The role of isoforms in the evolution of cryptic coloration in Peromyscus mice
Abstract
A central goal of evolutionary biology is to understand the molecular mechanisms underlying phenotypic adaptation. While the contribution of protein-coding and cis-regulatory mutations to adaptive traits has been well documented, additional sources of variation – such as the production of alternative RNA transcripts from a single gene, or isoforms – have been understudied. Here, we focus on the pigmentation gene Agouti, known to express multiple alternative transcripts, to investigate the role of isoform usage in the evolution of cryptic colour phenotypes in deer mice (genus Peromyscus). We first characterize the Agouti isoforms expressed in the Peromyscus skin and find two novel isoforms not previously identified in Mus. Next, we show that a locally adapted light-coloured population of P. maniculatus living on the Nebraska Sand Hills shows an upregulation of a single Agouti isoform, termed 1C, compared with their ancestral dark-coloured conspecifics. Using in vitro assays, we show that this preference for isoform 1C may be driven by isoform-specific differences in translation. In addition, using an admixed population of wild-caught mice, we find that variation in overall Agouti expression maps to a region near exon 1C, which also has patterns of nucleotide variation consistent with strong positive selection. Finally, we show that the independent evolution of cryptic light pigmentation in a different species, P. polionotus, has been driven by a preference for the same Agouti isoform. Together, these findings present an example of the role of alternative transcript processing in adaptation and demonstrate molecular convergence at the level of isoform regulation.
Introduction
Understanding the molecular basis of adaptation is one of the principal goals of evolutionary biology. Considerable efforts have been focused on the contributions of protein-coding and cis-regulatory mutations, and their relative importance, to phenotypic change (Carroll 2005; Hoekstra & Coyne 2007; Stern & Orgogozo 2008). In contrast, the production of multiple isoforms through the inclusion of different exons in mRNA – known as alternative mRNA processing – is a major source of genetic variation whose role in phenotypic adaptation has been comparatively understudied. A single protein-coding gene can produce multiple isoforms either through alternative splicing, which results in transcripts with different combinations of exons, or through the use of alternative transcription initiation or termination sites, which generate mRNAs that differ at the 5′ or 3′ untranslated regions (UTRs). Alternative processing is ubiquitous throughout eukaryotic evolution but is most prevalent in higher eukaryotes, with mammals having the highest genome-wide rate of alternative splicing events (Barbosa-Morais et al. 2012; Merkin et al. 2012) and alternative promoter usage (Landry et al. 2003; Baek et al. 2007; Shabalina et al. 2014). In addition, recent studies of mammalian gene expression show that transcripts from the majority of protein-coding genes, especially in primates, undergo alternative processing and generate different isoforms (Blencowe 2006; Kim et al. 2008). Thus, by increasing transcriptomic, and hence proteomic diversity, the production of multiple isoforms through alternative processing has been widely regarded as a key mechanism for generating phenotypic diversity and organismic complexity, particularly at large taxonomic scales (Barbosa-Morais et al. 2012; Merkin et al. 2012).
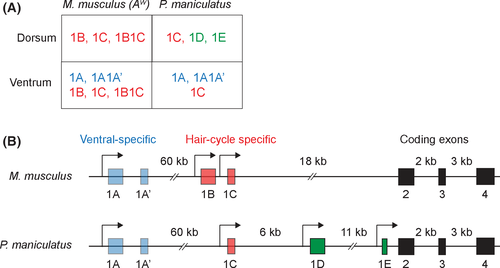
To investigate the role of alternative processing in adaptation between closely related species, we studied the pigmentation locus Agouti, which has been used as a model for isoform regulation in development (Vrieling et al. 1994) and has been repeatedly implicated in the evolution of natural variation in pigmentation (e.g. Rieder et al. 2001; Schmutz & Berryere 2007; Seo et al. 2007; Steiner et al. 2007; Linnen et al. 2009, 2013). Agouti encodes a secreted paracrine factor that induces pigment-producing cells (melanocytes) in hair follicles to switch from the synthesis of black pigment (eumelanin) to yellow pigment (phaeomelanin) during hair growth (Jackson 1994). Experiments performed in laboratory mouse (Mus musculus) neonates found that the Agouti locus comprises three constitutively transcribed coding exons and four upstream, alternatively transcribed 5′ UTRs, also called noncoding exons (Vrieling et al. 1994). Thus, in M. musculus, Agouti mRNAs are found as four different isoforms, each containing one to two noncoding exons upstream of the coding sequence. The ‘ventral-specific’ noncoding exons 1A and 1A’ are expressed in the ventral mesenchyme during embryonic development, whereas the ‘hair cycle-specific’ noncoding exons 1B and 1C are expressed during hair growth across all regions of the body (Vrieling et al. 1994). Thus, alternative promoters of Agouti allow for differences in the spatial and temporal deployment of a single coding sequence.
