Comparative population genomics reveals key barriers to dispersal in Southern Ocean penguins
Abstract
The mechanisms that determine patterns of species dispersal are important factors in the production and maintenance of biodiversity. Understanding these mechanisms helps to forecast the responses of species to environmental change. Here, we used a comparative framework and genomewide data obtained through RAD-Seq to compare the patterns of connectivity among breeding colonies for five penguin species with shared ancestry, overlapping distributions and differing ecological niches, allowing an examination of the intrinsic and extrinsic barriers governing dispersal patterns. Our findings show that at-sea range and oceanography underlie patterns of dispersal in these penguins. The pelagic niche of emperor (Aptenodytes forsteri), king (A. patagonicus), Adélie (Pygoscelis adeliae) and chinstrap (P. antarctica) penguins facilitates gene flow over thousands of kilometres. In contrast, the coastal niche of gentoo penguins (P. papua) limits dispersal, resulting in population divergences. Oceanographic fronts also act as dispersal barriers to some extent. We recommend that forecasts of extinction risk incorporate dispersal and that management units are defined by at-sea range and oceanography in species lacking genetic data.
1 INTRODUCTION
During the last 2.58 million years, periods of global warming and cooling have shaped the evolutionary trajectories of species around the globe (Hewitt, 2004), with those inhabiting the polar regions having been subject to the most extreme climatic shifts. The Anthropocene (Lewis & Maslin, 2015) will be characterized by changes outside of this natural variability, contributing to the sixth global mass extinction event (Barnosky et al., 2011) and altering the evolutionary pressures acting on species. The movement of individuals among populations within species can distribute adaptive genetic variants, potentially aiding resilience to changing conditions, and preventing populations from diverging through genetic drift (Slatkin, 1987). Barriers to dispersal can isolate populations, resulting in allopatric speciation if both populations are viable, or extirpation if they are not. For successful biodiversity conservation, understanding dispersal is key to distinguishing breeding populations and defining management units (Funk, McKay, Hohenlohe, & Allendorf, 2012), and to enable accurate forecasts of local or global extinction risk in response to habitat change.
Seabirds are highly threatened (Croxall et al., 2012), mobile and capable of dispersing large distances with few apparent abiotic barriers to movement, yet most are characterized by high levels of philopatry (Coulson, 2002). This appears to be the case for Antarctic and sub-Antarctic penguins, but they present a significant logistical challenge for studying dispersal (defined here as movement away from natal colonies to alternate breeding sites), as the vast majority of colonies are in remote locations. Banding studies initially suggested a high degree of philopatry in many species (Weimerskirch, Jouventin, Mougin, Stahl, & Van, 1985), and, until recently (Jenouvrier, Garnier, Patout, & Desvillettes, 2017), forecasts of extinction risk had not considered the potential buffering effect of dispersal (Cimino, Lynch, Saba, & Oliver, 2016; Jenouvrier et al., 2014). Genetic analyses (Clucas, Younger et al., 2016; Freer et al., 2015; Roeder et al., 2001; Younger, Clucas, et al., 2015, 2017), observations of colony movements (LaRue, Kooyman, Lynch, & Fretwell, 2015) and fluctuations in colony size (Kooyman & Ponganis, 2017) indicate that dispersal may be common. However, hydrographic features are thought to act as barriers to dispersal in a handful of sub-Antarctic and temperate penguin species (see Munro & Burg, 2017, for a review). A comprehensive study of dispersal barriers in Antarctic and sub-Antarctic penguins is therefore needed to clarify their potential evolutionary responses to increasing threats (Trathan et al., 2015), improve the accuracy of estimates of extinction risk, identify effective management strategies for their conservation and shed light upon the mechanisms that prevent or promote dispersal in these charismatic seabirds.
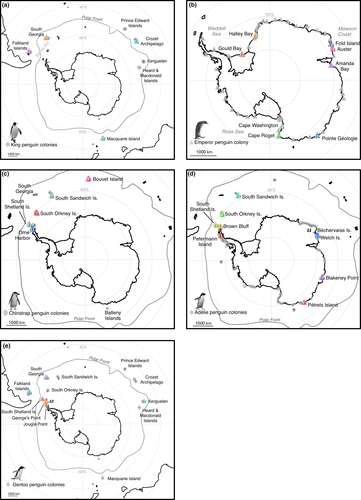
Here, we examine the relative importance of different ecological and evolutionary factors in determining dispersal and population differentiation in the Aptenodytes and Pygoscelis genera using a comparative population genomic framework. By comparing range-wide patterns of genetic differentiation in ecologically divergent species with overlapping distributions and shared ancestry, we aim to tease apart the mechanisms which have led to the distributions of genetic diversity that we see today. Many factors are known to influence the patterns of dispersal in seabirds (Friesen, Burg, & McCoy, 2007), and our comparative framework is designed to examine the relative importance of several of these factors to dispersal: oceanographic fronts, ephemerality of breeding habitat, geographic continuity of breeding habitat, and at-sea range. We generated robust single nucleotide polymorphism (SNP) data sets for five species of penguin, covering the majority of each species’ range (Figure 1), and compared the levels of dispersal within each species.
2 METHODS
2.1 Study species
Our study focused on Aptenodytes and Pygoscelis, which are sister genera within the penguin (Spheniscidae) family (Gavryushkina et al., 2017). We included all species within these genera: emperor (Aptenodytes forsteri), king (A. patagonicus), Adélie (Pygoscelis adeliae), chinstrap (P. antarctica), and gentoo penguins (P. papua). The breeding distributions of all species are shown in Figure 1. Emperor penguins are restricted to the Antarctic continent and breed primarily on sea ice, with a relatively continuous breeding distribution around the continent (Fretwell et al., 2012). Adélie penguins have a breeding distribution that encompasses ice-free areas of the Antarctic continent's coastline, along with several islands, all south of the Antarctic Polar Front (Schwaller, Southwell, & Emmerson, 2013). Chinstrap penguins similarly breed on ice-free areas but are not widespread on the Antarctic continent—colonies are found only at the Antarctic Peninsula and various islands south of the Antarctic Polar Front (Borboroglu & Boersma, 2013). Gentoo and king penguins have more northerly distributions, and both of these species have colonies both north and south of the Antarctic Polar Front, a potential dispersal barrier (Clucas, Younger et al., 2016; Friesen, 2015; Munro & Burg, 2017). King penguins are found exclusively on islands, whereas gentoo penguins also breed on the Antarctic Peninsula (Borboroglu & Boersma, 2013). We aimed to encompass as much of these breeding ranges as possible in our study design (Figure 1).
