Stepping into the past to conserve the future: Archived skin swabs from extant and extirpated populations inform genetic management of an endangered amphibian
Abstract
Moving animals on a landscape through translocations and reintroductions is an important management tool used in the recovery of endangered species, particularly for the maintenance of population genetic diversity and structure. Management of imperiled amphibian species rely heavily on translocations and reintroductions, especially for species that have been brought to the brink of extinction by habitat loss, introduced species, and disease. One striking example of amphibian declines and associated management efforts is in California's Sequoia and Kings Canyon National Parks with the mountain yellow-legged frog species complex (Rana sierrae/muscosa). Mountain yellow-legged frogs have been extirpated from more than 93% of their historic range, and limited knowledge of their population genetics has made long-term conservation planning difficult. To address this, we used 598 archived skin swabs from both extant and extirpated populations across 48 lake basins to generate a robust Illumina-based nuclear amplicon data set. We found that samples grouped into three main genetic clusters, concordant with watershed boundaries. We also found evidence for historical gene flow across watershed boundaries with a north-to-south axis of migration. Finally, our results indicate that genetic diversity is not significantly different between populations with different disease histories. Our study offers specific management recommendations for imperiled mountain yellow-legged frogs and, more broadly, provides a population genetic framework for leveraging minimally invasive samples for the conservation of threatened species.
1 INTRODUCTION
Translocations and reintroductions are fundamental management actions used in the recovery of threatened and endangered species (Armstrong & Seddon, 2008; Germano & Bishop, 2009; Griffith, Scott, Carpenter, & Reed, 1989; Seddon, Armstrong, & Maloney, 2007). While translocations and reintroductions have been successful for some animal populations (Dodd & Seigel, 1991; Fischer & Lindenmayer, 2000; Seigel & Dodd, 2002), they also present major challenges, especially in certain taxonomic groups, such as amphibians (Dodd & Seigel, 1991; Germano & Bishop, 2009; Seigel & Dodd, 2002). Amphibians are one of the most imperiled lineages worldwide, with greater than 30% of known species currently threatened with extinction (Stuart et al., 2004). Translocations and reintroductions are an important tool in amphibian conservation given local extirpations in many species around the world (Griffiths & Pavajeau, 2008; Harding, Griffiths, & Pavajeau, 2016). However, these approaches to combat amphibian declines have had variable success (Garner et al., 2016; Kriger & Hero, 2009; Woodhams et al., 2011). Amphibian translocation and reintroduction programs can be hindered by many factors such as complex life histories (Germano & Bishop, 2009), limited dispersal paired with high site fidelity (Reinert, 1991), insufficient natural history information (Germano & Bishop, 2009; Harding et al., 2016), and continued presence of unmitigated threats at release sites (Griffiths & Pavajeau, 2008; Seigel & Dodd, 2002; Woodhams et al., 2011). Even in the face of these challenges, translocations and reintroductions may be the only conservation tool available to restore many amphibian populations.
An emblematic example of amphibian declines and associated recovery efforts is the mountain yellow-legged frog (MYLF) species complex. The mountain yellow-legged frog (Rana muscosa) was split into the Sierra Nevada yellow-legged frog (Rana sierrae) and southern mountain yellow-legged frog (Rana muscosa) based on genetic, morphologic, and acoustic data (Vredenburg et al., 2007). In the Sierra Nevada mountains of California, both species inhabit mid and high elevation lakes, ponds, and streams (Stebbins, 2003). Once the most abundant amphibian in the Sierra Nevada (Grinnell & Storer, 1924), MYLFs have disappeared from >93% of their historical ranges despite the majority of their habitat being on federally protected lands (Vredenburg et al., 2007). Currently, both R. sierrae and R. muscosa are state and federally listed as threatened or endangered species (California Fish and Game Commission, 2011; US Fish & Wildlife Service, 2014). Primary causes of these declines include the widespread introduction of non-native trout into previously fishless water bodies (Bradford, Tabatabai, & Graber, 1993; Knapp, 2005; Knapp, Boiano, & Vredenburg, 2007; Knapp & Matthews, 2000; Vredenburg, 2004) and the spread of the amphibian chytrid fungus (Batrachochytrium dendrobatidis, hereinafter “Bd”) (Vredenburg, Knapp, Tunstall, & Briggs, 2010). Bd is a recently emerged and highly virulent fungal pathogen that attacks amphibian skin, causes the disease chytridiomycosis, and can rapidly lead to mortality in susceptible species. Bd currently threatens hundreds of amphibians species worldwide (Lips, 2016; Skerratt et al., 2007), and MYLFs are particularly susceptible.
