Independent domestication events in the blue-cheese fungus Penicillium roqueforti
Abstract
Domestication provides an excellent framework for studying adaptive divergence. Using population genomics and phenotypic assays, we reconstructed the domestication history of the blue cheese mould Penicillium roqueforti. We showed that this fungus was domesticated twice independently. The population used in Roquefort originated from an old domestication event associated with weak bottlenecks and exhibited traits beneficial for pre-industrial cheese production (slower growth in cheese and greater spore production on bread, the traditional multiplication medium). The other cheese population originated more recently from the selection of a single clonal lineage, was associated with all types of blue cheese worldwide except Roquefort, and displayed phenotypes more suited for industrial cheese production (high lipolytic activity, efficient cheese cavity colonization ability and salt tolerance). We detected genomic regions affected by recent positive selection and putative horizontal gene transfers. This study sheds light on the processes of rapid adaptation and raises questions about genetic resource conservation.
1 INTRODUCTION
What are the mechanisms of adaptive divergence (population differentiation under selection) is a key question in evolutionary biology for understanding how organisms adapt to their environment and how biodiversity arises. Domestication is a special case of adaptive divergence, involving strong and recent selection for traits that can be easily identified. Furthermore, closely related nondomesticated populations are often available, making it possible to contrast their traits and genomes with those of domesticated populations. Studying domestication can therefore provide a deeper understanding of the mechanisms of adaptive divergence. This approach has proved to be powerful for reconstructing the history of divergence and the genetic architecture of traits selected by humans when applied to maize and teosinte or to dog breeds and wolves (Albert et al., 2012; Axelsson et al., 2013; Freedman, Lohmueller, & Wayne, 2016; Hake & Ross-Ibarra, 2015; Li et al., 2016; Wang, Studer, Zhao, Meeley, & Doebley, 2015). Comparisons of domesticated varieties selected for different phenotypes have also proved to be a powerful approach for elucidating the mechanisms of adaptation, for example in dog breeds and pigeons (Parker, Harris, Dreger, Davis, & Ostrander, 2017; Shapiro et al., 2013). Studies on genetic diversity and subdivision in domesticated organisms also provide crucial information for the conservation of genetic resources. Indeed, recent breeding programmes have resulted in a massive loss of genetic diversity in crops and breeds, potentially jeopardizing adaptive potential for improvement (Gouyon, Leriche, Civard, Reeves, & Hulot, 2010; Harlan, 1992; Vavilov, 1992).
Fungi are interesting eukaryotic models for adaptive divergence studies, with their small genomes, easy access to the haploid phase and experimental tractability for in vitro experiments (Giraud, Koskella, & Laine, 2017; Gladieux et al., 2014). Many fungi are used as food sources (Dupont et al., 2016) and some have been domesticated for food production. Propagation of the latter is controlled by humans, and this has resulted in genetic differentiation from wild populations (Almeida, Barbosa, Bensasson, Gonçalves, & Sampaio, 2017; Almeida et al., 2014; Gallone et al., 2016; Gibbons et al., 2012; Gonçalves et al., 2016; Peter et al., 2018) and the evolution of specific phenotypes beneficial for humans (Dupont et al., 2016; Gallone et al., 2016; Gibbons & Rinker, 2015; Gibbons et al., 2012; Marsit et al., 2015). Saccharomyces cerevisiae yeasts domesticated for fermentation have provided important insight into adaptive divergence mechanisms, with different yeast lineages independently domesticated for different usages (Borneman et al., 2011; Gonçalves et al., 2016; Peter et al., 2018). Studies of yeast adaptation for alcohol and cheese production have highlighted the proximal genomic mechanisms involved, including horizontal gene transfer, selective sweep, hybridization and introgression (Legras et al., 2018; Marsit et al., 2015; Morales & Dujon, 2012; Novo et al., 2009; Peter et al., 2018).
Penicillium roqueforti, a filamentous fungus used in the dairy industry to impart the typical veins and flavour of blue cheeses, has recently emerged as an excellent model for studying adaptive divergence (Cheeseman et al., 2014; Ropars et al., 2015). Blue cheeses, including Roquefort, Gorgonzola and Stilton, are highly emblematic foods that have been produced for centuries (Vabre, 2015). The strongest genetic subdivision reported in P. roqueforti concerns the differentiation of a cheese-specific population that has acquired faster growth in cheese than other populations and better excludes competitors, thanks to very recent horizontal gene transfers, at the expense of slower growth on minimal medium (Gillot et al., 2015; Ropars, López-Villavicencio, Snirc, Lacoste, & Giraud, 2017; Ropars et al., 2015). Such genetic differentiation and recent acquisition of traits beneficial to cheesemaking in P. roqueforti suggests genuine domestication (i.e., adaptation under selection by humans for traits beneficial for food production). A second population identified in P. roqueforti and lacking the horizontally transferred regions includes strains isolated from cheese and other environments, such as silage, lumber and spoiled food (Gillot et al., 2015; Ropars et al., 2014, 2017). Penicillium roqueforti is the main contaminant of silage, spoilage typically occurring following breaks in plastic or after opening the stack for cattle feeding. In this context, it can produce harmful mycotoxins causing health disorders in cattle (Malekinejad, Aghazadeh-Attari, Rezabakhsh, Sattari, & Ghasemsoltani-Momtaz, 2015). In addition, P. roqueforti is one of the most common Penicillium species in spoiled food, where it is also responsible for mycotoxin production (Rundberget, Skaar, & Flåøyen, 2004). The existence of further genetic subdivision separating populations according to the original environment, or protected designation of origin (PDO) for cheese strains has been suggested, but, because it was based only on a few microsatellite markers, the resolution power was low and it was unclear what genetic subdivision was the most relevant (Gillot et al., 2015; Ropars et al., 2014, 2017). Secondary metabolite production (aroma compounds and mycotoxins) and proteolysis activity have been shown to differ between strains from different PDOs (Gillot et al., 2017). Of note, a high-quality P. roqueforti genome reference has been available since 2014 (Cheeseman et al., 2014), allowing more powerful analyses based on population genomics.
Another asset of P. roqueforti as an evolutionary model is the availability of vast collections of cheese strains and of historical records concerning cheesemaking (Aussibal, 1983; Labbe & Serres, 2009, 2004; Marre, 1906; Marres, 1935; Vabre, 2010). While the presence of P. roqueforti in cheese was initially fortuitous, since the end of the 19th century, milk or curd has been inoculated with the spores of this fungus for Roquefort cheese production. Spores were initially multiplied on bread, before the advent of more controlled in vitro culture techniques in the 20th century (Aussibal, 1983; Labbe & Serres, 2009, 2004; Marre, 1906; Marres, 1935; Vabre, 2010). Bread was inoculated by recycling spores from the best cheeses from the previous production (i.e., back-slopping) (Aussibal, 1983; Labbe & Serres, 2009, 2004; Marre, 1906; Marres, 1935; Vabre, 2010). This corresponds to yearly selection events since the 19th century until ~20 years ago when strains were stored in freezers. After World War II, strains were isolated in the laboratory for industrial use and selected based on their technological and organoleptic impact in cheeses, and produced compounds (Besana, D’Errico, & Ghezzi, 2017) that have probably accelerated domestication. This history further suggests that there may have been genuine domestication. Unintentional selection may also have been exerted on other traits, including growth and spore production on bread, the traditional multiplication substrate.
By sequencing multiple P. roqueforti genomes from different environments and analysing large collections of cheese strains, we provide evidence for adaptive divergence. We identified four genetically differentiated populations, two including only cheese strains and two other populations including silage- and food-spoiling strains. We inferred that the two cheese populations corresponded to two independent domestication events. The first cheese population corresponded to strains used for Roquefort production and arose through a weaker and older domestication event, with multiple strains probably originating from different cultures on local farms in the PDO area, presumably initially selected for slow growth before the invention of refrigeration systems. The second cheese population experienced an independent and more recent domestication event associated with a stronger genetic bottleneck. The “non-Roquefort” population showed beneficial traits for modern industrial production of cheese (e.g., faster growth in salted cheese, more efficient cheese cavity colonization and faster lipid degradation activities), while the Roquefort cheese population showed greater spore production on bread, the traditional medium for spore production. The four populations further showed differences in proteolysis activities, with a higher variance in the cheese populations. The two cheese populations also had different volatile compound profiles, with probable effects on cheese flavour. These phenotypic differences might be associated with genomic regions affected by recent positive selection and genomic islands specific to a single cheese population. Some of these genomic regions may have been acquired by horizontal gene transfers and have putative functions in the biochemical pathways leading to the development of cheese flavour.