2.2 Sampling and sequencing
Blood or tissue samples were collected from up to 16 individuals per colony across a large part of the range of each of the study species (Figure 1). We sampled a single representative colony for those islands or archipelagos with multiple colonies. Colony names, collection dates and tissue types are provided in Supporting information Table S1. Further details of the tissues collected from Adélie penguins at Béchervaise Island, Welch Island, Blakeney Point and Pétrels Island can be found in Ref. (Younger, Emmerson, Southwell, Lelliott, & Miller, 2015). Details of the tissue samples collected from emperor penguins at Halley Bay, Fold Island, Auster, Amanda Bay and Pointe Géologie can be found in Ref. (Younger, van den Hoff, Wienecke, Hindell, & Miller, 2016; Younger, Clucas, et al., 2015). All other samples were blood samples. To take blood, penguins were held with the flippers restrained and the head placed under the arm of the handler, or they were wrapped in cushioned material covering the head and preventing movement, to minimize stress during handling (Le Maho et al., 1992). A second handler took up to 1 ml blood from the brachial, intertarsal or jugular vein using a 25-G or 23-G needle and 1-ml syringe, after cleaning the area with an alcohol swab. Total restraint time was generally 2 to 3 min. All field activities were conducted under appropriate permits and were subject to independent ethical review. Samples were either stored frozen for transport, stored frozen in ethanol or Queen's Lysis buffer (Seutin, White, & Boag, 1991) for transport, or stored in RNAlater (Life Technologies) and transported at ambient temperature. All samples were then stored frozen at −20°C in Australia or the UK.
DNA was extracted from blood and tissue samples using QIAGEN DNeasy Blood and Tissue Kits. The digestion step was modified to include 40 μL proteinase K (at 20 mg/ml) and extended to 3 hr for blood samples. Details of the modifications made to the protocols for tissue samples are available in Younger, Emmerson, et al. (2015) and Younger, Clucas, et al. (2015). All samples were treated with 1 μL RiboShredder (Epicentre) to reduce RNA contamination, and DNA was visualized on a 1% agarose gel to confirm high molecular DNA was present. DNA concentration and purity were measured on a Qubit and NanoDrop (Thermo Fisher Scientific), respectively.
To identify genomewide SNPs for each species, we used standard restriction site-associated DNA sequencing (RAD-Seq) (Baird et al., 2008) with individual barcoding and the Sbf1 restriction enzyme. The NERC Biomolecular Analysis Facility at Edinburgh Genomics (https://genomics.ed.ac.uk) performed the library preparation and sequencing as described by Gonen et al. (2014) following Etter, Bassham, Hohenlohe, Johnson, and Cresko (2011). In short, 250 ng of DNA per individual was digested with Sbf1-HF (NEB) and then ligated to barcoded P1 adapters. Individuals were multiplexed into 18 libraries consisting of 19–23 barcoded individuals and were sheared into fragments of <300–400 bp. The number of individuals to multiplex into each lane was estimated using the cutting frequency of the Sbf1 enzyme and a genome size of 1.2 Gb, as per the published Adélie and Emperor penguin reference genomes at the time of sequencing. Subsequent analyses have shown their genomes are likely to be 1.25 and 1.39 Gb, respectively (Li et al., 2014). Size selection was performed by gel electrophoresis. Libraries were blunt-ended (NEB Quick Blunting Kit) and A-tailed before P2 adapters (IDT) were ligated. Enrichment PCR and purification with AMPure beads were performed before libraries were checked for size and quantity using Qubit and qPCR assays. Each library was then sequenced in a lane of an Illumina HiSeq 2500 using 125 base paired-end reads in high output mode (v4 chemistry).
2.3 Bioinformatics
Read quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Demultiplexing, removal of reads with adapter contamination and trimming to 113 bp were performed with process_radtags from the stacks pipeline v1.35 (Catchen, Amores, Hohenlohe, Cresko, & Postlethwait, 2011; Catchen, Hohenlohe, Bassham, Amores, & Cresko, 2013). We also used process_radtags to remove any read pairs with uncalled bases, a low quality score and/or a barcode or cut site with more than one mismatch. King and emperor penguin reads were aligned to the published emperor penguin reference genome (http://gigadb.org/dataset/100005; scaffold-level assembly) while Adélie, chinstrap and gentoo penguin reads were aligned to the published Adélie penguin reference genome (http://gigadb.org/dataset/100006; scaffold-level assembly) using bwa-mem (Li, 2013). Terminal alignments were prevented by enforcing a clipping penalty of 100, and reads with more than five mismatches, multiple alignments and/or more than two indels were removed using a custom python script (filter.py, available from https://doi.org/10.5061/dryad.7c0q8). PCR duplicates were removed with Picard Tools (http://broadinstitute.github.io/picard).
We called and filtered SNPs separately for each species using the Stacks pipeline, following many of the suggestions outlined in Benestan et al. (2016). All of the settings and filters were applied in the same way for each species. We ran the modules of the pipeline separately: pstacks – cstacks – sstacks – rxstacks – cstacks – sstacks – populations. Briefly, in pstacks we required a minimum stack depth (-m) of six reads mapping to the same location and used the bounded SNP model (significance level of α = 0.05, upper bound = 0.1, lower bound = 0.00041 corresponding to the highest sequencing error rate recorded by phiX spikes). We found that setting the stack depth to six gave sufficient numbers of polymorphic loci in all species after downstream steps had been performed, while maintaining sufficient (≥ 6X) depth at each locus to reliably call heterozygotes. In cstacks, we used all the individuals in each species to build the catalog of loci. In rxstacks, we removed confounded loci with a conservative confidence limit of 0.25, removed excess haplotypes from individuals and also removed any loci with a mean log likelihood <−10. We filtered SNPs in the populations module to retain a single random SNP per RAD-tag, remove any SNPs with a minor allele frequency (MAF) < 0.01 or heterozygosity > 0.5, remove any loci that were not present in all colonies and remove any SNPs that were not genotyped in at least 80% of individuals per colony. In the adegenet package (Jombart, 2008; Jombart & Ahmed, 2011) in R (R Core Team, 2013), we calculated whether SNPs were in Hardy–Weinberg equilibrium (HWE) in each colony and used vcftools v0.1.13 (Danecek et al., 2011) to calculate mean coverage for each SNP. SNPs were removed if they were out of HWE in > 50% of the colonies or had a mean coverage greater than twice the standard deviation for the species. For the number of SNPs before and after filtering, please refer to Supporting information Table S3. pgdspider v2.0.8.2 (Lischer & Excoffier, 2012) was used to convert the vcf file into other formats for further analyses.
2.4 Outlier loci detection
We identified SNPs that were potentially under selection in each species using the FST outlier method in bayescan v2.1 (Foll & Gaggiotti, 2008). These loci were removed prior to coalescent-based or population genetic analyses that assume loci are evolving neutrally. With this aim, the high false-positive rate associated with BayeScan (Lotterhos & Whitlock, 2014) was not a concern, and its power to detect loci genuinely under selection under a range of demographic scenarios was advantageous (Lotterhos & Whitlock, 2014). We set a conservative prior on the odds of neutrality (for every five loci, our prior expectation is that one is under selection) to identify all loci that could potentially be under selection. We deemed q-values <0.1 to be significant, meaning that one in ten loci identified was expected to be a false-positive neutral locus (Lotterhos & Whitlock, 2014; Storey & Tibshirani, 2003). SNPs that were identified to be putatively under selection were removed using VCFtools. We refer to the remaining SNPs as “neutral SNP data sets,” and those with the full complement before outliers were removed as the “total SNP data set,” although these definitions are applicable only to gentoo penguins, as these were the only species in which a large number of outliers were identified (Table 1).