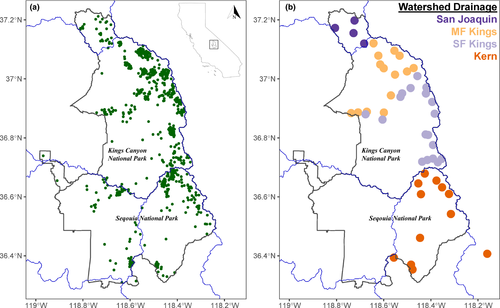
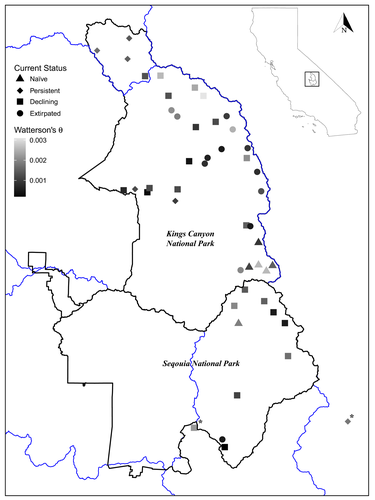
In response to the threat of MYLF extirpations in Sequoia and Kings Canyon National Parks (SEKI), populations in this jurisdiction are currently the focus of intensive conservation efforts. MYLFs historically occupied all major watersheds in SEKI but have declined precipitously over the past four decades (Bradford, 1991; Bradford, Graber, & Tabatabai, 1994; Rachowicz et al., 2006; Vredenburg et al., 2010), often due to the arrival of Bd. These Bd-caused declines have left over half of historically occupied lake basins empty of MYLFs (see all historical lakes once occupied by frogs in Figure 1a). However, some MYLF populations remain in SEKI, many of which are naïve to Bd and a few that are persisting or even recovering despite ongoing Bd infection. Persisting populations are important sources of frogs for restoring the species complex across its native range (Brown, Hayes, Green, & Macfarlane, 2014). Bd-naïve populations are probably highly susceptible to imminent infections and are therefore not currently used in translocations or reintroductions. With few conservation tools left for managers to pursue other than non-native trout eradication, MYLF conservation actions across SEKI have focused on using translocations and reintroductions to bolster extant populations or recover extirpated populations.
One of the main limitations in SEKI recovery and management efforts is designating effective conservation management units. Our current understanding of genetic variation in MYLFs is based on a 13-year-old study that used a single mitochondrial marker to describe genetic structure across the entire species range with 91 total individuals and limited sampling from SEKI (n = 39) (Vredenburg et al., 2007). This study identified a species-level split (between R. muscosa and R. sierrae) within SEKI park boundaries. The 2007 assessment has served as an important guide to MYLF conservation for over a decade, but a finer-scale study of spatial genetic variation in SEKI is urgently needed to better inform conservation efforts. Specifically, higher resolution genetic data can help with species delimitation, identifying management units, and aid in maintaining historical genetic structure in the face of ongoing threats.
To address the need for higher resolution genetic data, our study combines a minimally invasive sampling methodology and robust nuclear amplicon sequencing to create a population genetic framework for future MYLF translocation and reintroduction efforts. Notably, our study includes skin swab samples from both extant and extirpated populations across both species, providing a critical understanding of historical and contemporary genetic variation in these endangered species. Our study addresses the following three questions: (a) What are the key MYLF genetic groups that can serve as management units in SEKI; (b) How much gene flow is observed within and across major watershed boundaries in SEKI; and (c) Does genetic diversity differ among populations that are Bd-naïve, and either declining, extirpated, or persisting following Bd outbreaks? Our results provide a clear and robust delineation of frog management units and highlight the importance of genetic data for effective species recovery planning.
2 MATERIALS AND METHODS
2.1 Sampling and DNA purification
We used 598 archived swab DNA samples (2005–2014) from 48 lake basins across four major watersheds in SEKI that were previously collected for Bd surveillance (Figure 1b). We sampled relatively evenly across both species (R. sierrae; n = 304, R. muscosa; n = 294). We defined lake basins as “populations” within major watersheds (at HUC8 scale, with Kings watershed divided by two major forks), but it is important to note that lake basins are subdivided into numerous lakes and streams (as shown in Figure 1a). Additionally, we included two lake basins outside park boundaries (identified with an asterisk in Figure 1b, Mulkey Meadows & Lower Bullfrog Lakes) as they represent important populations for future frog recovery. Each individual frog was swabbed 30 times on ventral skin surfaces. DNA was extracted from swab samples using PrepMan Ultra Reagent according to manufacturer's protocol. Typically, minimally invasive samples contain many PCR inhibitors that can interfere with downstream data quality for DNA sequencing, so we used an isopropanol precipitation to purify swab extracts (Poorten, Knapp, & Rosenblum, 2017). We applied 1 µl of DNA per extract towards amplicon preparation and sequencing.
2.2 DNA sequencing
Using 50 amplicon markers previously developed for MYLFs (Poorten et al., 2017), we applied a microfluidic PCR approach to generate nuclear amplicons. Briefly, the Fluidigm Access Array and Juno platforms allowed for high throughput amplification of either 48 or 192 samples, respectively, across all markers, and produced PCR products ready for amplicon library preparation. Using this type of assay provides a relatively affordable (~$25 per sample) method to obtain robust results from lower DNA quality samples (Byrne et al., 2017). Given the small amount of DNA available from skin swabs versus traditional DNA sources, we used a preamplification step following the manufacturer's protocol (Fluidigm, South San Francisco, CA, USA). This initial PCR (with forward and reverse primers without tagged barcodes) increased amplification success of target regions. We then removed other potential PCR inhibitors such as excess primers and unincorporated nucleases from PCR products using ExoSAP-IT and diluted 1:5 in nuclease-free water.
Following preamplification, we applied a microfluidic PCR method to amplify target regions. Each well contained a preamplified PCR product for each sample and multiplexed primer pools which was loaded onto an Access Array or Juno platform. Following microfluidic PCR, samples were combined into an Illumina library prep which included a barcoded tag of each amplicon and each sample. Illumina libraries were run on ¼ MiSeq plate with 2 × 300 bp paired-end reads, resulting in ~4.5 million reads with ~290x coverage per amplicon (unique combinations of samples and amplicons) at the University of Idaho IBEST Genomics Resources Core. Our data set ran in two phases, 237 swabs samples on Fluidigm Access Array 48 × 48, followed by 361 samples on Fluidigm Juno 192 × 24. The two data sets were combined for sequence preprocessing and SNP genotyping.
