among insects, a shift to longer wavelength sensitivity is associated with shifts in diet and body colour (Martínez-Harms et al., 2012; van der Kooi et al., 2021). In the marine environment, green and brown algae broadly reflect in the green-red range (~500–650 nm) with a secondary, chlorophyll-generated peak in the far-red (> 700 nm, Figure 1b[i] and Figure S2a[i]). Simulations show that to see algae against a typical coral reef background, a reef fish may get away with two photoreceptors sensitive at 510 and 580 nm λmax (Marshall et al., 2003b). Herbivorous damselfishes seem to approximate this optimum with a RH2A and LWS-based pigment pair that are sensitive to 520 and 560 nm λmax, respectively (Marshall et al., 2006; Stieb et al., 2016). Indeed, damselfish visual models, including those constructed here, also indicate that the addition of an LWS-pigment increases the detection of algae against diverse backgrounds (Figure 1c[i] and Figure S2b[i]). However, long-wavelength-sensitivity may not only facilitate algal detection but may be beneficial for benthic feeding more generally. For example, lws is highly expressed in several blennies that feed predominantly on detritus or benthic algae (Cortesi et al., 2019).
4.2 Opsin gene expression tuned to fish coloration: Lws associated with orange/red
In addition to feeding ecology, we found that orange/red coloration in damselfish was associated with a change in double cone gene expression; lws was increased while rh2a reduced in damselfish species (Figures 1a[ii] and Figure S2, Table S6). Similarly, for butterfly- and angelfishes, lws was only expressed in species with orange/red coloration and lws expression was enhanced in the orange-tailed bicolour blenny, Ecsenius bicolor, and the ringtail surgeonfish (it has a yellow/orange blotch behind the eye), Acanthurus blochii (Figure 3, Table 1).
The function of orange/red coloration for intraspecific communication and mate choice is known for some freshwater and marine fishes. In Lake Victoria cichlids, different light regimes are associated with divergent visual sensitivities of lws, ultimately contributing to speciation through sensory drive based on sexually selected (red) male breeding coloration (Maan & Sefc, 2013; Seehausen et al., 2008). Across guppy populations, lws coding and expression are associated with red coloration in males (Sandkam et al., 2015), and knocking out lws in medaka (Oryzias latipes) reduces grey-orange colour distinction (Kamijo et al., 2018). All labrids investigated so far express lws with some species expressing up to five copies of the gene (Table 1 and Cortesi et al., 2021; Musilova et al., 2019; Phillips et al., 2016). Since many labrids show complex patterns of coloration dominated by green, red, and far-red components (Marshall et al., 2003b) and some species display sexual dimorphism in (red) coloration (Hodge et al., 2020), long-wavelength-sensitivity in this group is likely to facilitate intraspecific communication at close-range (Marshall, 2000a; Michiels et al., 2008).
In the reef environment, orange/red can become highly conspicuous against water or coral backgrounds near the surface, at least for short-distance viewing, but may help to camouflage against the background or within a group of similarly coloured fishes over longer distances (Cortesi, Feeney, et al., 2015; Marshall, 2000b; Marshall et al., 2003a; Marshall et al., 2019). Our damselfish visual models show that long-wavelength-sensitivity (given by the expression of LWS) increases the colour contrast of orange/red colours when perceived against other fish colours, the ambient illuminant, or specific backgrounds (Figure 1c[ii] and Figure S2b[ii]), which may be advantageous for species recognition and/or mate choice. Colour patterning has been shown to often be a trustworthy signal for intraspecific communication (Marshall et al., 2006; Sibeaux et al., 2019). Amphiprion biaculatus and Chrysiptera cyanea both exhibit colour patterns visualized with increased contrast when adding long-wavelength sensitivity (Figure 1c[ii] and Figure S2b[ii]). The orange tail of male Chrysiptera cyanea has been related to sexual selection and mating success in this species (Wacker et al., 2016). Anemonefish have a striking appearance with white stripes and orange/red body colorations and show the highest lws expression among damselfishes (Table S2, Figure 3). Having a cone type containing a long-wavelength-sensitive pigment combined with a relatively short single cone photoreceptor (Stieb et al., 2019) seems to increase the colour contrast of the striped pattern and thus may be important for conspecific detection and recognition. But seen from a distance through the eyes of (perhaps relatively red-blind) predators, orange-to-red anemonefishes may blend in with their anemone, which is also often red or orange (Figure S2a[i]).