| Species | N | Total SNP count | Median SNP depth | Number outlier loci | Neutral SNP count | Mean MAF (std dev) | FST (95% CI) | p | Significant pairwise FST |
|---|---|---|---|---|---|---|---|---|---|
| King | 64 | 5,154 | 27.7 | 0 | 5,154 | 0.075 (0.10) | 0.002 (0.001–0.003) | 0.001 | 4/6 (67%) |
| Emperor | 110 | 4,600 | 32.1 | 4 | 4,596 | 0.078 (0.09) | 0.003 (0.002–0.003) | 0.001 | 17/28 (61%) |
| Chinstrap | 44 | 12,921 | 26.7 | 0 | 12,921 | 0.078 (0.10) | 0.002 (0.001–0.003) | 0.003 | 3/10 (30%) |
| Adélie | 87 | 3,872 | 27.9 | 0 | 3,872 | 0.062 (0.09) | 0.002 (0.001–0.004) | 0.002 | 7/36 (19%) |
| Gentoo | 69 | 10,560 | 27.0 | 452 | 10,108 | 0.091 (0.11) | 0.217 (0.213–0.221) | 0.001 | 15/15 (100%) |
- FST: AMOVA-based FST estimates from the neutral data set with 95% confidence interval; MAF: mean minor allele frequency of the neutral data set; N: number of individuals; p: p-value for the FST estimate; Significant pairwise FST: proportion of significant pairwise FST comparisons between colonies for each species.
2.5 Contemporary population structure and summary statistics
The number of private alleles in each colony was calculated with the populations module in Stacks, using the total SNP data sets for each species. We calculated the observed (HO) and expected (HS) heterozygosity for each colony with the neutral SNP data sets using genodive v2.0b27 (Meirmans & Van Tienderen, 2004).
We also used GenoDive to calculate AMOVA-based FST estimates for each species using 999 permutations to calculate significance, and the Weir and Cockerham unbiased weighted FST estimator (Weir & Cockerham, 1984) between all pairs of colonies within each species. This measure has been shown to be robust to small sample sizes when FST is low (Willing, Dreyer, & van Oosterhout, 2012). Significance was calculated using 10,000 permutations of the data and corrected for multiple tests using the sequential goodness of fit method (SGoF+) (Carvajal-Rodriguez & de Uña-Alvarez, 2011). For gentoo colonies, we measured FST using both the neutral SNP data set and the total SNP data set, whereas for the other four species, the neutral data sets were used, since the number of outlier loci was very small (Table 1).
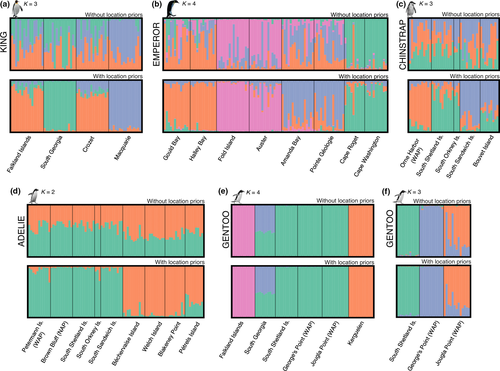
We used three different clustering methods for each species using the neutral SNP data sets: the Bayesian clustering algorithm employed by the program structure v2.3.4 (Pritchard, Stephens, & Donnelly, 2000), principal component analysis (PCA), and discriminant analysis of principal components (DAPC) (Jombart, Devillard, & Balloux, 2010). structure uses a Bayesian clustering approach with a Markov chain Monte Carlo (MCMC) sampling procedure, which results in estimates of the membership coefficients of each individual to each of the inferred clusters, effectively identifying genetic populations and then assigning individuals to those populations. For all taxa, we used the admixture model with correlated allele frequencies and ran the model both with and without supplying sampling locations as priors, to detect subtle versus strong population structure. In each case, we first ran the model for 100,000 generations, discarding the first 50,000 as burn-in, setting K (the number of clusters) to one but allowing lambda to vary in order to estimate the species-specific value of lambda to use. For subsequent runs, the species-specific value of lambda was set and the number of clusters was allowed to vary from K = 1 to K = N, where N was the number of colonies sampled for that species. Each analysis was run for 150,000 generations, discarding the first 50,000 as burn-in, and repeated ten times from a different random seed. We used structure harvester web v0.6.94 (Earl, 2012) to compare replicates and prepare files for clumpp v1.1.2 (Jakobsson & Rosenberg, 2007), which aligns the results from replicate runs of structure to check for multimodality and calculates the average membership coefficients of each individual to each cluster, ready for visualization with distruct v1.1 (Rosenberg, 2004). Mostly, we did not use the results from the Evanno method for estimating the “true” number of clusters in the data (Evanno, Regnaut, & Goudet, 2005), as it is not defined for K = 1 and it was often hard to find biological meaning in the results. This is not unexpected, as the Evanno method has been shown to perform poorly for scenarios of moderate to low genetic differentiation (Waples & Gaggiotti, 2006). There is a large body of literature suggesting that it is unrealistic to expect that a “true” value of K exists for any given data set (Benestan et al., 2016; Gilbert et al., 2012; Janes et al., 2017) and that in order to gain insight into different levels of genetic structure, it is better to view and report multiple K-values. Therefore, we discuss our results for multiple values of K for each taxon, to fully understand the levels of structure in the data.
Secondly, we performed PCAs for each species using the neutral SNP data sets. Allele frequencies were scaled and centred, and missing values were replaced with the species’ mean allele frequency using the scaleGen function from adegenet. PCA was computed with the dudi.pca function from the ade4 v1.7-11 package. The first three PCs were plotted against one another, but only the first two are shown, as population structure was not visible beyond PC2 in all species.
Finally, we used DAPC, which can be used to describe genetic clusters by creating synthetic variables (discriminant functions) that maximize the variance among clusters while minimizing the variance within them. When genetic differentiation is moderate to strong, individuals can be assigned to clusters using successive K-means clustering—the find.clusters function in adegenet (Jombart, 2008; Jombart & Ahmed, 2011)—before DAPC, thus negating the a priori assignment of individuals to groups determined by their sampling location. However, in all species other than gentoo penguins, the successive K-means clustering suggested K = 1 was most likely (the Bayesian Inference Criterion was at its minimum at K = 1) and so DAPC was performed when individuals were grouped by their colony of origin. Cross-validation with 1,000 replicates was used to determine the appropriate number of PCs to retain, and the posterior membership probability of each individual to each colony was plotted to determine how well individuals were assigned back to their colony of origin.
2.6 Phylogenetics and species delimitation of gentoo penguins
To investigate the phylogeographic relationships among gentoo penguin colonies, we used the coalescent species tree approach implemented in the snapp package (Bryant, Bouckaert, Felsenstein, Rosenberg, & RoyChoudhury, 2012) in beast v.2.4.0 (Bouckaert et al., 2014). SNAPP infers species trees from unlinked biallelic markers, such as SNPs. The method calculates species tree likelihoods directly from the data by estimating the probability of allele frequency change across nodes, thus avoiding the necessity of finding and combining individual gene trees. Nevertheless, the method is highly computationally demanding; therefore, we selected two random individuals (i.e., four haplotypes) per colony to include in the analysis and repeated the analysis twice with different individuals to ensure reproducibility. The neutral data set was used, and loci that were no longer polymorphic in the reduced set of individuals were removed, leaving 6,868 and 6,754 SNPs. The forward and backward mutation rates (u and v) were calculated from the data rather than estimated as part of the MCMC (note that in SNAPP analyses, the mutation rate (μ) is fixed at 1 and divergence times are not estimated). The MCMCs were run for three million generations with the first 10% discarded as burn-in. We monitored the traces for convergence using tracer v1.6 (Rambaut & Drummond, 2007), and when ESSs for all parameters were large (> 300) and the traces had reached stationarity, we concluded the analyses. densitree v2.0.1 was used to visualize the posterior distributions of topologies as cloudograms, hence allowing for a clear depiction of uncertainty in the topology.