2 MATERIAL AND METHODS
2.1 Isolation attempts of Penicillium roqueforti in ripening cellar and dairy environments
We sampled spores from the air in an artisanal cheese dairy company (GAEC Le Lévejac, Saint Georges de Lévejac, France, ~60 km from Roquefort-sur-Soulzon, producing no blue cheese), and sampling was performed in the sheepfold, milking parlour, cheese dairy and ripening cellar. We also sampled spores from the air in an abandoned ripening cellar in the town of Meyrueis (~70 km from Roquefort-sous-Soulzon) where Roquefort cheeses used to be produced and stored in the early 19th century. In total, 55 Petri dishes containing malt (2% cristomalt, Difal) and 3% ampicillin were left open for 6 days as traps for airborne spores (35 Petri dishes in the abandoned ripening cellar and 20 Petri dishes in the artisanal cheese dairy company). Numerous fungal colonies were obtained on the Petri dishes. One monospore was isolated from each of the 22 Penicillium-like colonies. DNA was extracted using the Nucleospin Soil Kit (Macherey-Nagel) and a fragment of the β-tubulin gene was amplified using the primer set Bt2a/Bt2b (Glass & Donaldson, 1995), and then sequenced. Sequences were blasted against the NCBI database to assign monospores to species. Based on β-tubulin sequences, 10 strains were assigned to P. solitum, six to P. brevicompactum, two to P. bialowienzense, one to P. echinulatum and two to the genus Cladosporium. No P. roqueforti strain could thus be isolated from this sampling procedure.
2.2 Genome sequencing and analysis
The genomic DNAs of cheesemaking strains obtained from public collections belonging to P. roqueforti, seven strains of P. paneum, one strain of P. carneum and one strain of P. psychrosexualis (Table S1) were extracted from fresh haploid mycelium after monospore isolation and growth for 5 days on malt agar using the Nucleospin Soil Kit. Sequencing was performed using the Illumina HiSeq 2500 paired-end technology (Illumina Inc.) with an average insert size of 400 bp at the GenoToul INRA platform and resulted in a 50–100× coverage. In addition, the genomes of four strains (LCP05885, LCP06096, LCP06097 and LCP06098) were used that had previously been sequenced using the ABI SOLID technology (Cheeseman et al., 2014). GenBank accession numbers are HG792015–HG792062.
Identification of presence/absence polymorphism of blocks larger than 10 kbp in genomes was performed based on coverage using mapping against the FM164 P. roqueforti reference genome. To identify genomic regions that would be lacking in the FM164 genome but present in other strains, we used a second assembled genome, that of the UASWS P. roqueforti strain collected from bread and belonging to the silage noncheese cluster, sequenced using Illumina HiSeq shotgun and displaying 428 contigs (GenNank accession numbers: JNNS01000420–JNNS01000428). Blocks larger than 10 kbp present in the UASWS genome and absent in the FM164 genome were identified using the nucmer program version 3.1 (Kurtz et al., 2004). Gene models for the UASWS genome were predicted with eugene following the same pipeline as for the FM164 genome (Cheeseman et al., 2014; Foissac et al., 2008). The presence/absence of these regions in the P. roqueforti genomes was then determined using the coverage obtained by mapping reads against the UASWS genome with the start/end positions identified by nucmer. The absence of regions was inferred when fewer than five reads were mapped. To determine their presence/absence in other Penicillium species, the sequences of these regions were blasted against nine Penicillium reference genomes (Table S1). PCR primer pairs were designed using primer3plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/) in the flanking sequences of these genomic regions in order to check their presence/absence in a broader collection of P. roqueforti strains based on PCR tests (Table S2). For each genomic island, two primer pairs were designed when possible (i.e., when sufficiently far from the ends of the scaffolds and not in repeated regions): one yielding a PCR product when the region was present and another one giving a band when the region was absent, in order to avoid relying only on lack of amplification for inferring the absence of a genomic region. PCRs were performed in a volume of 25 µl, containing 12,5 µl template DNA (10-fold diluted), 0.625 U Taq DNA Polymerase (MP Biomedicals), 2.5 µl 10× PCR buffer, 1 µl of 2.5 mm dNTPs and 1 µl of each of 10 µm primer. Amplification was performed using the following programme: 5 min at 94°C and 30 cycles of 30 s at 94°C, 30 s at 60°C and 1 min at 72°C, followed by a final extension of 5 min at 72°C. PCR products were visualized using stained agarose gel electrophoresis. Data were deposited at the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession number PRJEB20132 for whole genome sequencing and PRJEB20413 for Sanger sequencing.
For each strain, reads were mapped using stampy version 1.0.21 (Lunter & Goodson, 2011) against the high-quality reference genome of the FM164 P. roqueforti strain (Cheeseman et al., 2014). To minimize the number of mismatches, reads were locally realigned using the genome analysis toolkit (GATK) indelrealigner version 3.2-2 (McKenna et al., 2010). Detection of single nucleotide polymorphisms (SNPs) was performed using the GATK Unified Genotyper (McKenna et al., 2010), based on the reference genome in which repeated sequences were detected using repeatmasker (Smit, Hubley, & Green, 2013) and masked, so that SNPs were not called in these regions. In total 483,831 bp were masked, corresponding to 1.67% of the FM164 genome sequence. The 1% and 99% quantiles of the distribution of coverage depth were assessed across each sequenced genome and SNPs called at positions where depth values fell in these extreme quantiles were removed from the data set. Only SNPs with less than 10% of missing data were kept. After filtering, a total of 115,544 SNPs were kept.
Population structure was assessed using a discriminant analysis of principal components (DAPC) with the adegenet R package (Jombart, 2008). The genetic structure was also inferred along the genome by clustering the strains according to similarities of their genotypes, in windows of 50 SNPs, using the Mclust function of the mclust R package (Fraley & Raftery, 2002; Fraley, Raftery, & Scrucca, 2012) with Gower's distance and a Gaussian mixture clustering with K = 7 (as the above analyses indicated the existence of four P. roqueforti populations and there were three outgroup species).
We performed a neighbor-net analysis using the network approach to visualize possible recombination events within and between populations with the phangorn R package (Schliep, 2010). The substitution model used for building the distance matrix was JC69 (Jukes & Cantor, 1969).
Genetic diversity was estimated using the parameters θπ and θw with the compute programs associated with libsequence version 1.8.9 (Thornton, 2003) on 1,145 sliding windows of 50 kb with 25 kb of overlap distributed along the longest 11 scaffolds of the FM164 assembly (>200 kb). Linkage disequilibrium per genetic cluster (i.e., non-Roquefort, Roquefort, lumber/food spoiler and silage/food spoiler) was estimated using the r2 statistic, with vcftools version 0.1.15 (Danecek et al., 2011) and the following parameters: --geno-r2 --ld-window-bp 15,000. Plots were generated using R.
To identify genes evolving under positive selection in P. roqueforti genomes, first, we used the method implemented in snipre (Eilertson, Booth, & Bustamante, 2012), a Bayesian generalization of the log-linear model underlying the McDonald–Kreitman test. This method detects genes in which amino-acid changes are more frequent than expected under neutrality, by contrasting synonymous and nonsynonymous SNPs, polymorphic or fixed in two groups, to account for gene-specific mutation rates. McDonald–Kreitman tests can only be used for contrasting two groups that show both fixed and private and few shared polymorphisms so not all comparisons could be run between the four populations. Second, we performed a scan of the divergence statistics dxy between the two cheese populations, calculated using a custom R script in 50-kbp windows overlapping over 25 kbp along the genome. We considered genes belonging to the 1% most divergent regions and the 5% least genetically diverse (π values) as under positive selection in one of the populations. We did not consider the other pairwise comparisons (i.e., using “lumber/food spoiler” and “silage/food spoiler” populations; Figures 1-5), because most SNPs in those populations were shared by several strains, as shown by high diversity, positive Dt and low FST values (Table 1). Consequently, islands of high divergence and low diversity were restricted to cheese populations that were already found using pairwise comparison between cheese populations. We performed Gene Ontology (GO) annotation enrichment tests using separate Fisher's exact tests on the three ontologies (BP: biological process; CC: cellular component; MF: metabolic function).