2.3 Sequence processing and SNP genotyping
Starting with raw sequence reads, we used the dbcAmplicons software (https://github.com/msettles/dbcAmplicons) to trim adapter and primer sequences. Paired-end reads were merged to build continuous reads that extended the length of the amplicon using flash2 (Magoč & Salzberg, 2011). Sequences were demultiplexed using the reduce_amplicons.R script from the dbcAmplicons repository. After demultiplexing, we used bwa (“mem” mode) software to align reads to our reference target regions. Using BAM files from alignments, we applied FreeBayes, a Bayesian genetic variant detector that identified haplotype-based SNP calls (Garrison & Marth, 2012). FreeBayes software removed singleton alleles and used phased haplotypes encoded as alleles. Following singleton removal and phasing, we used default FreeBayes parameters and limited SNP calls to within our 50 amplicon regions. The resulting data set was a raw VCF file that we used for subsequent SNP filtering. We filtered SNPs using standard quality control parameters through vcftools (removing alignment mapping quality less than 30, supporting base quality less than 20, minimum supporting allele quality sum = 0, and proportion of genotypes called <60) (Danecek et al., 2011). Finally, we removed samples from downstream analyses that contained a high proportion of missing data (>50%), which left 385 samples in the data set for downstream analyses.
2.4 Inferring population genetic structure
Before inferring population structure, we assessed potential pseudoreplication and associated biases in our data set due to the physical linkage between SNPs in each of our amplicons. To do so, we first randomly subsampled one SNP per amplicon locus and conducted a principal component analysis (PCA) on that data subset. We repeated this procedure 500 times at both the basin level and the major drainage level to explore the consistency of inferred genetic relationships. We used a Procrustes transformation, implemented in R package vegan 2.5–6 (Oksanen et al., 2019), to keep a consistent orientation between PC plots for each random subset. We found some effect of sub-setting on inferred genetic relationships, but patterns of relatedness were generally consistent across random subsamples, and we found no directional biases (Results, Figure S1). After assessing potential biases, we used multiple methods to investigate genetic structure within our SNP data set. Using the full SNP data set, we examined differentiation at a coarse scale by comparing FST between major watersheds and conducting a PCA, both implemented in adegenet (Jombart, 2008). We tested for departures of FST from 0 through Monte-Carlo test of 1,000 simulations with pairwise FST values implemented in hierfstat (Goudet, 2005). For the PCA, we evaluated the first two principal components to visualize genetic structure at the watershed drainage scale. To more explicitly explore population structure and potential admixture among lake basins, we applied STRUCTURE (v. 2.3.4) to our multilocus genotypes. We ran an admixture model five times for each potential value of K (=1–6) with 10,000 steps burnin and 100,000 MCMC steps. The maximum value of K was chosen as double the number of populations at the watershed scale compared to previous genetic work (Vredenburg et al., 2007). By using a range of K values, we evaluated all biologically reasonable groupings rather than using a single K value from a model comparison approach. Additionally, we investigated substructure using similar STRUCTURE model parameters within each drainage. Paired with our STRUCTURE analyses, we used conStruct v1.03 (https://CRAN.R-project.org/package=conStruct), which models both continuous and discrete patterns of genetic differentiation (Bradburd, Coop, & Ralph, 2018). Briefly, conStruct accounts for patterns of isolation-by-distance by estimating ancestry proportions from samples while simultaneously estimating the decay of relatedness within a population due to distance across a landscape. We ran three replicate runs of conStruct for values of K between 1 and 7, each for 3,000 iterations. For each analysis, we compared models across different values of K by calculating the “layer contributions” – the amounts of total covariance explained by each discrete group in the model and rejecting values of K that resulted in negligible layer contributions. Finally, we applied an AMOVA to test for hierarchical structure between lake basin and watershed scales using the poppr R package (Kamvar, Tabima, & Grünwald, 2014).
2.5 Measuring gene flow
We also investigated patterns of migration among major watersheds. We applied TreeMix v. 1.13 (Pickrell & Pritchard, 2012), which uses a maximum likelihood approach to identify patterns of population splitting and admixture across all samples. Using the four watersheds as major population groups, we simulated 2–10 migration events (−m flag), generated bootstrap replicates to ensure confidence in our inferred tree of admixture events, and chose the best fit tree based on maximum likelihood values.
2.6 Patterns of historical genetic diversity in extant and extirpated populations
Lastly, we calculated standard measures of historical genetic diversity among all 48 lake basins. In this case, we define historical as samples collected before the detection of Bd from qPCR of skin swabs. Bd epizootics in MYLF populations cause mass die-offs and many populations in SEKI were extirpated within several years of such outbreaks (Vredenburg et al., 2010). Bd has now been detected across nearly all of SEKI, and, as a result, robust populations are rare (R. A. Knapp & D. M. Boiano, unpublished data). Using repeated surveys of frog populations conducted over the past 20 years (Jani, Knapp, & Briggs, 2017; R. A. Knapp, unpublished data; Vredenburg et al., 2010) and associated Bd surveillance, we classified the sampled lake basins into four frog population status categories (“Status” in Table 1). Of the sampled lake basins, a small number remain Bd-naïve (termed “naïve” [n = 6]). In addition, a few basins contain populations that are persisting or recovering following Bd-caused declines (termed “persistent” [n = 6]). A larger number of basins contain populations that declined following the arrival of Bd and are trending toward extirpation due to a lack of recruitment of animals into the adult size class (termed “declining” [n = 23]). The three categories of naïve, persistent, and declining are collectively referred to as “extant”. Finally, many basins contain sites from which MYLFs are entirely extirpated following Bd-caused declines (termed “extirpated” [n = 13]). Especially for recently declined or extirpated lake basins, historical genetic diversity can give context for how diversity was once distributed on the landscape. We compared historical genetic diversity of frogs across the four basin categories, and calculated Watterson's θ and observed heterozygosity using a custom R script and the adegenet R package, respectively (Jombart, 2008).