Deer mice (genus Peromyscus) populations vary tremendously in both colour and pattern. Across populations, there is often a close correspondence between the colour of mouse fur and the local soil, suggesting that colour matching is important for survival (Dice 1940, 1941; Haldane 1948). In both Nebraska and Florida, dark-coloured mice have colonized extreme light substrate environments that appeared in the last 10 000 years – the Sand Hills in Nebraska (Ahlbrandt & Fryberger 1980; Loope & Swinehart 2000) and the coastal islands in Florida (Campbell 1985; Stapor & Mathews 1991). In both cases, mice have independently evolved significantly lighter coats than mice inhabiting darker surrounding substrates (Fig. 1A and B), and there is experimental evidence that visually hunting predators generate strong selection favouring substrate matching (Dice 1941; Vignieri et al. 2010; Linnen et al. 2013).
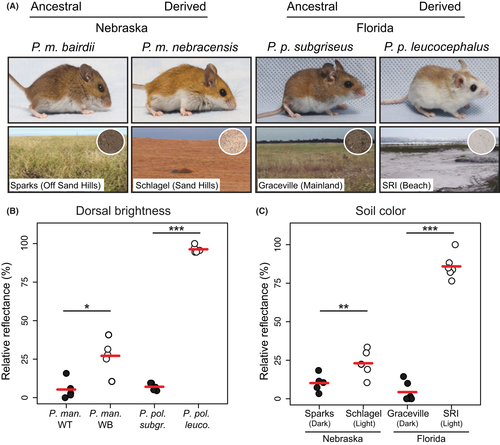
In addition to phenotypic convergence, the same gene, Agouti, has been shown to play a significant role in mediating this colour adaptation in both Nebraska and Florida populations (Steiner et al. 2007; Linnen et al. 2009, 2013; Manceau et al. 2011). Specifically, in light-coloured P. maniculatus inhabiting the Nebraska Sand Hills, referred to as ‘wideband’ mice, Agouti is expressed at higher levels and for a longer period during hair growth than in their ancestral dark-coloured counterparts (‘wild-type’ mice), which gives rise to a lighter and wider pheomelanic band on individual hairs and overall lighter appearance (Linnen et al. 2009). Furthermore, wideband mice display marked differences in additional pigment traits (e.g. dorso-ventral boundary, ventral colour and tail stripe) that are significantly associated with variants in the Agouti locus (Linnen et al. 2013). In Florida, changes in Agouti and two other pigmentation loci are responsible for producing the lighter pigmentation phenotypes displayed by a derived population of beach mice (P. polionotus leucocephalus) inhabiting the coastal sand dunes, relative to their ancestral mainland conspecifics (P. p. subgriseus) (Hoekstra et al. 2006; Steiner et al. 2007; Mullen & Hoekstra 2008). Like wideband mice from the Nebraska Sand Hills, Florida beach mice express Agouti at higher levels and in extended spatial domains compared with mainland mice (Manceau et al. 2011).
The implication of Agouti in the repeated evolution of cryptic coloration in Peromyscus, coupled with its regulatory architecture of alternative 5′ noncoding exons expressed in a region- and temporal-specific pattern, makes this an excellent system in which to study the role of isoform regulation in evolution and adaptation. Here, we first characterize the Agouti isoforms present in Peromyscus skin and examine their patterns of expression during hair growth. We then use in vitro experiments to study transcript-specific functional differences. Next, we use a phenotypically variable population in the Sand Hills to map variation in overall Agouti mRNA expression to the Agouti locus and to test for signatures of selection on the different isoforms. Finally, we study the expression of Agouti isoforms in Florida P. polionotus to establish whether the changes in isoform regulation seen in Nebraska P. maniculatus are shared with its sister species that independently evolved light coloration as an adaptation to a similar light-coloured sandy environment.