| (a) | Number of segregating sites per kilobase | π per site | Watterson's θ per site | D t | H f |
|---|---|---|---|---|---|
| Silage/food spoiler | 2.28 | 0.00098 | 0.00084 | 0.75689 | 0.00001 |
| Lumber/food spoiler | 1.59 | 0.00078 | 0.00070 | 0.77300 | −0.00004 |
| Non-Roquefort | 0.25 | 0.00008 | 0.00011 | −1.27191 | −0.00021 |
| Roquefort | 1.03 | 0.00043 | 0.00040 | 0.56090 | −0.00007 |
| Penicillium roqueforti | 2.75 | 0.00107 | 0.00070 | 1.80833 | 0.48170 |
| (b) | Silage/food spoiler | Lumber/food spoiler | Non-Roquefort |
|---|---|---|---|
| Roquefort | 0.21 | 0.27 | 0.62 |
| Non-Roquefort | 0.38 | 0.49 | |
| Lumber/food spoiler | 0.08 | ||
| (c) | |||
| Roquefort | 0.25/0.67/0.09 | 0.31/0.66/0.03 | 0.03/0.57/0.40 |
| Non-Roquefort | 0.07/0.82/0.11 | 0.03/0.69/0.28 | |
| Lumber/food spoiler | 0.58/0.42/0.00 |
2.3 Strain genotyping
By searching for the most polymorphic genomic regions capable of differentiating populations, we identified two genomic regions with multiple diagnostic SNPs allowing discrimination of the two cheese clusters. Two PCR primer pairs were designed (Table S2) to sequence these regions in order to assign the 65 strains (Table S1) that can be purchased at the Laboratoire Interprofessionnel de Production d’Aurillac (LIP) (the main French supplier of P. roqueforti spores for artisanal and industrial cheesemakers; https://www.lip-sas.fr/) to the identified clusters. PCR products were then purified and sequenced at Eurofins (France). Because one of the cheese clusters included strains carrying the Wallaby and CheesyTer genomic islands while the second cluster strains lacked these genomic regions (Ropars et al., 2015), we used previously developed primer pairs to check for the presence/absence of CheesyTer and Wallaby (Ropars et al., 2015).
Sequences were first aligned together with those extracted from sequenced genomes, allowing assignation of LIP strains to one of the two cheese populations using mafft software (Katoh & Standley, 2013) and then the alignments were visually checked. A tree reconstruction was then made using raxml version 7.0.3 following the GTRCAT substitution model, using two partitions corresponding to the two fragments and a 1,000 bootstrap tree was generated (Stamatakis, 2006).
The strain tree was also inferred by maximum likelihood using raxml (Stamatakis, 2006) under the GTRCAT model using 6,905 concatenated genes. To consider possible differences in nucleotide substitution rates, the data set was divided into two partitions, one including the 1st and 2nd codon positions and one including the 3rd codon positions. To assess node confidence, 1,000 bootstraps were computed.
2.4 Strain phenotyping
As we could not use all the strains in the experiments, a similar number of strains were chosen at random in each group to perform the experiments, in order to have a balanced design with no ascertainment bias. Experimental cheeses were produced in an artisanal dairy company (GAEC Le Lévejac). The same ewe curd was used for all produced cheeses. Seven P. roqueforti strains were used for inoculation (two from each of the “Roquefort,” “non-Roquefort” and “silage/food” spoiler clusters, and one from the “lumber/food” spoiler cluster; their identity is given in Table S1) using 17.8 mg of lyophilized spores. Three cheeses were produced for each strain in cheese strainers (in oval pots with opposite diameters of 8 and 9 cm, respectively), as well as a control cheese without inoculation. After 48 hr of draining, cheeses were salted (by surface scrubbing with coarse salt), pierced and placed in a maturing cellar for 4 weeks at 11°C. Cheeses were then sliced into six equal pieces and a picture of each slice was taken using a Nikon D7000 (zoom lens: Nikon 18–105 mm f:3.5–5.6G). Pictures were analysed using the geospatial image processing software envi (Harris Geospatial Solution) (Figure 6c). This software enables pixel classification according to their level of blue, red, green and grey into two to four classes depending on the analysed image. This classification allowed us to assign pixels to two classes corresponding to the inner white part and the cavities of the cheese, respectively (Figure 6c). For each picture, the percentage of pixels corresponding to the cavities was then quantified. Because the software could not reliably assign pixels to the presence versus absence of the fungus in cavities, we visually determined the cavity areas that were colonized by P. roqueforti using images. This allowed us to calculate a cheese cavity colonization rate. Because Penicillium spores have a high dispersal ability which could cause contaminations, we confirmed strain identity present in cheeses by performing Sanger sequencing of four diagnostic markers designed based on SNPs and specific to each strain (Table S2). For each cheese, three random monospore isolates were genotyped, and no contamination was detected (i.e., all the sequences obtained corresponded to the inoculated strains).
To compare the growth rates of the different P. roqueforti clusters on bread (i.e., the traditional multiplication medium), 24 strains were used (eight from each of the “Roquefort” and “non-Roquefort” clusters, five from the “silage/food spoiler cluster”, and three from the “lumber/food spoiler” cluster; the identities of the strains are shown in Table S1). Each strain was inoculated in a central point in three Petri dishes by depositing 10 µl of a standardized spore suspension (0.7 × 109 spores/ml). Petri dishes contained agar (2%) and crushed organic cereal bread including rye (200 g/L). After 3 days at 25°C in the dark, two perpendicular diameters were measured for each colony to assess colony size.
The lipolytic and proteolytic activities of P. roqueforti strains were measured as follows: standardized spore suspensions (2,500 spores/inoculation) for each strain (n = 47:15 from the “Roquefort” cluster, 15 from the “non-Roquefort” cheese cluster, 10 from the “silage/food spoiler” cluster and seven from the “lumber/food spoiler” cluster, identity in Table S1) were inoculated on the top of a test tube containing agar and tributyrin for lipolytic activity measure (10 ml/L, ACROS Organics) or semiskimmed milk for the proteolytic activity measure (40 g/L, from large retailers). The lipolytic and proteolytic activities were estimated by the degradation degree of the compounds, which changes the media from opaque to translucent. For each medium, three independent experiments were conducted. For each strain, duplicates were performed in each experiment and the limit of translucency/opaqueness in the medium was recorded. Measures were highly repeatable between the two replicates (Pearson's product-moment correlation coefficient of 0.93 in pairwise comparison between replicates, p < .0001). We measured the distance between the initial mark and the hydrolysis, translucent front, after 7, 14, 21 and 28 days of growth at 20°C in the dark.
A total of 47 strains were used to compare spore production between the four P. roqueforti clusters (Table S1), 15 belonging to the “non-Roquefort” cluster, 15 to the “Roquefort” cluster, 10 to the “silage/food spoiler” cluster and seven to the “lumber/food” spoiler cluster. After 7 days of growth on malt agar in Petri dishes of 60 mm diameter at room temperature, we scraped all the fungal material by adding 5 ml of tween water 0.005%. We counted the number of spores per millilitre in the solution with a Malassez haemocytometer (mean of four squares per strain) for calibrating spore solution. We spread 50 µl of the calibrated spore solution (i.e., 7 × 106 spores/ml) for each strain on Petri dishes of 60 mm diameter containing three different media, namely malt, cheese and bread agar (organic “La Vie Claire” bread mixed with agar), in duplicates (two plates per medium and per strain). After 8 days of growth at room temperature, we took off a circular plug of medium with spores and mycelium at the top, using Falcon 15-ml canonical centrifuge tubes (diameter of 15 mm). We inserted the plugs into 5-ml Eppendorf tubes containing 2 ml of tween water 0.005% and vortexed for 15 s to detach spores from the medium. Using a plate spectrophotometer, we measured the optical density (OD) at 600 nm for each culture in the supernatant after a four-fold dilution (Table S4).
To compare salt tolerance between P. roqueforti clusters, 26 strains were used (eight from the Roquefort cluster, ten from the non-Roquefort cluster, three from the silage/food spoiler cluster and five from the lumber/food spoiler cluster; strain identities are shown in Table S1). For each strain and each medium, three Petri dishes were inoculated by depositing 10 µl of standardized spore suspension (0.7 × 109 spores/ml) on Petri dishes containing either only malt (20 g/L), malt and salt (8% NaCl, which corresponds to the salt concentration used before fridge use to avoid contaminants in blue cheeses), only goat cheese, or goat cheese and salt (8% NaCl). The goat cheese medium was prepared as described in a previous study (Ropars et al., 2015). Strains were grown at 25°C and colony size was measured daily for 24 days.
Volatile production assays were performed on 16 Roquefort strains and 19 non-Roquefort cheese strains grown on model cheeses as previously described (Gillot et al., 2017). Briefly, model cheeses were prepared in Petri dishes and incubated for 14 days at 25°C before removing three 10-mm-diameter plugs (equivalent to approximately 1 g). The plugs were then placed into 22-ml Perkin Elmer vials that were tightly closed with polytetrafluorethylene (PTFE)/silicone septa and stored at − 80°C prior to analyses (Gillot et al., 2017). Analyses and data processing were carried out by headspace trap-gas chromatography-mass spectrometry (HS-trap-GC-MS) using a Perkin Elmer turbomatrix HS-40 trap sampler, a Clarus 680 gas chromatograph coupled to a Clarus 600T quadrupole MS (Perkin Elmer), and the open source xcms package of the R software (http://www.r-project.org/), respectively, as previously described (Pogačić et al., 2015).