| Basin | N | Major Watershed | Species | Status | Watterson's θ | H (Nei's) |
|---|---|---|---|---|---|---|
| LeConte Divide | 9 | San Joaquin | Rana sierrae | Persistent | 0.0017 | 0.0015 |
| McGee Basin | 9 | San Joaquin | Rana sierrae | Persistent | 0.0015 | 0.0022 |
| Darwin Bench | 8 | San Joaquin | Rana sierrae | Persistent | 0.0014 | 0.0018 |
| Evolution Basin | 8 | San Joaquin | Rana sierrae | Declining | 0.0013 | 0.0029 |
| Barrett Basin | 27 | MF Kings | Rana sierrae | Declining | 0.0031 | 0.0061 |
| Black Giant Basin | 13 | MF Kings | Rana sierrae | Declining | 0.0026 | 0.0023 |
| Dusy Basin | 20 | MF Kings | Rana sierrae | Declining | 0.0024 | 0.0026 |
| Rambaud Basin | 16 | MF Kings | Rana sierrae | Extirpated | 0.0021 | 0.0014 |
| Devils Crag Basin | 9 | MF Kings | Rana sierrae | Extirpated | 0.0019 | 0.0011 |
| Black Divide | 3 | MF Kings | Rana sierrae | Declining | 0.0013 | 0.001 |
| Amphitheater Basin | 13 | MF Kings | Rana sierrae | Declining | 0.0012 | 0.0018 |
| Volcanic Basin | 10 | MF Kings | Rana sierrae | Declining | 0.0012 | 0.0024 |
| Slide Basin | 8 | MF Kings | Rana sierrae | Declining | 0.0012 | 0.0017 |
| Swamp Basin | 11 | MF Kings | Rana sierrae | Persistent | 0.0012 | 0.0016 |
| Palisade Basin | 3 | MF Kings | Rana sierrae | Extirpated | 0.001 | 0.0012 |
| Observation Basin | 13 | MF Kings | Rana sierrae | Declining | 0.0009 | 0.0009 |
| Gorge Basin | 2 | MF Kings | Rana sierrae | Declining | 0.0007 | 0 |
| Horseshoe Basin | 4 | MF Kings | Rana sierrae | Declining | 0.0004 | 0.0004 |
| Spur Basin | 15 | SF Kings | Rana muscosa | Naïve | 0.0026 | 0.005 |
| Forester Basin | 9 | SF Kings | Rana muscosa | Naïve | 0.0026 | 0.0027 |
| Upper Basin | 15 | SF Kings | Rana muscosa | Extirpated | 0.0025 | 0.0061 |
| Marjorie Basin | 14 | SF Kings | Rana muscosa | Declining | 0.0021 | 0.0045 |
| Reflection Basin | 11 | SF Kings | Rana muscosa | Extirpated | 0.002 | 0.004 |
| Center Basin | 4 | SF Kings | Rana muscosa | Naïve | 0.0015 | 0.0014 |
| Sixty Lake Basin | 20 | SF Kings | Rana muscosa | Declining | 0.0015 | 0.0049 |
| Woods Basin | 1 | SF Kings | Rana muscosa | Extirpated | 0.0013 | 0 |
| Vidette Basin | 6 | SF Kings | Rana muscosa | Naïve | 0.0011 | 0.0026 |
| Granite Basin | 3 | SF Kings | Rana muscosa | Persistent | 0.001 | 0.0042 |
| Bullfrog Basin | 1 | SF Kings | Rana muscosa | Naïve | 0.0009 | 0 |
| Striped Basin | 1 | SF Kings | Rana muscosa | Extirpated | 0.0009 | 0 |
| Muro Blanco Basin | 12 | SF Kings | Rana muscosa | Extirpated | 0.0008 | 0.0041 |
| Pinchot Basin | 3 | SF Kings | Rana muscosa | Extirpated | 0.0008 | 0.0037 |
| Marion Basin | 2 | SF Kings | Rana muscosa | Extirpated | 0.0006 | 0.0007 |
| Rae Basin | 3 | SF Kings | Rana muscosa | Extirpated | 0.0005 | 0.0019 |
| Cartridge Basin | 2 | SF Kings | Rana muscosa | Extirpated | 0.0005 | 0.0014 |
| Lewis Basin | 2 | SF Kings | Rana muscosa | Declining | 0.0003 | 0 |
| Lower Bullfrog Lake * | 1 | Kern | Rana muscosa | Declining | 0.0022 | 0 |
| Milestone Basin | 19 | Kern | Rana muscosa | Declining | 0.002 | 0.0055 |
| Kern Bench | 9 | Kern | Rana muscosa | Naïve | 0.0019 | 0.0027 |
| Mulkey Meadows * | 6 | Kern | Rana muscosa | Persistent | 0.0018 | 0.0027 |
| Whitney Basin | 5 | Kern | Rana muscosa | Declining | 0.0017 | 0.0053 |
| Tyndall Basin | 4 | Kern | Rana muscosa | Declining | 0.0015 | 0.0044 |
| Upper Kern Basin | 15 | Kern | Rana muscosa | Declining | 0.0014 | 0.0042 |
| Sky Parlor Basin | 2 | Kern | Rana muscosa | Declining | 0.0011 | 0.0007 |
| Wright Basin | 3 | Kern | Rana muscosa | Declining | 0.0005 | 0.0016 |
| Wallace Basin | 3 | Kern | Rana muscosa | Declining | 0.0005 | 0 |
| Laurel Basin | 2 | Kern | Rana muscosa | Extirpated | 0.0005 | 0.0007 |
| Coyote Basin | 6 | Kern | Rana muscosa | Declining | 0.0002 | 0.0015 |
Note
- Genetic diversity calculated as Watterson's θ and Nei's unbiased gene diversity. Population status divided into four categories: naïve, persistent, declining, and extirpated. (*) Mulkey Meadows and Lower Bullfrog Lake lie outside park boundaries but represent important populations for Kern Watershed lake basins.