Materials and methods
Mice
Laboratory mouse strains
We originally acquired wild-derived P. m. bairdii strains from the Peromyscus Genetic Stock Center (University of South Carolina) and now maintain them at Harvard University. As representatives of the mice found in Nebraska, we used P. maniculatus wild-type (a+/a+) and P. maniculatus heterozygous for the wideband allele and a non-Agouti allele (awb/a−). The non-Agouti allele has a 125-kb deletion that removes the entire regulatory region and first coding exon, resulting in a complete loss of expression (Kingsley et al. 2009); thus, P. maniculatus awb/a− are effectively hemizygous for the dominant wideband allele. For many of the experiments, however, we also generated wideband mice homozygous for the wideband Agouti allele (awb/awb). As representatives of mice in Florida, we used P. polionotus subgriseus mainland mice, obtained from the Peromyscus Genetic Stock Center, and P. p. leucocephalus, trapped in Santa Rosa Island. All experiments described here were evaluated and approved by Harvard University's IACUC committee and performed following their established guidelines and regulations.
Wild-caught mice
Mice used for the association and selection studies were collected as described in Linnen et al. (2009) from two Sand Hills sites located <15 km apart in Cherry County, Nebraska (site 1: Schlagel Creek Wildlife Management Area, N = 62; site 2: Ballard's Marsh Wildlife Management Area, N = 29).
Quantification of phenotype and soil coloration
We prepared flat skins of mice from the strains mentioned above using standard museum protocols, which were then deposited in the Mammal Department of the Museum of Comparative Zoology at Harvard University. For all the specimens, we quantified dorsal coloration using a USB4000 spectrophotometer and a PX-2 pulsed xenon light source and recorded readings using the program SpectraSuite (Ocean Optics). Using a reflectance probe with a shield cut at a 45° angle (to minimize diffuse reflection), we took and averaged three measurements of the dorsal skin of each mouse. We trimmed the data to 300–700 nm range, which represents the visible spectrum of most visual predators (Bennet & Lamoreux 2003), and extracted seven colour summary statistics using the program CLRvars (Montgomerie 2008): B2 (mean brightness), B3 (intensity), S3 (chroma), S5c (chroma), S6 (contrast, amplitude), H3 (hue) and H4c (hue). After performing a normal-quantile transformation on each variable, we performed a principal components analysis (PCA) on the transformed data in r (R Core Team, 2014) using prcomp.
To quantify variation in soil colour, we collected soil samples from four different localities representing the natural habitats of the mice used in this study: Schlagel Creek Wildlife Management Area, Cherry County, Nebraska (P. maniculatus wideband mice); Sparks, Cherry County, Nebraska (P. maniculatus wild-type mice); Santa Rosa Island, Okaloosa County, Florida (P. p. leucocephalus beach mice); and Graceville, Jackson County, Florida (P. p. subgriseus mainland mice). We quantified soil reflectance with a spectrophotometer as described above.
Rapid Amplification of cDNA Ends (RACE)
We sampled dorsal and ventral skin of P. maniculatus wideband (awb/a−) and wild-type strains and used 5′ rapid amplification of cDNA ends (RACE) to identify the Agouti isoforms present in Peromyscus. To explore and characterize the full diversity of Agouti isoforms, we sampled tissue from pups [postnatal day 4 (P4)], as previously done in Mus (Vrieling et al. 1994), and from adults (>30 days), because the pelage in adult Peromyscus differs from juveniles (Fig. S1, Supporting information; Golley et al. 1966), and therefore may contain additional transcripts not found during the initial hair cycle.
For each mouse strain, we extracted RNA from skin taken from 1 to 4 individuals. We dissected skin immediately after sacrifice and stored it in RNAlater (Qiagen) at 4 °C. We extracted total RNA using the Qiagen RNeasy Fibrous Tissue Kit. To maximize tissue disruption and RNA yield, we performed two 2-min homogenizations at 50 Hz using a 5-mm steel ball and a Tissuelyser LT (Qiagen). Following extraction, we quantified RNA using a Quant-iT RNA Kit and a Qubit fluorometer (Invitrogen). From total RNA, we then purified mRNA using NucleoTrap mRNA kits (Clontech) and quantified mRNA using fluorescence as described above.