All phenotypic measures are reported in Table S4. Statistical analyses for testing differences in phenotypes between populations and/or media (Table S5) were performed with R software (http://ww.r-project.org). Because phenotypes have not been assessed using the same strains, principal components analysis (PCA) on all phenotypes was performed using Bayesian missing data correction in the pcamethods R package (Stacklies, Redestig, Scholz, Walther, & Selbig, 2007).
Differences in volatile profiles among the two P. roqueforti cheese populations were analysed using a supervised multivariate analysis method, orthogonal partial least squares discriminant analysis (OPLS-DA). OPLS is an extension of PCA that is more powerful when the number of explained variables (Y) is much higher than the number of explanatory variables (X). PCA is an unsupervised method maximizing the variance explained in Y, while partial least squares (PLS) maximizes the covariance between X and Y(s). OPLS is a supervised method that aims at discriminating samples. It is a variant of PLS which uses orthogonal (uncorrelated) signal correction to maximize the explained covariance between X and Y on the first latent variable, and components > 1 capture variance in X which is orthogonal (uncorrelated) to Y. The optimal number of latent variables was evaluated by cross-validation (Pierre et al., 2011). Finally, to identify the volatile compounds that were produced in significantly different quantities between the two populations, a jackknife resampling on PLS regression coefficient was performed followed by a t test using the plsr() function in the R software (http://ww.r-project.org).
2.5 Demographic modelling using approximate Bayesian computation (ABC)
The likelihoods of 11 demographic scenarios for the P. roqueforti populations were compared using ABC (Beaumont, 2010; Lopes & Beaumont, 2010). The scenarios differed in the order of demographic events, and included 21 parameters to be estimated (Table S3A). A total of 262 fragments, ranging from 5 to 15 kb, were generated from observed SNPs by compiling in a fragment all adjacent SNPs in complete linkage disequilibrium. The population mutation rate θ (the product of the mutation rate and the effective population size) used for coalescent simulations was obtained from data using θw (Watterson's estimator). Simulated data were generated using the same fragment number and sizes as the SNP data set generated from the genomes. Priors were sampled in a log-uniform distribution (Table S3A). For each scenario, one million coalescent simulations were run and the following summary statistics were calculated on observed and simulated data using msABC (Pavlidis, Laurent, & Stephan, 2010): the number of segregating sites, the estimators π (Nei, 1987) and θw (Watterson, 1975) of nucleotide diversity, Tajima's D (Tajima, 1989), the intragenic linkage disequilibrium coefficient ZnS (Kelly, 1997), FST (Hudson, Slatkin, & Maddison, 1992), the percentage of shared polymorphisms between populations, the percentage of private SNPs for each population, the percentage of fixed SNPs in each population, Fay and Wu's H (Fay & Wu, 2000), the number of haplotypes (Depaulis & Veuille, 1998) and haplotype diversity (Depaulis & Veuille, 1998). For each summary statistic, both average and variance values across simulated fragments were calculated. The choice of summary statistics to estimate posterior parameters is a crucial step in ABC (Csilléry, Blum, Gaggiotti, & François, 2010). Summary statistics were selected using the AS.select() function with the neuralnet method in the “abctools” R package (Nunes & Prangle, 2015). In total, 101 summary statistics were kept for subsequent analyses. Cross validation was run with the neuralnet method using 100 samples and a tolerance of 0.01 (Figure S4C). Model selection was performed using four tolerance rates ranging from 0.005 to 0.1 and rejection, logistic regression and neural network methods. Because there was still uncertainty in the choice between scenarios 4 and 5 after model selection (i.e., whether it was the “non-Roquefort” or “Roquefort” population that diverged first from the ancestral population) (Table S3), an extra one million simulations were run for each of those two scenarios and model selection was performed again. All tolerance rates and methods favoured scenario 4 over scenario 5 with an absolute confidence of 1.000.
The posterior probability distributions of the parameters, the goodness of fit for each model and model selection (Table S3) were calculated using a rejection–regression procedure (Beaumont, 2010). Acceptance values of 0.005 were used for all analyses. Regression analyses was performed using the “abc” R package (Csilléry, François, & Blum, 2012) (http://cran.rproject.org/web/packages/abc/index.html).
2.6 Estimate of time since domestication
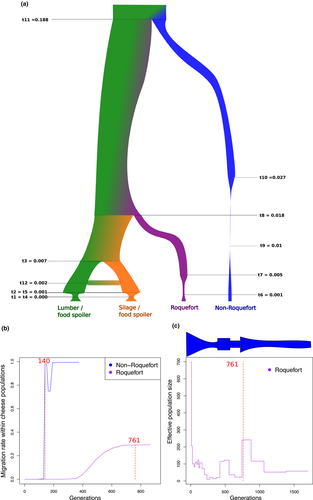
The multiple sequentially Markovian coalescent (MSMC) software was used to estimate the domestication times of cheese populations (Schiffels & Durbin, 2014). The estimate of the last time gene flow occurred within each cheese population was taken as a proxy of time since domestication as it also corresponds in such methods to bottleneck date estimates and is more precisely estimated. Recombination rate was set at zero because sexual reproduction has probably not occurred since domestication in cheese populations. Segments were set to 21 * 1 + 1 * 2 + 1 * 3 for the “Roquefort” population, which contains three haplotypes (Figure 2), and to 10 * 1 + 15 * 2 for the “non-Roquefort” population, which contains two closely related haplotypes (Figure 2). In both cases, MSMC was run for 15 iterations and otherwise default parameters. The mutation rate was set to 10–8.
3 RESULTS
3.1 Two out of four populations are used for cheesemaking: one specific to the Roquefort PDO and a worldwide clonal population
We sequenced the genomes of 34 P. roqueforti strains from public collections (Ropars et al., 2017), including 17 isolated from blue cheeses (e.g., Roquefort, Gorgonzola, Stilton), 17 isolated from noncheese environments (mainly spoiled food, silage and lumber), and 11 outgroup genomes from three Penicillium species closely related to P. roqueforti (Table S1). After data filtering, we identified a total of 115,544 SNPs from the reads mapped against the reference P. roqueforti FM164 genome (29 × 106 bp, 48 scaffolds).
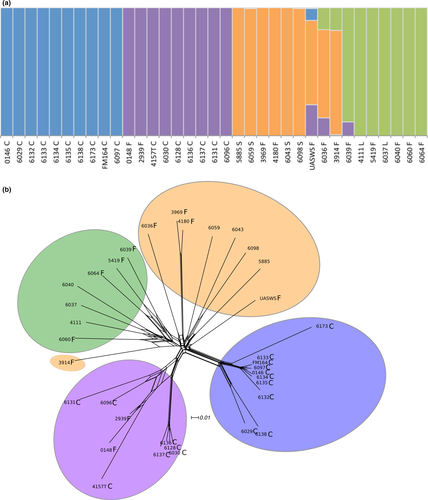
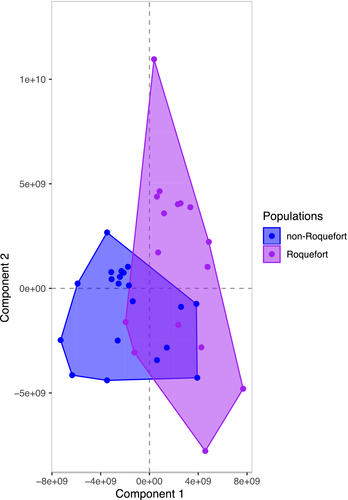
We used faststructure (Figure 1a; Figure S1A) as well as three clustering methods free from assumptions about mating system and mode of reproduction, based on genetic differences: a DAPC (Figure S1B), a SplitsTree (Figure 1b) and a clustering based on similarities between genotypes along the genomes in 50 SNP-windows (Figure 2a). A maximum likelihood tree based on single copy orthologous genes also retrieved the same four genetic groups (Figure 2b). All the methods separated the P. roqueforti strains into four genetic clusters (Figures 1 and 2; Figure S1), two of which almost exclusively contained cheese strains (n = 10 and n = 9 respectively); the only exceptions were two strains, isolated from a brewery (LCP00148) and brioche (LCP02939), respectively, clustering with cheese strains, thus probably corresponding to feral strains (i.e., strains from domesticated clusters living in noncheese environments [Figures 1 and 2]). A third cluster contained both silage strains (n = 4) and food-spoiling strains (n = 4), while the fourth one contained mostly food-spoiling strains (n = 5) plus strains from lumber (n = 2) (Figures 1 and 2; Table S1). Of note, these two latter clusters corresponding to strains from other environments did not include a single cheese strain. The two cheese clusters were not the most closely related one to each other, as shown by clustering (Figure 2a), the maximum likelihood tree (Figure 2b), DAPC (Figure S1B) and the highest FST value (Table 1), suggesting independent domestication events. Moreover, cheese clusters displayed much lower genetic diversity than noncheese clusters, as shown by their small ϴ values (corresponding to 4Neμ, i.e., the product of the effective population size and the mutation rate) and more homogeneous colours in distance-based clustering (Table 1 and Figure 2a). One of the two cheese clusters displayed a particularly low level of genetic diversity (Table 1 and Figure 2) with only 0.03% polymorphic sites (i.e., only ~7,000 SNPs segregating within this cluster across the 30-Mb genome), and a lack of recombination footprints (i.e., a higher level of linkage disequilibrium, as shown by the more gradual decay of r2 values (Figure S2), and by the large single-colour blocks along the genomes, Figure 2a,b). These findings suggest that the second cheese population is a single clonal lineage, in which a low level of polymorphism has been generated by mutations. The other cheese population also appears to lack recombination footprints, while including several clonal lineages (Figure 2). Given such a lack of recombination footprints, clustering methods free of assumptions on modes of recombination were better suited to analyse the data set. The faststructure software, which assumes random mating, nevertheless yielded similar results (Figure 1a).