3 RESULTS
3.1 Genetic structure
After filtering, SNP genotyping, and phasing, our data set included 385 individuals and 1,447 SNPs. From the original 598 samples, our 385 samples for downstream analysis resulted in a 64% success rate. Percent success sequencing from swabs was similar across both species (R. muscosa: 67.7% [n = 199], R. sierrae: 61.2% [n = 186]); across contemporary and historical sampling periods (extant: 65.3% [n = 305], extirpated: 67.7% [n = 80]); and across disease status groups (naïve: 55.7% [n = 44], persistent: 74.2% [n = 46], declining: 66.0% [n = 215], extirpated 67.8% [n = 80]). The average number of SNPs per contig was 31 ± 8 SD and the average length of contig was 359 ± 60 bp SD. Inferred population genetic structure indicated that samples largely clustered by major watershed drainage (Figure 2). Our PCA analyses formed three groups across four watersheds with PC loadings strongly correlated with latitude or watershed (PC 1) and longitude (PC 2). STRUCTURE and conStruct results suggest three clusters forming 2–4 different groupings (Figures 3, 4). AMOVA results were consistent with major genetic groupings, with the majority of genetic variation (58.45%,) partitioned between major watersheds and remaining genetic variation partitioned among lake basins within drainages, and among all samples (38.96%, 2.58% respectively). Permutation significance testing for AMOVA showed significant differences among major watersheds (p < .001) and among samples within major watersheds (p < .001). Within watersheds, however, we found no substructuring from both STRUCTURE and ConStruct. Thus, the four sampled watershed basins could be described as three genetic groups, with samples from San Joaquin and Middle Fork (MF) Kings representing a northernmost cluster, samples from South Fork (SF) Kings representing a central cluster, and samples from Kern representing a southern cluster. Notably, both STRUCTURE and conStruct indicated some admixture among basins, particularly between the MF and SF Kings watersheds. The three genetic groupings we found are not entirely concordant with the previous split described between R. sierrae and R. muscosa (Vredenburg et al., 2007). Although we did find that R. sierrae and R. muscosa samples segregated in largely distinct clusters, we also found some admixture between the named species (notably between the MF and SF watersheds) and found additional genetic discontinuities within named species (notably between the SF and Kern watersheds).
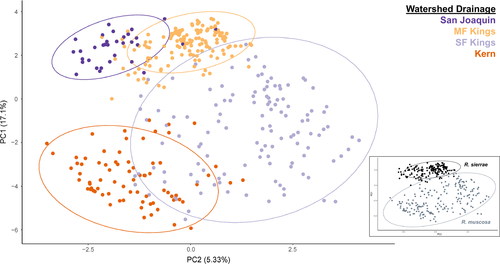
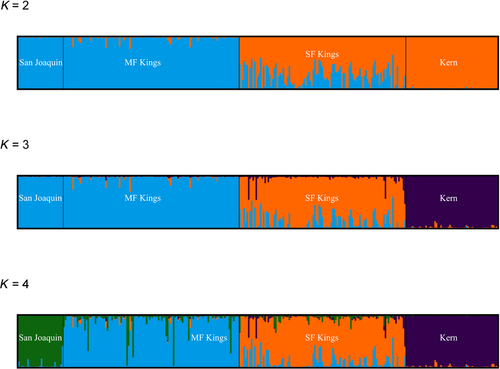
To examine possible impacts of pseudoreplication on our results due to physical linkage between SNPs on the same amplicon, we tested for biases introduced by using the complete data set. Using randomly subsetted SNP data sets (retaining only a single, randomly selected SNP per amplicon), we found some effect on inferred genetic relationships but no directional bias (Figure S1). Pseudoreplication due to linkage should artificially increase our certainty, but not introduce bias, in our results. Our results were broadly comparable across PCA, STRUCTURE, and ConStruct groupings (Figure S2). Finally, we considered a range of possible K values given the issues with identifying a single “optimal” K (Meirmans, 2015). Overall, our results were highly consistent across approaches, so we describe biogeographic patterns based on K = 3, which appears supported across methods and is biologically the most relevant.
Levels of differentiation based on FST among the four sampled watersheds were also consistent with clustering results (Table S1). The San Joaquin and MF Kings watersheds, which can be interpreted as constituting a single genetic cluster, exhibited the most limited differentiation (FST = 0.05). Admixture between MF and SF Kings was similarly reflected by low cross-basin differentiation (FST = 0.06). Consistent with a less porous genetic break between SF Kings and Kern, we observed greater differentiation between these basins (FST = 0.13). As expected, FST between nonadjacent basins was higher [MF Kings-Kern (FST = 0.17), and San Joaquin-Kern watersheds (FST = 0.21)]. Simulations for departures of FST showed significant differentiation between major watersheds (Monte-Carlo test, nsim = 1,000, p < .001).