To investigate whether described (Stonehouse, 1970) and putative (de Dinechin et al., 2012) subspecies of gentoo penguins are reciprocally monophyletic, and therefore taxonomically valid, we used raxml v8.2.7 (Stamatakis, 2014) to infer maximum-likelihood phylogenies among the full complement of gentoo penguin individuals using the total SNP data set. An ascertainment bias correction was applied to the likelihood calculations, as recommended when using SNPs to account for the lack of invariant sites (Leaché, Banbury, Felsenstein, de Oca, & Stamatakis, 2015). When using an ascertainment bias correction, all potentially invariant sites must be removed from the data set. An alignment site consisting of only heterozygotes and homozygotes for a single allele (e.g., Rs and As with no Gs) is considered potentially invariant by RAxML; therefore, we filtered out such sites using the Phrynomics R script (https://rstudio.stat.washington.edu/shiny/phrynomics/). After this filtering step, 5,871 SNPs remained in the data set. We conducted 20 independent maximum-likelihood tree inferences and then drew bootstrap supports from 1,000 replicates onto the best scoring topology. All searches were conducted under the GTRGAMMA nucleotide substitution model.
To further investigate the validity of the proposed subspecies divisions within gentoo penguins (de Dinechin et al., 2012; Stonehouse, 1970), we compared species delimitation models using the Bayes factor delimitation method BFD* (Leaché, Fujita, Minin, & Bouckaert, 2014) as implemented within the snapp package (Bryant et al., 2012) in BEAST 2.4.3 (Bouckaert et al., 2014). The BFD* method estimates the marginal likelihoods of competing species delimitation models using path sampling, such that the models can be ranked by their marginal likelihoods, with Bayes factors then used to assess support for the model rankings. A total of 16 individuals were included in the analysis, such that each putative subspecies unit within our species delimitation models had a minimum of four representatives. Therefore, we included four individuals from each of the following: (a) the Falkland Islands, (b) Kerguelen, (c) South Georgia and (d) the South Shetland Islands/western Antarctic Peninsula. Four species delimitation models were examined: (a) the currently recognized taxonomy of northern (Pygoscelis papua papua) and southern gentoos (P. p. ellsworthii); (b) the taxonomy suggested by mitochondrial DNA studies (Clucas et al., 2014; de Dinechin et al., 2012; Vianna et al., 2017), in which the Falkland Islands and Kerguelen are split whereas South Georgia and the Antarctic Peninsula are grouped together; (c) the four groups suggested by our DAPC and Structure analyses; and (d) all colonies grouped except for Kerguelen. We used the neutral SNP data set for the BFD* analysis, as required for coalescent-based methods. SNPs that were no longer polymorphic within the reduced data set of individuals were removed, leaving 8,116 SNPs. In SNAPP, we specified a gamma prior distribution for the theta parameter, with alpha = 1 and beta = 1000, for a prior mean of 0.001 on theta. This value was chosen to reflect the 0.1% sequence divergence observed among gentoo penguin alleles known to belong to a single subspecies. The speciation rate, lambda, was fixed at 495, as calculated from the tree height and number of tips using the Python script yule.py (https://github.com/joaks1/pyule). Initial exploratory path sampling analyses were conducted to determine an appropriate number of steps to produce stability of the marginal likelihood, with 72 deemed more than sufficient. Path sampling analyses of 72 steps were then conducted for each of four species delimitation models with 100,000 MCMC generations following 10,000 pre-burn-in.
Finally, to estimate divergence times among the major gentoo lineages, we performed time-calibrated Bayesian phylogenetic analyses of published mitochondrial hypervariable region (HVR) sequences for 47 gentoo penguins, with the topology constrained to that inferred using the SNP data set, using BEAST 2.4.4 (Bouckaert et al., 2014). Representatives from Kerguelen (n = 7), the Falkland Islands (n = 10), South Georgia (n = 10), the South Shetlands (n = 10) and the West Antarctic Peninsula (n = 10) were included, along with chinstrap penguins (n = 3) as the outgroup. Accession numbers for these sequences can be found in the Supplementary Information. The nucleotide substitution model was specified as HKY with four gamma categories. We used the Yule tree prior with a strict molecular clock calibrated with the divergence of chinstrap and gentoo penguins, estimated at 3.17 Ma (95% HPD: 1.69 - 4.94) (Gavryushkina et al., 2017). The topology was constrained to that resolved by our RAxML and SNAPP analyses of the full data set. Two independent analyses, from different random number seeds, were performed to ensure reproducibility of the posterior distribution. The MCMCs were run for 150 million generations and convergence of the posteriors confirmed using tracer v1.6 (Rambaut & Drummond, 2007). A maximum clade credibility tree with mean node heights was estimated from each posterior after removing the first 10% of samples as burn-in.
3 RESULTS
3.1 Genotyping
We genotyped 376 individuals across five penguin species, representing 32 breeding colonies (Figure 1). To ease interspecific comparisons, we refer to island or archipelago names rather than colonies (Supporting information Tables S1, S2). RAD-Seq yielded an average of 11.6 million reads per individual, with 97.1% retained after quality control. Alignment to reference genomes, SNP calling and filtering generated high-coverage, neutral SNP data sets (Table 1). Median sequencing depth per SNP ranged from 26.7 in chinstrap penguins to 32.1 in emperor penguins (Table 1, Supporting information Figure S1). Sequencing depth of individuals was similar among species (Supporting information Figure S2, Table S3), as was the distribution of minor allele frequencies (Supporting information Figure S3). There was no evidence for lane effects in our data: Sequencing depth did not vary significantly among lanes, the locations of SNPs called within reads were even and similar across lanes, and the patterns of population structure that we found showed no relationship to the lanes on which individuals were sequenced.
Given the large number of outliers in the gentoo penguin data set, we investigated whether loci putatively under selection were influencing patterns of genetic differentiation by comparing pairwise FST values from both the neutral and total SNP data sets (Supporting information Tables S4, S5). The differentiation patterns were the same; therefore, we used the neutral SNP data set for all subsequent analyses.
3.2 Levels of intraspecific variation among species
The mean minor allele frequency (MAF) did not differ among species (Table 1). Measures of genetic differentiation based on FST were therefore comparable (Jakobsson, Edge, & Rosenberg, 2013). It should be noted that the data set MAFs were low (0.062–0.091), as expected for biallelic SNP data sets generated from RAD-Seq. The maximum values of FST were therefore mathematically constrained to small values (Jakobsson et al., 2013), and relatively low, yet statistically significant, observations of FST should not be misinterpreted as a lack of genetic differentiation.