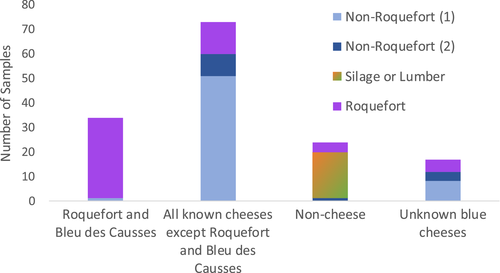
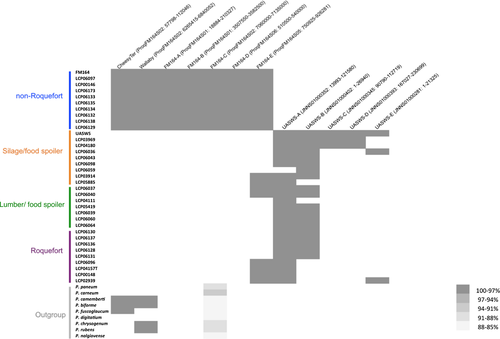
We used genome sequences to design genetic markers (Table S2) for assigning a collection of 65 strains provided by the main French supplier of P. roqueforti spores for artisanal and industrial cheesemakers, 18 additional strains from the National History Museum collection in Paris (LCP) and 31 strains from the collection of the Université de Bretagne Occidentale (UBOCC, Table S1) to the four genetic clusters. Of these 148 strains, 55 were assigned to the more genetically diverse of the two cheese clusters. The majority of these strains included strains used for Roquefort PDO cheese production (n = 30); three strains originated from Bleu des Causses cheeses (Figure 3; Table S1 and Figure S3), produced in the same area as Roquefort and using similarly long storage in caves. The remaining strains of this cluster included samples from other blue cheeses (n = 13), unknown blue cheeses (n = 5) or other environments (n = 4), the last probably associated with feral strains. Because of its main usage in Roquefort production, we refer to this cluster hereafter as the “Roquefort” population. Of the remaining 95 strains, 60 belonged to the second cheese cluster, which was less genetically diverse and contained mainly commercial strains used to produce a wide range of blue cheeses (Figure 3; Table S1 and Figure S3). This cluster was therefore named the “non-Roquefort” population. A single strain (LCP00146) in this “non-Roquefort” population had probably been sampled from a Roquefort cheese, but it did not appear phenotypically different from other strains in its genetic group; the “Roquefort” origin may, however, be dubious as no brand was recorded for this strain from an old collection. The “Roquefort” population also included 13 strains used to inoculate other types of blue cheese (e.g., Gorgonzola or Bleu d’Auvergne), but strains from these types of cheeses were more common in the “non-Roquefort” population. Of note, all the strains from the “non-Roquefort” cluster harboured Wallaby and CheesyTer (Figure 4), two large genomic regions recently shown to have been transferred horizontally between different Penicillium species from the cheese environment and conferring faster growth on cheese (Cheeseman et al., 2014; Ropars et al., 2015), whereas all the strains in the “Roquefort” cluster lacked those regions.
3.2 Two independent domestication events in Penicillium roqueforti for cheesemaking
We compared 11 demographic scenarios with ABC, simulating either a single domestication event (the most recent divergence event then separating the two cheese populations) or two independent domestication events, with different population tree topologies and with or without gene flow (Figure S4). Parameters in the scenarios modelled corresponded to the divergence dates, the strength and dates of bottlenecks and population growth, and rates of gene flow. ABC simulates sequence evolution under the various scenarios using the coalescent theory framework and compares various population statistics under a Bayesian framework between the simulation outputs and the observed data to identify the most likely scenario (Beaumont, Zhang, & Balding, 2002). The ABC results showed that the two P. roqueforti cheese populations (“Roquefort” and “non-Roquefort”) resulted from two independent domestication events (Figure 5a). The highest posterior probabilities were indeed obtained for the S4 scenario, in which the two cheese populations formed two lineages independently derived from the common ancestral population of all P. roqueforti strains (Figure 5a, model choice and parameter estimates in Figure S4 and Table S3). We inferred much stronger bottlenecks in the two cheese populations than in the noncheese populations, with the most severe bottleneck found in the “non-Roquefort” population. Some gene flow (m = 0.1) was inferred between the two noncheese populations but none with cheese populations. The bottleneck date estimates in ABC had too large credibility intervals to allow inferring domestication dates (Table S3). We therefore used the MSMC method to estimate times since domestication, considering that they corresponded to the last time there was gene flow between genotypes within populations, given the lack of recombination footprints in cheese population and the mode of conservation and clonal growth of cheese strains by humans, and given that this also corresponds to bottleneck date estimates in coalescence. The domestication for the “Roquefort” population was inferred seven times longer ago than for the “non-Roquefort” population, both domestication events being recent (~760 vs. 140 generations ago, Figure 5b,c). Unfortunately, generation time, and even generation definition, are too uncertain in the clonal P. roqueforti populations to infer domestication dates in years. In addition, the MSMC analysis detected two bottlenecks in the history of the “Roquefort” population (Figure 5c).
3.3 Isolation attempts of Penicillium roqueforti in ripening cellars or dairy environments
To investigate whether a wild P. roqueforti population occurred in ripening cellars or dairy environments that could be at the origin of the observed cheese populations, we sampled spores from the air in an artisanal cheese dairy company, ~60 km from Roquefort-sur-Soulzon, producing no blue cheese to avoid feral strains (i.e., dispersal from inoculated cheeses). We also sampled spores from the air in an abandoned ripening cellar ~70 km from Roquefort-sous-Soulzon, where Roquefort cheeses used to be produced and stored in the early 19th century. In total, 55 Petri dishes containing malt and ampicillin were left open for 6 days as traps for airborne spores. One monospore was isolated from each of the 22 Penicillium-like colonies, cultivated and identified using a taxonomically relevant marker of Penicillium species (i.e., a fragment sequence of the β-tubulin gene). No P. roqueforti strain could be identified, indicating that this species is not frequent in these environments.