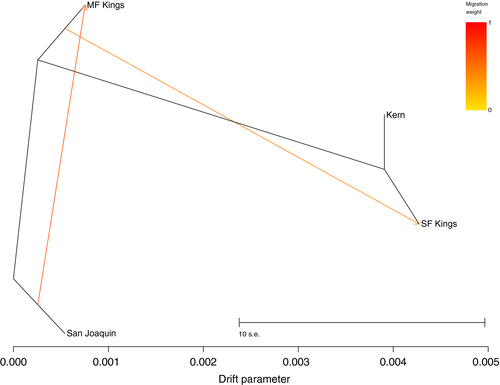
3.2 Gene flow
Given patterns of admixture observed across watershed boundaries, we estimated relative weights of migration among watersheds. The highest likelihood tree from our TreeMix analysis inferred two migration events. Using a two-migration event tree, the strength and directionality of migration was greatest from San Joaquin to MF Kings (which together form a single genetic cluster) followed by MF Kings to SF Kings (Figure 5). While SF Kings and Kern cluster closely in topology, TreeMix support our structuring results that there is still a major barrier to migration between these two watersheds. It is important to note that the TreeMix model has several assumptions about the processes of gene flow. Mainly, migration is modelled as occurring in a single time point as opposed to ongoing long-term gene flow (Pickrell & Pritchard, 2012). This assumption is probably violated in our case, since there is probably ongoing gene flow given our admixture, but the topology did not change by adding migration events and matches our genetic groupings.
3.3 Genetic diversity of populations differing in Bd exposure history and outcome
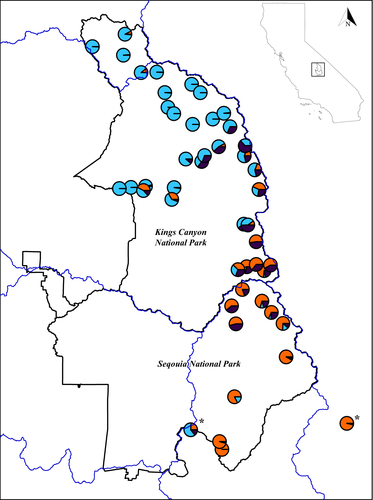
To examine the extent to which historical genetic diversity is distributed among frog populations with different Bd-related histories, we compared mean Watterson's θ for samples of four different types of populations (assigned at the lake basin scale): naïve, persistent, declining, and extirpated (Table 1). Historical genetic diversity was highest in naïve basins (0.002 ± 0.0007 SD) followed by persistent (0.0014 ± 0.0003 SD) and declining (0.0014 ± 0.0008 SD) basins. Extirpated basins (0.0012 ± 0.0007 SD) harboured the least historical genetic diversity of our status groups, but differences in genetic diversity between basin types were not significant (ANOVA, F = 1.32, p = .281). Within lake basins that still have frogs (all extant, n = 35), mean historical genetic diversity was highest in Barrett (0.0031, MF Kings) while Coyote basin (0.0002, Kern) exhibited the lowest historical genetic diversity (Table 1, Figure 6).
4 DISCUSSION
The planning of effective translocations and reintroductions requires a baseline understanding of genetic diversity and structure for the species of interest. In cases of rapid species declines, archived samples may be the only opportunity to provide genetic context for recovery actions. Therefore, our study leveraged archived swab samples from both extant and extirpated populations of an endangered frog species complex within an actively managed protected area. Using amplicon-based Illumina sequencing, we addressed three main objectives: identifying mountain yellow-legged frog management units within SEKI, refining our understanding of gene flow across major watershed boundaries, and assessing historical genetic diversity among extant (naïve, persistent, and declining) and extirpated lake basins to identify what diversity was present in SEKI before the arrival of Bd. Overall, we found that frog populations in SEKI structured into three genetic clusters with evidence for some gene flow between the clusters. Additionally, we found that genetic diversity did not differ between populations with different disease histories. Our findings provide finer spatial and genomic resolution across the remaining frog localities in SEKI. Broadly, we demonstrate the power of combining samples from extant and extirpated populations and suggest how they can inform translocations and reintroductions for conservation.
4.1 Factors influencing frog population structure in SEKI
Our tests for genetic structure used a variety of methods (PCA, STRUCTURE, conStruct, and AMOVA) and recovered similar genetic clusters. Samples from the San Joaquin and MF Kings watersheds together composed one genetic cluster, samples from the SF Kings watershed created a second cluster, and samples from the Kern watershed comprised a third (Figure 2-4). While we identified three genetic groupings, we recovered some admixture between basins. Not only did we find evidence of significant gene flow between San Joaquin-MF Kings samples (which together comprise a single genetic group), but we also inferred more limited gene flow between the remaining adjacent watersheds (MF Kings-SF Kings and SF Kings-Kern) (Figures 3 and 4). Our model-based analyses suggested that a two-migration event scenario was the best fit for the data, with migration probably strongest between San Joaquin-MF Kings and MF Kings-SF Kings (Figure 5). In summary, there is evidence for differentiation across watershed boundaries in SEKI MYLFs, but some boundaries have been more porous to gene flow over time than others.