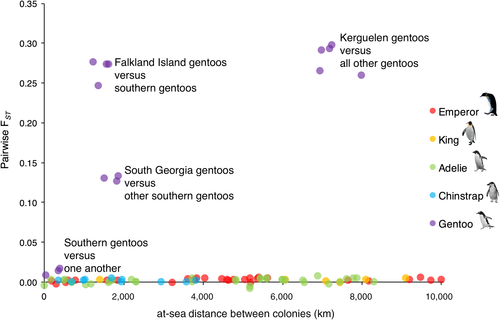
AMOVA-based estimates of FST were small but significant for king, emperor, chinstrap and Adélie penguins (FST = 0.002–0.003, Table 1; Figure 2), suggesting that genetic differentiation is present across the range of each species. The proportion of significantly differentiated pairs of colonies within these species ranged from 19% (Adélies) to 67% (kings), indicating there is likely ongoing gene flow among colonies for all pelagic species, but at insufficient levels to result in panmixia (Table 1). In comparison, overall FST was two orders of magnitude greater in gentoo penguins (FST = 0.217, p = 0.001; Figure 2), and every pair of colonies was significantly differentiated from one another, indicating minimal gene flow among colonies of gentoo penguins. The relationship between FST and geographic distance across species is striking. King, emperor, chinstrap and Adélie penguins exhibit very low levels of differentiation even when colonies are separated by thousands of kilometres, whereas the gentoo penguin colonies are highly differentiated, even when separated by less than 100 km (Figure 2).
3.3 King penguins
Genetic differentiation among king penguin colonies at the Falkland Islands, South Georgia, the Crozet Islands and Macquarie Island (Figure 1a) was subtle, despite being separated by thousands of kilometres of open ocean (Clucas, Younger et al., 2016). Most surprisingly, the Falkland Islands were subtly genetically differentiated from nearby South Georgia, ca. 1,400 km away but on the opposite side of the Polar Front, but indistinguishable from the Crozet Islands ca. 7,500 km away and on the same side of the Polar Front. Our analyses showed South Georgia to be the most divergent population, followed by Macquarie Island (Figure 4a, Supporting information Figure S5; Table S6; see also Clucas, Younger et al., 2016). Genetic differentiation across the range of the king penguin was subtle, as evidenced by small pairwise FST values (Supporting information Table S6); invariant genetic diversity among colonies (Supporting information Table S2); the selection of K = 1 as the best clustering solution by both the successive K-means clustering algorithm and structure (Supporting information Figure S4e); subtle clustering in our PCA and DAPC analyses (Figure 4a, Supporting information Figures S5a, S11a); and a low level of population differentiation when location priors were not used in structure (Figure 4a, Supporting information Figure S6).
3.4 Emperor penguins
Among the eight sampled emperor penguin colonies (Figure 1b), there are four genetically differentiated metapopulations (Younger et al., 2017). The Ross Sea metapopulation appears the most divergent from the rest, consistent with findings from mitochondrial DNA (Younger, Clucas, et al., 2015). The four metapopulations are apparent in the structure results with and without location priors, with colonies clustered by geographic region: (a) Ross Sea, (b) Mawson Coast, (c) Weddell Sea and (d) Amanda Bay and Point Géologie (Figure 4b). DAPC shows the distinctness of the Ross Sea colonies (Cape Roget, Cape Washington) and the slight differentiation of the Mawson Coast colonies (Fold Island, Auster) (Figure 3b). Differentiation between the Weddell Sea and Amanda Bay/Pointe Géologie was not discernible in either the DAPC (Figure 3b) or the structure analysis at K = 3 (Supporting information Figure S7), suggesting these metapopulations are only slightly distinct. Overall, the observed differentiation among emperor penguins was subtle—successive K-means clustering could not detect clusters among individuals, and PCA did not show highly distinct clusters (Supporting information Figure S5b)—however, pairwise FST values between colonies were statistically significant for 17 out of 28 comparisons (Supporting information Table S7) and posterior membership probabilities of individuals to their colony of origin were relatively high with DAPC (Supporting information Figure S11b).

3.5 Chinstrap penguins
Population differentiation among chinstrap penguin colonies across their range was extremely low. Only three of ten pairwise FSTs were significant (Supporting information Table S8), successive K-means clustering could not detect any clusters, and structure had the highest mean posterior probability when K = 1 (Supporting information Figure S4c) and could not discern clusters without location priors (Figure 4c, Supporting information Figure S8). With location priors, Orne Harbor appeared subtly differentiated, the South Sandwich Islands and Bouvet Island clustered together, as did the South Orkney Islands and South Shetland Islands (Figure 4c, Supporting information Figure S8). This differentiation was not as obvious with DAPC (Figure 3, Supporting information Figure S11c) or PCA (Supporting information Figure S5c), suggesting that they are only subtly differentiated. It should be noted that at the South Orkney Islands, we only sampled three individuals and conclusions regarding that colony should be considered preliminary.
3.6 Adélie penguins
The Adélie penguin colonies sampled can be divided into the “western colonies” of the Antarctic Peninsula and Scotia Arc, and the “eastern colonies” along the Mawson Coast and East Antarctica (Figure 1d). We uncovered subtle genetic differentiation coincident with this geographic division (Figure 3d, 4d). Despite separations in excess of 4,000 km, the genetic divergence was very slight, suggesting ongoing gene flow. Successive K-means clustering could not detect any genetic differentiation and the highest posterior mean log likelihood was achieved at K = 1, both with and without location priors in structure (Supporting information Figure S4d). Without location priors, the signal of two geographic clusters was lost at K = 2 (Figure 4d) but was visible at higher values of K (Supporting information Figure S9). Pairwise FST comparisons were significant for only 6 out of 20 pairs of colonies across the regions (Supporting information Table S9), and PCA showed only subtle differentiation between eastern and western colonies (Supporting information Figure S5). Finally, a high level of admixture among colonies was visible when we plotted individual membership probabilities to each colony with DAPC (Supporting information Figure S11d).
3.7 Gentoo penguins
For gentoo penguins, we designed our sampling scheme (Figure 1e) to include the two currently recognized subspecies: the northern gentoo (the nominate subspecies, Pygoscelis papua papua (Forster, 1781)), which is formally distributed north of 60°S; and the southern gentoo (Pygoscelis papua ellsworthii), formally distributed on the Antarctic Peninsula and maritime Antarctic islands south of 60°S (Clements et al., 2017; Murphy, 1947; Stonehouse, 1970); and the putative Indian Ocean subspecies (de Dinechin et al., 2012), which is still formally regarded as P. p. papua (Clements et al., 2017).
Our analyses showed four distinct groupings of gentoo penguins. Both the maximum-likelihood phylogeny (Figure 5a) and the Bayesian coalescent-based species tree (Figure 5b) gave 100% support for four clades, corresponding to (1) the Falkland Islands (northern gentoo), (2) Kerguelen (Indian Ocean gentoo), (3) South Shetland Islands and western Antarctic Peninsula (southern gentoo) and (4) South Georgia (currently designated as northern gentoo). Bayes factor species delimitation overwhelmingly supported the scenario of four distinct lineages, yielding a Bayes factor of 17,595 when compared to the current taxonomy, and of 1,231 over the next most supported model (the three taxa mitochondrial DNA hypothesis of Falkland Islands vs. Kerguelen vs. South Georgia, South Shetlands, Antarctic Peninsula), where a Bayes factor of 10 is considered decisive (Kass & Raftery, 1995) (Supporting information Table S10).