4.3 The evolution of red sensitivity, body coloration, and herbivory in reef fish
Supporting a scenario in which fish vision, coloration, and trophic ecology are co-evolving, we discovered repeated evolutionary shifts in spectral sensitivity to longer wavelengths (given by increased lws expression) that appeared to be adaptations to algal feeding. Further, we found that orange/red coloration in damselfish is only present in benthic algal feeding species (Figure 1a[iii], Figure 2c). Where ancestral nodes are reconstructed as lacking orange/red coloration, such coloration may then have evolved independently in several extant omnivorous or algivorous species, as well as in the ancestors of the herbivorous Stegastinae and omnivorous Amphiprioninae (Figure 2c). However, there are ancestral state reconstructions that are less clear than others (e.g., basal nodes in Figure 2a,c) or result in different scenarios of transition rates (Figure 2a,c compared to Figures S7a,c).
The sensory drive hypothesis (Endler, 1992; Ryan & Cummings, 2013) suggests that long-wavelength-sensitivity evolved initially in adaptation to feeding strategy, but is now also coevolving with red social signals. It has been suggested, for example, that in primates and guppies, a visual system tuned to find red coloured food is likely to have predated female preference for males displaying red coloration (Endler, 1983; Fernandez & Morris, 2017; Grether et al., 2005; Rodd et al., 2002). In damselfishes, long-wavelength-sensitivity is evolutionarily associated with orange/red coloration and algal feeding, but never with zooplanktivory. Also, long-wavelength-sensitivity can evolve in the absence of orange/red coloration but never in the absence of algal feeding, and benthic algal feeding species have the highest lws expression. Finally, orange/red coloration is exclusively emerging with and most likely after algal feeding. Hence, ancestral state reconstructions suggest that a sensory bias is at play with the following scenario: (1) feeding on algae (but not zooplankton) favours (2) the evolution of long-wavelength-sensitivity (higher lws expression) that (3) is now leading to the evolution of orange/red social signals, which in turn causes selection for (4) increasing lws expression even further.
5 CONCLUSIONS
Our results indicate that variation among coral reef fishes in sensitivity to light of longer wavelengths may have evolved as an adaptation to feeding ecology and that, once evolved, it subsequently facilitated the evolution of orange/red fish colour elements, possibly facilitating social signalling. Our study reveals how the evolutionary feedbacks between variation in ecology, here foraging strategy, and social signalling, mediated by variation in visual sensitivity, might help explain the astonishing colour diversity of reef fishes. We call for more behavioural studies to test the predictions that our hypothesis makes with regard to feeding performance and behavioural interactions.
AUTHOR CONTRIBUTIONS
Sara M. Stieb, Karen L. Carleton, Fabio Cortesi, Ole Seehausen and N. Justin Marshall designed the study. Sara M. Stieb, Fabio Cortesi and N. Justin Marshall caught the specimens, and took body reflectance measurements. Sara M. Stieb and Fabio Cortesi prepared retinal tissue for RNA sequencing and analysed the data. Ole Seehausen, Luiz Jardim de Queiroz and Sara M. Stieb performed ancestral state reconstructions. Karen L. Carleton and Sara M. Stieb performed visual models. All authors contributed to writing the manuscript and approved the final version.
ACKNOWLEDGEMENTS
We would like to thank the staff at the Lizard Island Research Station for support during fieldwork and acknowledge the Dingaal, Ngurrumungu and Thanhil peoples as traditional owners of the lands and waters of the Lizard Island region. We also thank Cairns Marine for supplying fish, and Janette Edson, Queensland Brain Institute Genomics Facility, for library preparation and sequencing. We are appreciative of the inputs from the Seehausen laboratory, the Fish Ecology and Evolution department meetings at EAWAG, and two anonymous reviewers.
FUNDING INFORMATION
This work was supported by the German Research Foundation (DFG) awarded to SMS, the Australian Research Council (ARC) Discovery Projects (DP150102710 and DP180102363), Sea World Research and Rescue Foundation to JM and FC, the AFOSR/AOARD to JM, UQ Development and ARC DECRA Fellowships (DE200100620) to FC, EAWAG's Academic Transition Grant (5221.00979.008.05 ATG21) to LJdQ, the National Institutes of Health (1R01EY024629) to KLC, and by EAWAG.
CONFLICT OF INTEREST
The authors declared no conflict of interest for this article.
Open Research
OPEN RESEARCH BADGES
This article has earned an Open Data badge for making publicly available the digitally-shareable data necessary to reproduce the reported results. The data is available at 10.5061/dryad.qv9s4mwgf.
DATA AVAILABILITY STATEMENT
Newly generated opsin gene sequences ([data set Stieb, 2021] OK350470–OK350614; see Table S6) and retinal transcriptomes ([data set Stieb & Cortesi, 2021] Bioproject PRJNA747115: SAMN21876388-SAMN21876434) have been deposited in GenBank. The R-code and all spectral measurements used for visual models, alignments and R-code used for ancestral state reconstructions and corresponding figures as well as alignments of newly sequenced opsin genes have been deposited to Dryad (doi: 10.5061/dryad.qv9s4mwgf).