Several factors probably contribute to patterns of drainage-level genetic variation in MYLFs. Certain environmental characteristics, such as topography and fluvial distances, are known to separate montane amphibian populations (Funk et al., 2005; Giordano, Ridenhour, & Storfer, 2007; Lowe, Likens, McPeek, & Buso, 2006; Murphy, Dezzani, Pilliod, & Storfer, 2010; Richards-Zawacki, 2009; Spear, Peterson, Matocq, & Storfer, 2005). Given the steep slopes and high ridges between drainages in this portion of the Sierra Nevada, the topographic isolation of lake basins, and the highly aquatic life history of MYLF, our admixture and gene flow results suggest similar characteristics could have shaped our observed genetic patterns across frog populations. These characteristics can be highlighted by the porous patterns of genetic variation between San Joaquin and MF Kings. Frog populations in these two watersheds have the least genetic differentiation between drainages (FST), and Muir Pass (elevation 3,644 m), which separates them, has a relatively smooth topographic gradient. As a result, lakes and streams are in close proximity to the pass and there are fewer barriers to frog movement. Other environmental and life history factors could also impact frog movement across the landscape. Such variables could include temperature-moisture regimes, habitat permeability, presence of non-native predatory trout, and frost-free periods between sites (Murphy et al., 2010). Future work would benefit from generating explicit models to correlate patterns of genetic variation with environmental variables and landscape features.
In addition to the potential contribution of geographic barriers to observed patterns of genetic diversity, we also found a general signal of isolation-by-distance both within and across watersheds. Moreover, we identified a general pattern of asymmetrical gene flow with frogs migrating preferentially north to south across our study area (from the San Joaquin to MF Kings and from MF Kings to SF Kings, Figure 5). TreeMix models are probably violated if there is ongoing gene flow, but we can cautiously interpret topologies and directionality of gene flow to understand relationships between major drainages. North-south axes of differentiation have also been observed in other Sierra Nevada herpetofaunal taxa, probably influenced by one or more broad vicariant events (e.g., climatic or glacial; Feldman & Spicer, 2006; Moritz, Schneider, & Wake, 1992; Recuero, Martínez-Solano, Parra-Olea, & García-París, 2006; Rissler, Hijmans, Graham, Moritz, & Wake, 2006; Shaffer, Fellers, Magee, & Voss, 2000; Shaffer, Pauly, Oliver, & Trenham, 2004; Vredenburg et al., 2007). It is important to note that patterns of population structure and gene flow inferred here do not reflect current migration, given the small number of remaining MYLFs in SEKI. Historically, high abundances and widespread localities of MYLFs across SEKI suggest that connectivity among populations within and between lake basins would have been much higher than at present (Figure 1a). Thus it is also possible that observed genetic patterns could partially be a geographic artefact of recently lost MYLF populations, for example if the full complement of historical populations created more genetic continuity across the landscape (Froufe, Alekseyev, Knizhin, Alexandrino, & Weiss, 2003; Lind, Spinks, Fellers, & Shaffer, 2011; Waters et al., 2007).
4.2 Genetic diversity in SEKI
Our analyses - using swab samples from both extant and extirpated lake basins - also provide insight into historical genetic diversity in SEKI MYLFs given dramatic recent declines. Analyzed skin swabs were collected over the last decade (before, during, and after population declines) and provide an opportunity to describe historical genetic diversity for the species (i.e., before the arrival of Bd). In terms of rank order, Bd-naïve basins harboured the most genetic diversity, while basins from which frogs have been extirpated harbored the least. Basins where frogs have survived a Bd-outbreak were intermediate in genetic diversity. Despite this rank order, differences were not statistically significant, probably due to low total numbers of lake basins with naïve and persisting populations. Overall, mean genetic diversity varied by two orders of magnitude across all basins (Table 1, Figure 6). Inferred genetic diversity (based on sampling conducted across 20 years) may be higher than current genetic diversity given ongoing Bd-related declines. Furthermore, because samples were limited, we needed to bin samples across years, constraining our ability to estimate and identify fluctuations in genetic diversity (Palm, Laikre, Jorde, & Ryman, 2003; Tessier & Bernatchez, 1999). However, given that many of the populations sampled represent the last remaining chance to describe historical MYLF diversity, our findings provide crucial data for translocation and reintroduction efforts by describing fine-scale patterns of diversity across the landscape.
4.3 Management implications for reintroductions and translocations
The vast majority of MYLF sites in SEKI have been extirpated in large part due to threats of non-native trout and disease, which are still present on the landscape. Only a handful of lake basins harbor frog populations that have not experienced Bd outbreaks or are persisting despite Bd presence. In our study, only twelve lake basins are considered “persistent” or “naïve” with regard to Bd. Of the twelve lake basins with persisting populations, eight had higher than average historical genetic diversity. These few basins represent the best remaining chance, if currently available genetic diversity is representative of historic levels, to bolster frog populations in SEKI. With an alarmingly small number of basins still harboring frogs, conservation managers have few options for translocations. However, even in the face of dwindling management options, our results can provide some guidance for moving frogs on the landscape.
At the broadest level, our results suggest that managing frogs by major genetic group within SEKI may be more productive than managing frogs solely based on the species-level split. Our observed patterns of genetic variation (based on multilocus nuclear data) are not entirely concordant with previous mtDNA results that indicated a species-level break at the MF-SF Kings watershed boundary (Vredenburg et al., 2007). Although we found that R. sierrae and R. muscosa samples segregated into largely distinct genetic clusters, we also found evidence for admixture between the named species (across the MF and SF watersheds). We also describe a genetic break within R. muscosa (between the SF and Kern watersheds). Such differences between mtDNA and nuclear DNA data sets are common (e.g., Toews & Brelsford, 2012), especially when one set of markers shows stronger (or different) genetic discontinuities than the other. Typically, named species are treated separately for management decisions (Mace, 2004). However, when species boundaries are unclear, genetic clusters might be better functional units for conservation decision making (Coates, Byrne, & Moritz, 2018). In this case, management in SEKI might better focus on the major genetic groups as management units rather than simply relying on species designations.