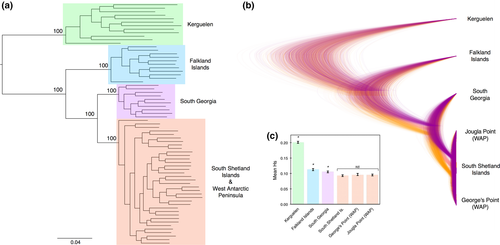
Both phylogenies showed Kerguelen to be the most distantly related clade, with South Georgia and the southern gentoo resolved as reciprocally monophyletic sister groups. Our estimates of lineage divergence times indicated that Kerguelen gentoos split from other gentoos ca. 0.91 Ma, followed by the divergence of the Falklands lineage ca. 0.60 Ma (Supporting information Table S11). It should be noted that our estimates of divergence time differ markedly from those based on mitochondrial data (Clucas et al., 2014; Levy et al., 2016), most likely because mitochondrial data alone were unable to resolve the topology completely, resulting in more recent coalescent events. The same pattern of relatedness was evident from our DAPC and PCA analyses (Figure 3e, Supporting information S5e). structure completely differentiated the Falkland Islands and Kerguelen populations from all other colonies, with no evidence of admixture among those three groups when K = 4 (Figure 4e). The significant divergence of the Falkland Islands population from South Georgia and the southern gentoos is notable, because the southern gentoo colonies on the South Shetland Islands and Antarctic Peninsula are geographically closer to the Falkland Islands than they are to South Georgia; however, the Falkland Islands is north of the Polar Front, whereas the other colonies all lie south of the Polar Front. structure also clearly differentiated South Georgia from the southern gentoo colonies when Kerguelen and Falkland Islands were removed from the analysis; the maximum posterior log likelihood occurred at K = 2; and individuals from South Georgia were fully assigned to a separate cluster (Supporting information Figure S10). For the results of all the hierarchical structure analyses for gentoo penguins, see Supporting information Figure S10. Pairwise FST values among the four clades ranged 0.127 to 0.298 and were all highly significant (p < 0.001, Supporting information Table S4). FST values were two orders of magnitude greater than observed within the other penguin species studied, even though geographic distances among colonies were similar (Figure 2). The genetic diversity of the four clades was significantly different and greatest for Kerguelen (Figure 5c).
The southern gentoo colonies on the South Shetland Islands and western Antarctic Peninsula were clearly differentiated from one another in a structure analysis with and without location priors (Figure 4f), by DAPC and PCA (Figure 4f, Supporting information S5f, S11e), and all pairwise FST comparisons were significant, with values exceeding those observed for all other species (range = 0.009–0.017, Supporting information Table S4; all other species range = −0.008 to 0.008, Supporting information Tables S6–S9). Given the geographic proximity of these colonies (50–400 km separations), this level of genetic differentiation is in stark contrast to the other species studied.
4 DISCUSSION
4.1 Factors influencing patterns of genetic variation in penguins
To identify key factors that influence dispersal in penguins, we compared patterns of intraspecific genetic differentiation across the global distributions of Aptenodytes and Pygoscelis using genomewide SNPs. Four out of five species (king, emperor, chinstrap and Adélie penguins) showed low levels of genetic differentiation over thousands of kilometres, whereas the fifth species, the gentoo penguin, had remarkably high levels of intraspecific genetic differentiation and deep phylogenetic splits.
The low levels of genetic differentiation observed in king, emperor, chinstrap, and Adélie penguins may be attributable to either gene flow among colonies, or incomplete lineage sorting following recent population divergences. While we cannot explicitly rule out incomplete lineage sorting, evidence from the ecological literature lends considerable support to the hypothesis that dispersal among colonies is facilitating gene flow. There have been several documented instances of migration of king penguins among colonies over distances up to 5,600 km (Weimerskirch et al., 1985; Woehler, 1989), and the recent formation of several new king penguin colonies (Delord, Barbraud, & Weimerskirch, 2004; van den Hoff, McMahon, & Field, 2009; Pistorius, Baylis, Crofts, & Pütz, 2012) provides direct evidence of dispersal for this species. For emperor penguins, there were six observations of colony establishment or relocation in a period of just five years, again providing direct evidence of dispersal (LaRue et al., 2015). Large fluctuations in abundance at individual colonies from year to year have been observed in both emperor (Kooyman & Ponganis, 2017) and Adélie penguins (Che-Castaldo et al., 2017; Dugger, Ainley, Lyver, Barton, & Ballard, 2010), indicating either dispersal or a high incidence of skipped breeding. Overall, there is a growing body of evidence indicating that dispersal in many species of penguins is a regular occurrence.
Many factors have been previously identified as potential drivers of dispersal patterns in seabirds (Friesen, 2015; Friesen et al., 2007), and we will discuss our genomic results with respect to the most relevant of these for Southern Ocean penguins.
4.1.1 At-sea range
The most important factor in determining patterns of intraspecific genetic variation in penguins appears to be their at-sea range. The four species for which we found evidence of dispersal over large spatial scales are all considered pelagic, spending at least a portion of their life history in the open ocean far from their colonies. Adult emperor, Adélie and chinstrap penguins travel up to 1,400 km (Ratcliffe & Trathan, 2012), 2,200 km (Dunn, Silk, & Trathan, 2011) and 3,900 km (Hinke et al., 2015) away from their colonies during the nonbreeding period, respectively. Juvenile emperor penguins are even more mobile than adults, with recorded journeys covering in excess of 7,000 km in just eight months (Thiebot, Lescroël, Barbraud, & Bost, 2013), and individuals documented in the vicinity of other breeding colonies (Kooyman, Kooyman, Horning, & Kooyman, 1996; Wienecke, Raymond, & Robertson, 2010). In the winter, king penguins travel up to 1,800 km to forage in the marginal ice zone (Bost, Charrassin, Clerquin, Ropert-Coudert, & Le Maho, 2004; Charrassin & Bost, 2001) and juveniles have been observed at breeding colonies up to 5,600 km from their natal colonies (Weimerskirch et al., 1985). With the exception of the king penguin, the at-sea ranges of these pelagic penguins exceed the average distances between colonies. This wide-ranging behaviour is likely to facilitate dispersal, as evidenced by overall low genetic differentiation within all the pelagic species.
In contrast, gentoo penguins have a coastal lifestyle, rarely ranging beyond the continental shelf, and forage inshore on locally available prey (Lescroël & Bost, 2005; Ratcliffe & Trathan, 2012) rather than making long journeys to exploit specific prey resources. Adult gentoo penguins typically forage within 40 km of colonies during the breeding period and are rarely seen more than 50 km offshore during the non-breeding period, although they have been tracked up to 380 km offshore of the Falkland Islands over the Patagonian shelf (M. Tierney pers. comm.). Juvenile gentoo penguins travel further, while still remaining over the continental shelf, and there is one documented instance of dispersal of 500 km between archipelagos in the Indian Ocean sector (Thiebot, Lescroël, Pinaud, Trathan, & Bost, 2011). The species’ tendency to stay in shelf waters may act as a barrier to dispersal in gentoo penguins by reducing mixing with individuals from distant colonies, contributing to the high degree of genetic differentiation we recorded.