3.4 Contrasting fitness traits between Penicillium roqueforti populations
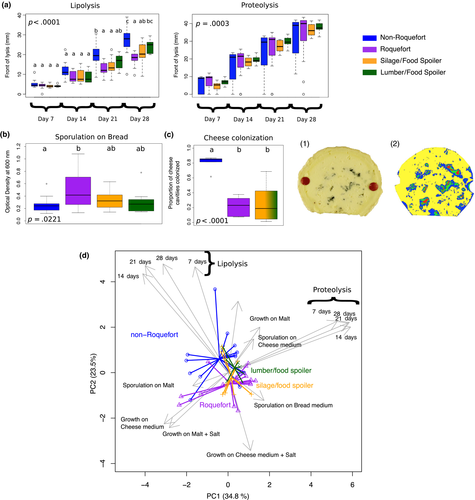
We tested whether different phenotypes relevant for cheesemaking had evolved in the two cheese clusters, relative to other populations (Figure 6; Tables S4 and S5, Figure S5). We first assessed lipolytic and proteolytic activities in the P. roqueforti populations. These activities are important for energy and nutrient uptake, as well as for cheese texture and the production of volatile compounds responsible for cheese flavours (Gillot et al., 2017; McSweeney, 2004). Lipolysis was significantly faster in the “non-Roquefort” population than in the “Roquefort” and silage/food spoiling populations (Figure 6a; Tables S4 and S5). A significant population effect was found for proteolytic activity (Figure 6a; Tables S4 and S5), with faster proteolysis activities in cheese populations; post hoc pairwise tests, however, did not have enough power to assess which pairs of populations had different proteolytic activities. Variances showed significant differences between populations (Levene test F-ratio = 5.97, df = 3, p < .0017), with the two cheese populations showing the highest variances, and with extreme values above and below those in noncheese populations (Figure 6a). Of note, proteolysis is a choice criterion for making different kinds of blue cheeses that is often showcased by culture producers (e.g., https://www.lip-sas.fr/index.php/nos-produits/penicillium-roquefortii/18-penicillium-roquefortii). This suggests that some cheese strains may have been selected for higher and others for lower proteolytic activity. Alternatively, selection could have been relaxed on this trait in the cheese populations, leading to some mutations decreasing and other increasing proteolysis in different strains, thus increasing variance in the populations.
The ability of P. roqueforti strains to produce spores may also have been selected by humans, both unwittingly, due to the collection of spores from mouldy bread, and deliberately, through the choice of inocula producing bluer cheeses. We detected no difference in spore production between the P. roqueforti populations grown on cheese medium or malt (Figure S5). However, we observed significant differences in spore production on bread medium (Figure 6b). The “Roquefort” population produced the highest number of spores and significantly more than the “non-Roquefort” population (Figure 6b; Tables S4 and S5).
Finally, we produced experimental cheeses inoculated with strains from the different P. roqueforti populations to assess their ability to colonize cheese cavities, a trait that may have been subject to human selection to choose inocula producing the most visually attractive blue cheeses. The fungus requires oxygen and can therefore sporulate only in the cheese cavities, its spores being responsible for the typical colour of blue-veined cheeses; the application of highly salted solutions followed by tin foil wrapping prevents sporulation on the surface of cheeses. Strains from the “non-Roquefort” population were the most efficient colonizers of cheese cavities (Figure 6c; Tables S4 and S5); no difference was detected between strains from the “Roquefort” and noncheese populations. Overall, cheese strains showed much larger phenotypic variation than the strains from other environments (Figure 6d).
As P. roqueforti strains were traditionally multiplied on bread loaves for cheese inoculation, they may have been subject to unintentional selection for faster growth on bread. However, growth rate on bread did not significantly differ between populations (Figure S5, Tables S4 and S5).
High salt concentrations have long been used in cheesemaking to prevent the growth of spoiler and pathogenic microorganisms. We found that the ability to grow on salted malt and cheese media decreased in all P. roqueforti populations (Figure S5, Tables S4 and S5). We found a significant interaction between salt and population factors, and post-hoc tests indicated that the “Roquefort” population was more affected by salt than the other populations (Figure S5, Tables S4 and S5).
Volatile compound production was also investigated in the two cheese populations, as these compounds are important for cheese flavour (McSweeney, 2004). We identified 52 volatile compounds, including several involved in cheese aroma properties, such as ketones, free fatty acids, sulphur compounds, alcohols, aldehydes, pyrazines, esters, lactones and phenols (Curioni & Bosset, 2002) (Figure 7). The two cheese populations presented significantly different volatile compound profiles, differing by three ketones, one alcohol and two pyrazines (Table S6). The “Roquefort” population produced the highest diversity of volatile compounds (Figure 7).
3.5 Detection of genomic regions population-specific or affected by recent positive selection
We searched nonexhaustively for footprints of additional horizontal gene transfers by mapping genomes on the two available high-quality genome assemblies, one from the “non-Roquefort” cluster and one from the silage cluster. We identified five regions present in the genomes of strains from the “non-Roquefort” population and absent from the other populations. We also detected five other genomic islands present in several P. roqueforti strains but absent from the “non-Roquefort” cheese strains (Figure 4). Nine of these 10 genomic regions were not found in the genomes of the outgroup Penicillium species analysed here and they displayed no genetic diversity in P. roqueforti. No SNPs were detected, even at synonymous sites or in noncoding regions, suggesting recent acquisitions, by horizontal gene transfer. The absence of the genomic islands in some populations and outgroups prevented running gene topology analyses designed for horizontal gene transfer analyses but were even stronger evidence for the existence of horizontal gene transfer. Only FM164-C, one of the genomic islands specific to the “non-Roquefort” population, was present in the outgroup genomes, in which it displayed variability, indicating a loss in the other lineages rather than a gain in the “non-Roquefort” population and the outgroup species (Figure 4). The closest hits in the NCBI database for genes in the 10 genomic islands were in Penicillium genomes. Most of the putative functions proposed for the genes within these genomic regions were related to lipolysis, carbohydrate or amino-acid catabolism and metabolite transport. Other putative functions concerned fungal development, including spore production and hyphal growth (Table S7). In the genomic regions specific to the “non-Roquefort” population, we also identified putative functions potentially relevant for competition against other microorganisms, such as phospholipases, proteins carrying peptidoglycan- or chitin-binding domains, and chitinases (Table S7) (Gooday, Zhu, & O’Donnell, 1992). Enrichment tests were nonsignificant, probably due to the small number of genes in these regions.
Footprints of positive selection in P. roqueforti genomes were first detected using an extension of the McDonald–Kreitman test, which identifies genes with more frequent amino-acid changes than expected under neutrality, neutral substitution rates being assessed by comparing the rates of synonymous and nonsynonymous substitutions within and between species or populations to account for gene-specific mutation rates. We ran the test with three hierarchical levels of population subdivision, contrasting two lineages having both fixed and private polymorphisms and few shared polymorphisms, as should be done in McDonald–Kreitman tests. First, no significant footprint of positive selection was detected for any gene in the whole P. roqueforti species by comparison with P. paneum. In a second test, we identified four genes as evolving under positive selection in the “non-Roquefort” population (Table S8). Two of these genes evolved under negative selection in pooled P. roqueforti populations and corresponded to a putative aromatic ring hydroxylase and a putative cyclin. Aromatic ring hydroxylases are known to be involved in the catabolism of aromatic amino acids, which are precursors of flavour compounds (Ardö, 2006; Yvon & Rijnen, 2001). In a third test, we identified a set of 15 genes as evolving under positive selection in the “Roquefort” population but not in either the noncheese P. roqueforti populations or the “non-Roquefort” population (Table S8). Interestingly, eight of these 15 genes clustered at the end of the largest scaffold (Figure 8a).
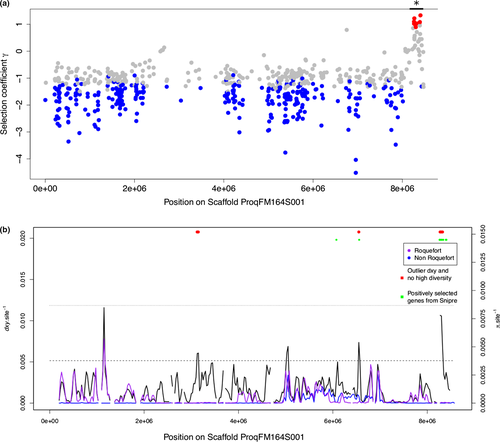
Second, we looked for regions of low diversity and high divergence between the two cheese populations as these are footprints of recent divergent selection (i.e., positive selection in one or both of the two cheese populations but for differentiated alleles). The identified regions showed a good overlap with those detected in the snipre analysis (Figure 8b); in particular, the same genomic island at the end of scaffold 1 stood out. In the regions of high divergence and low diversity, we found a significant enrichment in transcription-related genes (GO: 0000981 RNA polymerase II transcription factor activity, sequence-specific DNA binding; Fisher's exact test p < .01; Figure S6). We found a particularly high divergence on the gene coding for RPB2 subunit of RNA polymerase II with a high number of fixed differences that were specific to the “Roquefort” population; fixed differences were synonymous, suggesting that important changes concern rather the regulation level than the protein itself.