A conservative management approach suggests that moving frogs between adjacent basins is more favourable than moving frogs over long distances between nonadjacent basins. Moving frogs between proximate lake basins increases the likelihood that translocated genotypes would have been historically present. Moving animals between nearby lake basins can also help maintain locally adapted alleles. Additionally, lack of genetic substructure within watersheds suggests that moving frogs within a basin will have little impact on overall genetic structure. Therefore, managers could move frogs within watersheds to reestablish MYLFs in lake basins from which they have been extirpated. Current population census data will also be critical for assessing which basins with adequate historical genetic diversity also have viable frog numbers. Similarly, specific threats on the landscape may change which lake basins will be the best source for donor individuals. For example, translocating frogs that have persisted in the face of Bd may be a high priority given the ongoing threat of Bd on the landscape (Joseph & Knapp, 2018). Some declining frog populations may retain high historical genetic diversity, but high Bd susceptibility and low recruitment (leading to potential loss of genetic diversity) may make them poor sources for translocations.
Our gene flow data also suggest that moving frogs from north to south would better maintain historical genetic patterns (Figure 5). This is less important within watersheds, where genetic substructure is not pronounced. Overall, it may be less ideal to move frogs between major watersheds, especially when they coincide with genetic breaks. However, given the low number of remaining MYLF populations in SEKI, cross-watershed translocations may be necessary. In these cases, the more conservative management action would be to maintain a north-south direction of genetic exchange.
Our recommendations prioritize maintaining historical population genetic structure and the potential for locally adapted alleles among lake basins. However, conservation managers confront complex tradeoffs, and therefore other strategies may be worth considering. For example, if reducing the threat of inbreeding depression and augmenting genetic diversity is a key concern (Moritz, 1999; Weeks et al., 2011), managers may consider moving frogs further distances than adjacent lake basins. Ultimately, translocations and reintroductions may be ineffective unless ongoing threats are mitigated. Given that Bd is still present on the landscape, introducing frogs from naïve lake basins that may be especially susceptible to chytridiomycosis increases the likelihood of recovery failure. Thus, identifying populations that are truly recovering after exposure to Bd will remain a critical objective for field research (Knapp et al., 2016). Lastly, coupling frog genetic data presented here with Bd genetic data across SEKI could illuminate whether different Bd genotypes exist among lake basins and help managers avoid moving Bd genotypes among susceptible individuals. We have recently developed a complementary Bd genotyping assay (Byrne et al., 2017) and can now leverage Bd positive skin swab samples to genotype Bd across SEKI and assess whether frog and Bd genotypes co-vary spatially.
Fine-scale studies such as this genetic assessment within SEKI and similar work in Yosemite National Park (Poorten et al., 2017) will be crucial for MYLF recovery in individual parks. However, remnant populations in the two national parks represent only a portion of the total MYLF range. A full rangewide analysis will be critical to resolve several outstanding issues about the species complex. Critically, additional work is required to refine our understanding of within and between species differentiation. Genetic management units identified in this study are relevant for SEKI, but a rangewide analysis would provide more clarity for conservation action on genetic variation across the range. An updated rangewide genetic assessment would increase resolution outside park boundaries (as there are many additional frog populations adjacent to the parks) and allow coordinated conservation actions across multiple jurisdictions and stakeholders. In addition, our assay could be expanded to include detection of SNPs that may be important not only for maintaining neutral processes but also candidate adaptive loci important for Bd-resistance.
In conclusion, our study highlights the power of archived genetic samples for current conservation decision-making. Especially in cases of rapid species declines, our study provides a framework to harness critical genetic information even as populations are being extirpated. We leveraged MYLF samples from lake basins whose frog populations have been all but lost from the landscape. These samples provide crucial baseline data for understanding historical population structure and genetic diversity in SEKI. Populations will probably continue to be extirpated as disease spreads through the remaining naïve populations. Nonetheless, with a clearer understanding of historical patterns of population structure, gene flow, and genetic diversity, conservation decisions can be guided more effectively for this imperiled species complex.
ACKNOWLEDGEMENTS
We thank Thomas Poorten for assistance in bioinformatics and field crews for collecting skin swabs. Data collection and analyses performed by the IBEST Genomics Resources Core at the University of Idaho were supported in part by NIH COBRE grant P30GM103324. All sample collections were authorized by research permits provided by SEKI and the Institutional Animal Care and Use Committee at University of California, Santa Barbara and University of California, Berkeley. Funding was provided by National Park Service, National Science Foundation LTREB DEB-1557190, and US Fish and Wildlife.
AUTHOR CONTRIBUTIONS
APR, RAK, DMB, and EBR designed research; APR, RAK, DMB, and CJB collected data and performed research; APR, and GB analyzed data; APR, EBR, RAK, GB, DMB, CJB, and EBR wrote the paper.
Open Research
DATA AVAILABILITY STATEMENT
All SNP VCF data and sample metadata are available in Dryad repository https://doi.org/10.6078/D1311X. SNP calling and scripts to reproduce figures are available at https://github.com/andrew-rothstein