In general, at-sea range may be an important determinant of dispersal patterns for marine taxa. Wide-ranging organisms have greater opportunity to come into contact with individuals from other populations and to prospect other breeding sites, both of which may facilitate dispersal. This has been demonstrated in many seabirds (see Friesen et al., 2007 for a review), as well as in other marine taxa. For example, humpback whales (Megaptera novaeangliae) have discrete breeding grounds (Clapham, 1996), but undertake long foraging journeys during the non-breeding season that bring them into contact with individuals from other breeding locations (Amaral et al., 2016). This appears to facilitate dispersal, with direct observations of individuals moving between breeding sites in different oceans (Stevick et al., 2011) and evidence of gene flow across ocean basins (Rosenbaum et al., 2009). Similarly, Atlantic Bluefin tuna (Thunnus thynnus) show site fidelity to two disparate spawning grounds on either side of the Atlantic Ocean, but individuals from both populations mix on their pelagic foraging grounds across the North Atlantic (Block et al., 2005). This migratory behaviour and intermixing of stocks likely facilitate gene flow, which may explain the low levels of genetic differentiation between the Gulf of Mexico and Mediterranean populations (Rooker et al., 2007).
However, the influence of wide-ranging behaviour on dispersal patterns cannot be generalized to all marine taxa. Atlantic salmon (Salmo salar) are highly migratory with individuals from both sides of the Atlantic mixing on foraging grounds off western Greenland, yet high levels of natal homing have led to genetic differences allowing individuals to be assigned back to their natal river with high success (King, Kalinowski, Schill, Spidle, & Lubinski, 2001). Equally, wide-ranging behaviour may not facilitate dispersal in other species of birds outside of the penguin order. For example, Wilson's Warblers (Cardellina pusilla) are migratory passerines with genetically distinct breeding populations in the east and west of North America that share overwintering habitat in Central America, yet there is no evidence of gene flow between breeding populations (Irwin, Irwin, & Smith, 2011; Ruegg et al., 2014).
4.1.2 Natal philopatry
We observed genetic differentiation between gentoo penguin colonies separated by less than 50 km. This finding cannot be explained solely by its coastal lifestyle, because the species is known to visit other colonies within this range (Ratcliffe & Trathan, 2012). Given the very small spatial scale over which population differentiation was observed, it is possible that natal philopatry also plays a role in limiting gene flow in gentoo penguins. On Possession Island (Crozet Archipelago), almost all the first breeding attempts occurred in a range of 2–5 km of the natal colony (C.A Bost, pers. comm.). Natal philopatry is thought to be common among seabirds (Coulson, 2002) and has been identified as a barrier to gene flow in other species, although it usually acts in combination with other isolating mechanisms (Friesen, 2015).
Paradoxically, the range of the gentoo penguin is expanding southwards coincident with sea ice decline (Lynch, Naveen, Trathan, & Fagan, 2012). The genetic differentiation found here would suggest that immigration rates at new colonies should be low. Instead, high rates of breeding success and recruitment may explain rapid colony growth after establishment.
4.1.3 Oceanographic fronts
Both gentoo and king penguins have breeding distributions spanning the Antarctic Polar Front (Figure 1a, e), and our results indicate that the front may be a barrier to dispersal in both species. The Antarctic Polar Front is the convergent boundary between cold Antarctic waters and warmer sub-Antarctic waters and constitutes an important feature for seabird communities (Bost et al., 2009). King penguins from South Georgia, which is the only breeding population situated south of the front in our data set, were the most genetically divergent, albeit subtly. Furthermore, king penguins from the Falkland Islands were genetically indistinguishable from those at Crozet, ca. 7,500 km away but also north of the Polar Front, whereas they were differentiated from those at South Georgia, only 1,400 km away but on the opposite side of the front. A similar pattern was evident in gentoo penguins. Our study included one gentoo penguin colony north of the Polar Front, at the Falkland Islands, whereas the other colonies were all distributed south of the front (Figure 1e). The Falkland Islands were genetically divergent from all other colonies. Compellingly, the colonies on the South Shetland Islands and Antarctic Peninsula were more closely related to the South Georgia colony, which is also to the south of the front, than they are to the Falkland Islands, which is more proximate but to the north of the front. This suggests that, in addition to their coastal lifestyle, the Polar Front is a barrier to gentoo penguin dispersal. Such a finding is consistent with studies on the genetic structure of several Antarctic marine vertebrates and invertebrates (see Rogers, 2012 for a review).
The oceanic regimes on either side of the Antarctic Polar Front differ in their physical and biological characteristics, including sea surface temperature and primary productivity, and hence exert different selective pressures. Oceanographic fronts may act as barriers to dispersal either by physically deterring dispersal or by reducing the fitness of immigrants from foreign oceanic regimes (Friesen, 2015). The role of oceanic regimes in the formation of genetically distinct populations has been shown in a broad range of highly mobile vertebrate taxa, including rockhopper (de Dinechin, Ottvall, Quillfeldt, & Jouventin, 2009) and yellow-eyed penguins (Boessenkool, Star, Waters, & Seddon, 2009), cetaceans (Fontaine et al., 2007), fish (Shaw, Arkhipkin, & Al-Khairulla, 2004) and various flying seabirds (Gómez-Díaz, González-Solís, & Peinado, 2009; Techow, Ryan, & O'Ryan, 2009). Our findings of population divergence in highly mobile marine taxa underline the importantce of recognizing extrinsic barriers to dispersal in the marine realm.
4.1.4 Breeding habitat quality, continuity and ephemerality
Emperor penguins have a relatively continuous distribution around Antarctica with most colonies being situated within the range of individuals foraging from adjacent colonies (Fretwell et al., 2012) (Figure 1b). Their fast-ice breeding habitat is highly ephemeral, leading to changes in colony locations over years (Fretwell, Trathan, Wienecke, & Kooyman, 2014; LaRue et al., 2015; Trathan, Fretwell, & Stonehouse, 2011) and millennia (Younger, Clucas, et al., 2015; Younger et al., 2016). The low levels of genetic differentiation among emperor colonies likely reflect the need for flexibility in breeding location. The case of the Adélie penguin is similar, in that its breeding habitat is somewhat ephemeral, with access blocked periodically by sea ice or icebergs (Dugger et al., 2010), and with several large discontinuities in its circumpolar range (Figure 1d) where ice-free habitat does not exist. We found that Adélie penguins were subtly genetically differentiated across a gap of several thousand kilometres in their breeding distribution (Figure 1d), suggesting that the lack of contiguous habitat suitable for breeding moderately impedes gene flow. In regions where Adélie penguins are distributed more or less continuously (e.g., between Béchervaise Island and Petrels Island), there was no evidence of genetic divergence over thousands of kilometres, indicative of dispersal consistent with the ephemerality of the breeding habitat and facilitated by its continuity.