Following mRNA purification, we used the SMARTer RACE cDNA Amplification Kit (Clontech) to prepare 5′-RACE ready cDNA and perform RACE PCRs. To increase the specificity of the 5′ RACE PCRs, we performed a nested PCR. For the first RACE PCR, we used the kit-provided UPM primer in conjunction with a primer located in the exon 4 of Agouti (‘GSP1’ sequence: GTTGAGTACGCGGCAGGAGCAGACG). For the nested PCR, we used the kit-provided NUPA primer in conjunction with a primer located upstream of GSP1 (‘NGSP1C’, sequence: TCTTCTTCAGTGCCACAATAGAAACAG). Following the second PCR, we gel-purified any obvious bands using a QIAquick Gel Extraction Kit (Qiagen). We then ligated the resulting DNA to vectors and transformed competent cells using the pGEM-T Easy Vector Kit (Promega).
Following transformation, we performed colony PCRs using Qiagen TAQ and M13F and M13R primers. PCR products were purified enzymatically using exonuclease I and shrimp alkaline phosphatase (USB). Purified PCR products were sequenced on an ABI 3730xl Genetic Analyzer at Harvard University's Genomics Core facility. In total, we obtained sequences from 24 to 89 Agouti clones from each of eight distinct genotype/stage/tissue combinations. Then, all Agouti clones were sequenced using primers placed at the 5′ end and with Agouti-Exon3-R (see Table S1, Supporting information) to confirm the presence of the Agouti coding sequence.
Quantitative PCR (qPCR)
Laboratory strains
We performed gene expression analyses in P. maniculatus wideband (awb/awb), P. maniculatus wild-type, P. p. subgriseus and P. p. leucocephalus mice. In all cases, we used age-matched mice to minimize the variability in Agouti expression introduced when hair follicles are at different stages of the hair cycle. We extracted RNA from dorsal and ventral tissue as indicated above, isolated mRNA from total RNA using the NucleoTrap mRNA Kit (Clontech) and directly synthesized cDNA using qScript cDNA SuperMix (Quanta BioSciences). We performed all reactions in triplicate using PerfeCta SYBR Green FastMix (Quanta BioSciences) and calculated relative expression between samples using the 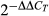 method (Livak & Schmittgen 2001), using β-actin as a reference gene.
method (Livak & Schmittgen 2001), using β-actin as a reference gene.
To measure levels of total Agouti, we used the primers Agouti-Exon2-F and Agouti-Exon3-R to amplify all transcripts containing the Agouti coding sequence. To measure levels of individual isoforms, we paired a forward primer within the noncoding exon (1C-F, 1D-F or 1E-F, respectively) with a reverse primer in the coding region (Agouti-Exon2-R). We amplified β-actin using the primers β-actin-Pero-F and β-actin-Pero-R. All primer sequences were designed from the Peromyscus maniculatus genomic reference (Pman_1.0, GenBank Accession no.: GCA_000500345.1) and can be found in Table S1 (Supporting information).
Wild-caught mice
We removed a 5-mm skin biopsy from the dorsum of recently sacrificed mice, preserved tissue in RNAlater and extracted RNA as indicated above. We synthesized cDNA and performed qPCRs to measure total Agouti and β-actin as described for laboratory strains, with the exception that the mRNA purification step was omitted.
Dorsal depilation
It is well established that Agouti expression is tightly linked to hair growth (Vrieling et al. 1994). Contrary to newborn pups, in which hair emerges simultaneously across the body, hair follicles of adult mice are desynchronized with respect to the hair cycle (Muller-Rover & Paus 2001). Thus, the quantification of Agouti expression in adult dorsal skin represents an average expression level across many follicles that are each at different stages in the cycle. To understand the detailed dynamics of Agouti isoform expression across the entire hair cycle in adults, we depilated the backs of adult wild-type and wideband mice, a procedure that resets and synchronizes the hair follicle program (Paus & Cotsarelis 1999), and compared isoform expression at different time points. We anesthetized adult P. maniculatus wideband and wild-type mice by performing intraperitoneal injections with a cocktail of ketamin/xylazine (0.1 mL/20 g mouse wt) and depilated a small patch (~1 cm2) in the dorsum using a melted beeswax/resin mixture. After the procedure, we applied topical lidocaine for pain relief every hour for the next 8 h.