4 DISCUSSION
We report here the genetic subdivision of Penicillium roqueforti, the fungus used worldwide for blue cheese production, with unprecedented resolution, providing insights into its domestication history. Population genomics studies on strains from various substrates, including a large collection of cheeses, identified four genetically differentiated populations, two of which being cheese populations probably originating from independent and recent domestication events. One P. roqueforti cheese population included all the genotyped strains except one used for PDO Roquefort cheeses, produced in the French town of Roquefort-sur-Soulzon, where blue cheeses have been made since at least the 15th century, and probably long before (Aussibal, 1983; Labbe & Serres, 2009, 2004; Marre, 1906; Marres, 1935; Vabre, 2015, 2010). The strains from this “Roquefort” population lack the horizontally transferred Wallaby and CheesyTer genomic islands, in contrast to the “non-Roquefort” population. We thus reveal that the previous main genetic structure found based on a few microsatellite markers separating populations with and without Wallaby and CheesyTer (Ropars et al., 2014) correspond to the subdivision between a Roquefort clonal lineage and all other populations. We also show that there are four main genetic clusters in P. roqueforti, while previous studies could not distinguish between two, three or six clusters (Gillot et al., 2015; Ropars et al., 2014, 2017). The noncheese strains had previously been found to be genetically differentiated from cheese strains, but only when considering six genetic clusters (Ropars et al., 2014), and we reveal here that the noncheese strains form two genetically differentiated clusters.
We observed that the two P. roqueforti cheese populations differed in several traits important for cheese production, probably corresponding to historical differences. Indeed, the “Roquefort” population has retained moderate genetic diversity, consistent with soft selection during pre-industrial times on multiple farms near Roquefort-sur-Soulzon, where specific strains were kept for several centuries. The “Roquefort” population grew slower in cheese (Ropars et al., 2015) and had weaker lipolytic activity. Slow maturation is particularly crucial for the storage of Roquefort cheeses for long periods in the absence of refrigeration (Marre, 1906) because they are made of ewe's milk, a product available only between February and July. During storage, cheeses could become over degraded by too high rates of lipolysis, thus probably explaining the low lipolysis activity in the “Roquefort” strains. By contrast, most other blue cheeses are produced from cow's milk, which is available year-round. The “Roquefort” population showed greater sporulation on bread than the “non-Roquefort” population, which is consistent with unconscious selection for this trait when strains were cultured on bread in Roquefort-sur-Soulzon farms before cheese inoculation during the end of the 19th and beginning of the 20th centuries.
Lipolytic activity is known to impact texture and the production of volatile compounds affecting cheese pungency (Alonso, Juarez, Ramose, & Martin-Alvarez, 1987; De Llano, Ramos, Polo, Sanz, & Martinez-Castro, 1990; De Llano, Ramos, Rodriguez, Montilla, & Juárez, 1992; Martín & Coton, 2016; Thierry et al., 2017; Woo & Lindsay, 1984). The “Roquefort” and “non-Roquefort” populations showed different volatile compound profiles, suggesting also different flavour profiles. It would be of interest to evaluate the aromatic profiles of the noncheese populations to evaluate which aromatic traits (e.g., methyl ketones) have been selected in both or either of the cheese populations. The discovery of different phenotypes in the two cheese populations, together with the availability of a protocol for inducing sexual reproduction in P. roqueforti (Ropars et al., 2014), paves the way for crosses to counteract degeneration after clonal multiplication and bottlenecks, for variety improvement and the generation of diversity.
Both cheese populations were found to have gone through bottlenecks. The cheese populations were the easiest to sample compared to other environments, where P. roqueforti is relatively rarely found. It therefore seems highly unlikely that the lower genetic diversity in the cheese populations would reflect sampling biases. In particular, the least diverse cheese population was the one including the highest numbers of countries and sampled cheese types, indicating a genuine strong bottleneck. There was no particular sampling bias regarding geography either (Tables S1 and S3). The bottleneck has probably been accelerated by the current use of cheese producers buying starter cultures from a handful of companies. Larger sampling in future studies may reveal further genetic diversity in cheese strains, but the inference of strong bottlenecks is likely to hold given our broad sampling. A previous study showed that these bottlenecks, together with clonal multiplication, decreased fertility, with different stages in sexual reproduction affected in the two populations identified here as the “Roquefort” and “non-Roquefort” lineages (Ropars, Lo, et al., 2016). The “non-Roquefort” population, despite suffering from a more severe and more recent bottleneck, was found to be used in the production of all types of blue cheese worldwide, including Gorgonzola, Bleu d’Auvergne, Stilton, Cabrales and Fourme d’Ambert. The “non-Roquefort” population grows more rapidly on cheese (Ropars et al., 2015), and exhibits a greater ability to colonize cheese cavities, higher salt tolerance and faster lipolysis than the “Roquefort” population. These characteristics are consistent with the “non-Roquefort” population resulting from a very recent strong selection of traits beneficial for modern and accelerated production of blue cheese using refrigeration techniques, followed by a worldwide dissemination for the production of all types of blue cheeses. Such drastic losses of genetic diversity in domesticated organisms are typical of strong selection for industrial use by a few international firms and raise concerns about the conservation of genetic resources, the loss of which may hinder future innovation. More generally in crops, the impoverishment in genetic diversity decreases the ability of cultivated populations to adapt to environmental and biotic changes to meet future needs (Gouyon et al., 2010; Harlan, 1992; Vavilov, 1992). The PDO label, which imposes the use of local strains, has probably contributed to the conservation of genetic diversity in the “Roquefort” population (see “Cahier des charges de l'appellation d'origine protégée Roquefort,” i.e., the technical specifications for Roquefort PDO). We inferred two bottlenecks in the “Roquefort” population, more ancient than in the “non-Roquefort” population, probably corresponding to a pre-industrial domestication event when multiple local farms multiplied their strains, followed by a second bottleneck when fewer strains were kept by the first industrial societies. For other blue cheeses, even if their production was also ancient, the performant “non-Roquefort” clonal lineage could have been recently chosen to fit modern industrial production demands due to the lack of PDO rules imposing the use of local strains. However, despite a much lower genome-wide diversity in domesticated populations, proteolysis and the diversity of volatile compounds was higher in cheese than in noncheese populations. In fact, different strains with more or less rapid proteolysis and lipolysis are sold for specific blue cheese types (e.g., milder or stronger), in particular by the French LIP company (https://www.lip-sas.fr/index.php/nos-produits/penicillium-roquefortii/18-penicillium-roquefortii). Such a high phenotypic diversity within the cheese populations is consistent with diversification of usage under domestication, and in particular when different characteristics are desired according to cheese type. This has already been observed in relation to the diversification of crop varieties or breeds in domesticated animals (Parker et al., 2017; Shapiro et al., 2013).
When studying adaptation in domesticated organisms, it is often useful to contrast traits and genomic variants between domesticated and closely related wild populations to determine the nature of the adaptive changes occurring under artificial selection (Swanson-Wagner et al., 2012; Xue, Bradbury, Casstevens, & Holland, 2016). The only known noncheese populations of P. roqueforti occur essentially in human-made environments (silage, food and lumber), consistent with the specific adaptation of these populations to these environments. The two noncheese populations were inferred to have diverged very recently, and displayed footprints of recombination and marked differentiation from the cheese populations. Domesticated populations are expected to be nested within their source population because they recently diverged from a subset of individuals within the source population (e.g., Matsuoka et al., 2002); in our case cheese populations were not nested within noncheese populations, suggesting that, despite our considerable sampling efforts, we have been so far unable to identify the wild population that is the most closely related to cheese strains. We have included in our study all the noncheese strains available in public collections worldwide and we have been unsuccessful at isolating further noncheese P. roqueforti strains other than in silage or spoiled food. Identifying and further sampling P. roqueforti strains in genuine wild environments would allow further inference but is highly challenging. The high level of diversity and inferred demographic history of P. roqueforti indicate that most food-spoiling strains belong to differentiated populations and are not feral cheese strains. In addition, no single cheese strain was found in the food-spoiling and silage populations. This was shown by both genome sequences and by the genotyping of a larger number of strains using a few selected markers, in the present study and based on microsatellite markers in a previous work (Ropars et al., 2017). Consequently, P. roqueforti spores from blue cheeses may, rarely, spoil food and food-spoiling and silage strains are not used for cheesemaking nor recombine with cheese strains. Such a lack of incoming gene flow into cheese populations has allowed trait differentiation in cheese strains as expected under domestication.