While the sub-Antarctic breeding habitat of the king and northern colonies of gentoo penguins experience climatic variability, in general, conditions are far more stable than those in the Antarctic, which are highly influenced by seasonal ice advance and retreat. The chinstrap penguin occupies a somewhat intermediate habitat in the maritime Antarctic, subject to variability in sea ice that may occasionally limit access to colonies. King, gentoo and chinstrap penguins also have patchy distributions, with breeding sites situated on archipelagos (Figure 1a, c, e). The patchiness and relative stability suggest that high natal philopatry and local adaptation may be selected for in these species. However, we find that is not the case, except for the gentoo penguin, for which other dispersal barriers have already been noted. The large at-sea distributions of chinstrap and king penguins may facilitate gene flow such that the dispersal barriers posed by their patchy distributions are overcome. Occasional, large-scale disruptions in breeding habitat may cause pulses of dispersal in some species, for example, chinstrap penguins may have been displaced from the South Sandwich Islands as a result of recent volcanic activity.
4.1.5 Implications for modelling studies
Our results show that pelagic penguins can, and do, disperse among colonies separated by thousands of kilometres. Dispersal can decouple the relationship between local climate and demographic rates (Jenouvrier et al., 2017), facilitate range shifts, furnish populations with potentially adaptive genetic variants, and bolster population stability by compensating for low birth rates or survival (Lowe & Allendorf, 2010). Modelling studies that forecast population trends for pelagic penguins under future climate change scenarios should incorporate the dispersal patterns that we have outlined here, as in a recent study of emperor penguins (Jenouvrier et al., 2017). The conclusions of modelling studies for pelagic penguins that do not incorporate dispersal (Abadi, Barbraud, & Gimenez, 2017; Cimino et al., 2016; Jenouvrier et al., 2014) should be treated with caution.
4.2 Cryptic diversity within gentoo penguins
The currently recognized taxonomy of gentoo penguins is for two subspecies, the northern (Pygoscelis papua papua) distributed north of 60°S, and the southern (Pygoscelis papua ellsworthii) distributed on the Antarctic Peninsula and maritime Antarctic islands south of 60°S (Clements et al., 2017; Forster, 1781; Murphy, 1947). Our data support the existing classification of a northern gentoo subspecies; however, contrary to current taxonomic limits (Clements et al., 2017), we found that South Georgian gentoos are more closely related to the southern subspecies than the northern, a conclusion that is supported by previous studies of morphology (de Dinechin et al., 2012), mitochondrial DNA (Clucas et al., 2014) and microsatellites (Levy et al., 2016). We recommend formal taxonomic revision of the boundary between northern and southern gentoo penguins to reflect this.
The degree of genetic divergence of gentoo penguins at Kerguelen points to a need for morphological and ecological study to determine whether these are a distinct species worthy of formal description. The case for revision has been based until now on mitochondrial DNA (Clucas et al., 2014; de Dinechin et al., 2012; Vianna et al., 2017) and microsatellites (Levy et al., 2016; Vianna et al., 2017), and we have now confirmed deep lineage divergences using genomewide data. In the light of these results, there is also an urgent need to characterize gentoo penguins breeding at other archipelagos using genomic data, particularly Crozet Archipelago and Macquarie Island, as there is likely to be more cryptic diversity. Accurate species boundaries and the recognition of cryptic species are crucial for the conservation of biodiversity, particularly in the light of the challenges (Trathan et al., 2015) that will face Southern Ocean biota in the Anthropocene. The three lineages of gentoo penguins are on separate evolutionary trajectories. By conserving their full spectrum of genetic variation, the evolutionary and adaptive potential of gentoo penguins can be maximized.
4.3 Predicting population structure
Understanding the mechanisms behind patterns of species dispersal has never been more important. Climate change is dramatically altering the marine environment, leading to changes in habitat availability, quality and ephemerality, as well as shifting oceanographic features. Understanding current barriers to dispersal is essential for forecasting how species might respond to changes in their environment and for implementing ecologically meaningful conservation strategies. Our findings show that at-sea range and oceanography are likely predictive of population structure in penguins. For species that journey into pelagic waters and range further than the average distance between colonies, we observed very little population differentiation. For colonies that are separated by oceanographic fronts, we observed greater genetic divergence than would be expected based on distance alone. We suggest that for colonies or species of penguins for which genetic data are unavailable, these predictive factors could be used to guide estimates of management units.
ACKNOWLEDGEMENTS
This study was funded by a NERC PhD studentship (1272500; GC), Australian Antarctic Science Program grants (4184; KM, JY, LE; 4087; CS, LE), a Holsworth Wildlife Research Endowment (JY), the Sea World Research and Rescue Foundation (JY, KM), an Endeavour Research Fellowship (JY), The Darwin Initiative (DPLUS 002; TH), Quark Expeditions (TH, GC), the French Polar Institute IPEV (394; CAB) and U.S. National Science Foundation (ANT-0739575, MP). We are very grateful to these individuals and organizations for contributions of penguin genetic material: Jerry Kooyman, Francoise Amelineau, Julie McInnes, Helen Achurch, Cecilia Carrea, Laura Morrissey, Thierry Raclot, Phil Trathan, Andy Black, Alex Corbeau, Joan Ferrer, Onno Huyser, the U.S. Antarctic Program and the U.S. Antarctic Marine Living Resource Program for logistical support at King George Island, the Government of South Georgia and the South Sandwich Islands and Quark Expeditions for support in sample collection around the Scotia Arc, and the British Antarctic Survey for logistical support in sample collecting at the South Orkney Islands. Thanks to the Oxford Advanced Research Computing (ARC) facility (https://doi.org/10.5281/zenodo.22558) and staff at Edinburgh Genomics, which is partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1).
DATA AVAILABILITY
The Illumina reads are available from the NCBI Sequence Read Archive: https://www.ncbi.nlm.nih.gov/bioproject/ PRJNA384210 for emperor penguins; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA343012 for king penguins; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA493660 for gentoo, Adélie, and chinstrap penguins. Additional materials are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.7c0q8 for python script filter.py and the king penguin SNP data set; https://doi.org/10.5061/dryad.4s7t3 for the emperor penguin SNP data set; https://doi.org/10.5061/dryad.bs30388 for Adélie, gentoo (neutral and full), and chinstrap penguin SNP data sets, plus input files for SNAPP, BFD*, and RAxML.
COMPETING FINANCIAL INTERESTS
The authors do not have any competing financial interests.
ETHICAL APPROVAL
Ethical approval for all research involving the handling of birds was provided by the following: the University of Oxford, the University of Western Australia, the Australian Antarctic Division's animal ethics committee, the British Antarctic Survey, the Auburn University Institutional Animal Care and Use Committee, University of North Carolina Wilmington Institutional Animal Care and Use Committee, and the Institut Polaire P. E. Victor. Permits for sampling activities were provided by the Falklands Island Government, the Government of South Georgia & the South Sandwich Islands, the US Antarctic Program, the US National Science Foundation, the UK Foreign & Commonwealth Office, the Tasmanian Parks Department and the Norwegian Polar Institute.
AUTHOR CONTRIBUTIONS
G.C. and J.Y. collected, analysed and interpreted the data; wrote the manuscript; and participated in study conception and design. D.K. participated in bioinformatics. L.E., C.S. and B.W. collected genetic samples and participated in interpreting the data. C.B., G.M., M.P., J.H., S.C., M.D. and R.P. participated in sample collection. P.L. carried out laboratory work. K.M., T.H. and A.R. conceived and designed the study.