mRNA stability assays
Our RACE experiment revealed that the dorsal skin in Peromyscus expresses three Agouti isoforms (1C, 1D and 1E) simultaneously, differing only in the first, noncoding exon. To determine whether there were differences in mRNA half-life, we cloned each of the three isoforms from P. maniculatus wild-type into a pHAGE-CMV-eGFP-W expression vector such that the noncoding exon and Agouti coding sequence replaced the eGFP coding sequence. We used Lipofectamine (Life Technologies) to transfect plasmids into human embryonic kidney (HEK293) cells and 40 h after transfection halted transcription by treating cells with 10 μg/mL actinomycin D (Sigma) in DMSO. HEK cells were used in these stability assays because they provide a suitable cellular environment for transcription of mRNA encoded by expression vectors; in addition, they do not express Agouti endogenously, so measurements performed (see below) accurately reflect the amount of transcript produced from the transfected expression vector alone. As a control, we treated cells with an equal volume of 100% DMSO. At 0, 2 or 4 h after actinomycin treatment, we collected cells in TRIzol reagent (Invitrogen) and extracted RNA using the Direct-zol RNA Mini Kit (Zymo Research). We then performed qPCR as described above, using primers common to all constructs (i.e. located in exons 2 and 3): Agouti-Exon2-F and Agouti-Exon3-R to amplify Agouti and β-actin-Human-F and β-actin-Human-R to amplify endogenous β-actin. Primer sequences can be found in Table S1 (Supporting information). We used three replicates for each condition and time point. Agouti expression in actinomycin-treated cells at each time point was calculated as a percentage of Agouti expression in DMSO-treated cells at the same time point using the 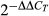 method. We then tested for significant differences between the half-lives of the isoforms by comparing the slopes of the respective trendlines, using one-way analysis of covariance (ancova).
method. We then tested for significant differences between the half-lives of the isoforms by comparing the slopes of the respective trendlines, using one-way analysis of covariance (ancova).
Luciferase assays
To establish whether the noncoding exons of the isoforms expressed in the dorsal skin of Peromyscus showed differences in translation, we generated luciferase reporter plasmids by cloning each of the three noncoding exons (1C, 1D or 1E) from both P. maniculatus wideband and wild-type strains into a 5′ UTR reporter vector (pLightSwitch_5UTR; Switchgear Genomics) upstream of the luciferase coding sequence and downstream of the ACTB promoter. We generated mutated 1D reporter plasmids from the wild-type 1D plasmid using the Q5 Site-Directed Mutagenesis Kit (New England BioLabs). We transfected plasmids into HEK293 cells using Lipofectamine (Invitrogen), with six replicate transfections per construct. Forty-eight hours after transfection, we measured luminescence as a readout of protein production using a microplate reader (SpectraMax L). We quantified transcription from each plasmid as follows: we collected six replicates of transfected cells in TRIzol reagent (Invitrogen) and extracted RNA using the Direct-zol RNA Mini Kit (Zymo Research). We then carried out qPCR as described above using the primer pairs Luciferase-F and Luciferase-R, and β-actin-Human-F and β-actin-Human-R. Primer sequences can be found in Table S1 (Supporting information).
Association tests
Using association mapping in a variable, natural population, previous work has shown that multiple mutations in Agouti are statistically associated with different aspects of coat colour (Linnen et al. 2013). These mutations may affect phenotype via measurable changes to Agouti expression level. To determine whether colour phenotypes and Agouti expression colocalize to the same Agouti region(s), we measured Agouti expression (overall Agouti and isoform 1C) as described above in 88 mice that had been genotyped and scored for colour traits as described in Linnen et al. (2013). To test for an association between Agouti genotype and expression, we performed single-SNP linear regressions in plink v1.07 (Purcell et al. 2007). To control for population structure, we genotyped individuals at 2077 unlinked SNPs as described in Linnen et al. (2013), and used smartpca to conduct a PCA, followed by twstats to evaluate the statistical significance of each principal component (Patterson et al. 2006). We detected four significant genetic principal components and included these as covariates in the association analysis. Prior to analysis, we performed a normal-quantile transformation on the phenotype and expression data in r to ensure that our phenotypic data were normally distributed (Guan & Stephens 2011). To correct for multiple testing, we used the step-up method for controlling the false discovery rate, which we set to a threshold of 10% (Benjamini & Hochberg 1995).