It came as a surprise that the two noncheese populations split more recently from each other than from the cheese lineages. In particular, the “non-Roquefort” population diverged the earliest from the unidentified ancestral population, and this has occurred long ago probably before blue cheese invention and therefore in another environment than cheese. Much more recently, in industrial times, cheesemakers and then spore producers have probably only kept the most performant clonal lineage of this population for cheesemaking, losing most of the initial diversity, as indicated by the very strong and recent bottleneck inferred in this lineage. This loss in genetic diversity has probably been accelerated by the current practice of cheesemakers buying spores from just a few companies. Possible scenarios to explain the existence of two separated clusters thriving in food and silage differentiated from cheese strains include the very recent adaptive differentiation of a population from silage on human food or vice versa. The finding that silage strains are only found in one cluster suggests an adaptation to this ecological niche, although experiments will be required to test this hypothesis. Food spoiling strains are, in contrast, found in three clusters and may thus not constitute a specific population adapted to this environment and may instead represent migrants from several populations belonging to other ecological niches. “Lumber/food spoiler” and “silage/food spoiler” clusters may alternatively represent populations thriving in as yet unidentified environments, dispersing to silage and food. Another hypothesis would be a single domestication event for cheesemaking before the divergence of the four lineages, followed by an escape and subsequent differentiation of the “lumber/food spoiler” and “silage/food spoiler” lineages in other human-related habitats. However, this hypothesis would not predict such high genetic diversity in “the lumber/food spoiler” and “silage/food spoiler” populations, and in particular the similar nucleotidic diversity levels in the two noncheese populations as in the P. carneum and P. paneum outgroups. Given the very low genetic diversity in the cheese populations, coalescence events occurred recently in the past, preventing tests of the occurrence of bottlenecks in the common ancestor of the four P. roqueforti populations.
The history of blue cheese production may provide circumstantial clues to the origin of P. roqueforti cheese populations. Indeed, the first blue cheeses probably resulted from the sporadic accidental contamination of cheese with spores from the environment, such as mouldy food. However, this would not be consistent with the demographic history inferred here for cheese and food-spoiling strains, as the cheese strains were not found to be nested within the food-spoiling strains, some of which originated from mouldy bread. Furthermore, old French texts suggest that the blue mould colonized the cheese from within (Labbe & Serres, 2009, 2004; Vabre, 2015), which would indicate that the milk or curd was contaminated. French cheese producers began to inoculate cheeses with P. roqueforti spores from mouldy rye bread at the end of the 19th century (Labbe & Serres, 2009, 2004; Vabre, 2015). Breads were specifically made with a 2:1 mixture of wheat and rye flour and were baked rapidly at high temperature (500°C), to yield a protective crust, around a moist, undercooked interior (Aussibal, 1983; Marre, 1906); the mould developed from the inside of the bread after 1–5 months in the Roquefort caves (Labbe & Serres, 2009, 2004; Vabre, 2015). Surveys of the microorganisms present in their caves (Chaptal, 1789; Marcorelle & Chaptal, 1833; Marre, 1906) and our unsuccessful attempts to obtain samples from a maturing cellar suggest that P. roqueforti spores did not originate from the caves, which were nevertheless crucial due to the ideal conditions provided for P. roqueforti development (Marre, 1906). Bread may have been colonized from the environment or from rye flour if the source P. roqueforti population was a rye endophyte or pathogen. This last hypothesis would be consistent with the lifestyle of many Penicillium species, which live in close association with plants, often acting as plant pathogens or necrotrophs (Ropars, Vega, et al., 2016), and with the occurrence of a P. roqueforti population in lumber and silage. In fact, a recent study reports the finding of P. roqueforti as an endophyte and could be inoculated on wheat (Ikram et al., 2018), although species identification should be checked with more powerful markers. If this hypothesis is correct, then cheeses may historically have become contaminated with P. roqueforti from fodder during milking.
Comparison between noncheese and cheese populations allowed us to identify specific traits and genes that have been under selection in cheese as opposed to other environments. Furthermore, the two independently domesticated P. roqueforti cheese populations, exhibiting different traits, represent a good model for studying the genomic processes involved in adaptation. We could not run analyses of selective sweep detection based on local decrease in genetic diversity in the genomes; indeed, because of the clonality of cheese populations, the whole genome will have hitchhiked with any selected locus. This effect has probably contributed to the strong bottlenecks. We were nevertheless able to identify candidate genes and evolutionary mechanisms potentially involved in adaptation to cheese in P. roqueforti. The horizontally transferred CheesyTer genomic island probably contributes to the faster growth of the strains identified here as constituting the “non-Roquefort” population (Ropars et al., 2015). Indeed, CheesyTer includes genes with putative functions involved in carbohydrate utilization (e.g., β-galactosidase and lactose permease genes) that are specifically expressed at the beginning of cheese maturation, when lactose and galactose are available. This horizontal gene transfer may thus have been involved in adaptation to recently developed industrial cheese production processes in the “non-Roquefort” population, conferring faster growth. We also identified additional genomic islands specific to the “non-Roquefort” population, probably acquired recently and including genes putatively involved in fungal growth and spore production. In the genomic islands specific to the cheese populations, several genes appeared to be involved in lipolysis, carbohydrate or amino-acid catabolism, and metabolite transport, all of which are important biochemical processes in the development of cheese flavour. In the “Roquefort” population, a genomic region harbouring genes with footprints for positive selection included several genes encoding proteins potentially involved in aromatic amino-acid catabolism corresponding to precursors of volatile compounds. Further studies are required to determine the role of these genes in cheese flavour development.
In conclusion, we show that P. roqueforti cheese populations represent genuine domestication. The domestication process in cheese fungi has been more recent and different from those in emblematic crops or animals. Nevertheless, we did observe strong genetic differentiation from noncheese populations, strong bottlenecks and trait differentiation with probable benefits for cheese production. This suggests genuine domestication, as has been reported previously in other fungi (Almeida et al., 2014; Baker et al., 2015; Gallone et al., 2016; Gibbons et al., 2012; Gonçalves et al., 2016; Libkind et al., 2011; Sicard & Legras, 2011), and defined as “the genetic modification of a species by breeding it in isolation from its ancestral population in an effort to enhance its utility to humans” (Gibbons & Rinker, 2015). Furthermore, a previous study has shown that the “non-Roquefort” strains have acquired genes conferring better growth in cheese (Ropars et al., 2015). Our study revealed genetic divergence of cheese populations from noncheese populations, as well as the evolution of specific traits, with beneficial characteristics for cheese production. These findings therefore indicate the occurrence of domestication, a special case of adaptive divergence. We found that gene flow was prevented by clonality of cheese lineages and lack of migration between cheese and noncheese populations, and that adaptation occurred on several traits beneficial for cheese production (lipolysis, proteolysis, spore production, volatile compound production, growth in salted cheese, cheese cavity colonization ability). Genomic footprints of adaptation were found in terms of rapid amino-acid changes and horizontal gene transfers. The two independent domestication events identified here interestingly represent adaptations to different production modes. Our findings concerning the history of P. roqueforti domestication thus shed light on the processes of adaptation to rapid environmental change, but they also have industrial implications and raise questions about the conservation of genetic resources in the agri-food context.
ACKNOWLEDGMENTS
This work was supported by the ERC starting grant GenomeFun 309403 awarded to T.G., the ANR FROMA-GEN grant (ANR-12-PDOC-0030) to A.B., an “Attractivité” grant from Paris-Sud University to A.B. and the ANR FUNGADAPT (ANR-19-CE20-0002) awarded to M.C., E.C., J.R., A.B. and T.G. We thank Kamel Soudani for help with image analysis and Aurélien Tellier for advice concerning ABC analyses. We are grateful to Coralie Benel and Francis Roujon of GAEC Le Lévejac for assistance with cheesemaking and Paul Villain for experimental help. Sequencing was performed at GenoToul INRA platform. We thank INRA and MNHN for granting access to four genomes sequenced with the help of Joëlle Dupont, Sandrine Lacoste, Yves Brygoo and Jeanne Ropars in the framework of the ANR “Food Microbiomes” project (ANR-08-ALIA-007-02) coordinated by Pierre Renault.
AUTHOR CONTRIBUTIONS
T.G. and A.B. acquired the funding, designed and supervised the study. S.L. and A.S. produced the genomes. E.D., A.B. and R.dlV. analysed the genomes. E.D., S.L., J.R., A.S., M.C., A.T., E.C., M.L.P. and D.R. performed the experiments. E.D., A.B. and T.G. analysed the data from the experiments. E.D., A.B. and A.F. performed ABC analyses. E.D. and T.G. wrote the manuscript with contributions from the other authors.
Open Research
DATA AVAILABILITY STATEMENT
Whole genome sequences: European Nucleotide Archive PRJEB20132. DNA fragments: European Nucleotide ArchivePRJEB20413. Description of the isolates used in this study: Dryad https://doi.org/10.5061/dryad.n5tb2rbrw. All phenotypic measures: Dryad https://doi.org/10.5061/dryad.n5tb2rbrw. Volatile compounds quantities: Dryad https://doi.org/10.5061/dryad.n5tb2rbrw. Primer sets designed in this study: Dryadhttps://doi.org/10.5061/dryad.n5tb2rbrw.