Results
Peromyscus expresses two Agouti isoforms not previously identified in Mus
RACE from P. maniculatus ventral samples revealed the presence of the ventral-specific isoforms 1A, and 1A1A’ (an isoform containing both the 1A and 1A’ exons), and the hair cycle-specific 1C, similar to what is seen in M. musculus (Vrieling et al. 1994). However, none of our clones contained sequences corresponding to exon 1B found in M. musculus (Vrieling et al. 1994) (Fig. 2A and Fig. S2, Supporting information). Our RACE experiment in dorsal skin revealed that, in addition to hair cycle-specific isoform 1C, P. maniculatus expressed two additional isoforms of Agouti, hereafter referred to as 1D and 1E, which had not been previously reported in M. musculus (Fig. 2A, B and Fig. S2, Supporting information). Like the previously described Agouti isoforms, 1D and 1E each consist of an alternate noncoding exon (exon 1D or 1E), followed by three protein-coding exons (exons 2, 3 and 4), which are shared by all Agouti isoforms. Both exons 1D and 1E are located downstream of exons 1A and 1C in the Agouti locus (Fig. 2B) and show polymorphisms between wideband and wild-type P. maniculatus (Fig. S2, Supporting information). To confirm the absence of isoform 1B in ventral/dorsal tissue and of isoforms 1D and 1E in ventral tissue, as indicated by our RACE experiments, we carried out qPCR using isoform-specific primers in the respective tissues and did not detect any expression of these transcripts. Together, our results show that P. maniculatus expresses some, but not all, of the Agouti isoforms previously described in M. musculus (the ventral-specific 1A, 1A1A’ and the hair cycle-specific 1C) and also contains two novel dorsal-specific isoforms that had not been described previously (1D and 1E).
Differences in Agouti mRNA levels between wideband and wild-type mice are driven by upregulation of isoform 1C
Wideband and wild-type mice differ markedly in their dorsal coloration; therefore, we focused here on analysing the isoforms present in the dorsum. Our RACE experiments demonstrated that the dorsal skin of P. maniculatus expresses at least three different Agouti transcripts (1C, 1D and 1E) simultaneously, but it remained unknown whether all three contribute to the increase in Agouti mRNA levels seen in wideband mice relative to wild-type ones or whether this difference is driven only by a subset. To answer this question, we used qPCR to measure the relative expression of total Agouti mRNA and each of its isoforms in dorsal skin of wideband and wild-type P. maniculatus. The expression of overall Agouti was approximately 22-fold higher in wideband mice than in wild-type mice (P = 0.0011, two-tailed t-test) (Fig. 3A). We then measured isoform-specific expression and found that isoform 1C was significantly higher in wideband than in wild-type mice (P = 0.0016, two-tailed t-test) (Fig. 3B). In contrast, we did not observe significant differences in mRNA levels between the two strains in expression of isoform 1D or 1E (P = 0.3080 and P = 0.6286, respectively; two-tailed t-tests) (Fig. 3B).
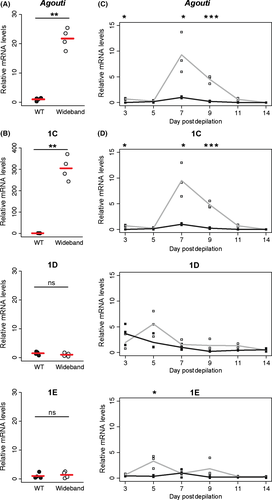
We next quantified total Agouti and isoform-specific expression at different time points following dorsal depilation to examine the dynamics of Agouti isoform expression during hair growth. In both strains, total Agouti is expressed initially at low levels, peaks at day 7 after depilation and then decreases (Fig. 3C), a pattern that mirrors the expression of Agouti during the first hair cycle in pups (Linnen et al. 2009). Total Agouti levels in wideband mice were higher than in wild-type mice at days 3, 7 and 9 after depilation (P = 0.0011, P = 0.0228 and P = 0.0006, respectively, two-tailed t-tests) (Fig. 3C). When we measured transcript-specific mRNA levels in wideband and wild-type mice, we found that the expression of isoform 1C closely matched the expression of overall Agouti, peaking at day 7, and differing from wild-type mice also at days 3, 7 and 9 (P = 0.0012, P = 0.0109 and P = 0.0003, respectively, two-tailed t-tests) (Fig. 3D). This marked similarity suggests that isoform 1C levels are primarily responsible for explaining overall Agouti levels. In contrast, isoforms 1D and 1E did not differ between wideband and wild-type mice at any time points, with one exception: expression of 1E was higher in wideband than in wild-type at day 5 (P = 0.0166, two-tailed t-test) (Fig. 3D).
To determine whether the same patterns occur in neonates, we measured isoform-specific mRNA levels at postnatal day 4, which corresponds to the stage in the first hair cycle when Agouti expression is highest (Linnen et al. 2009). Quantitative PCR confirmed that wideband mice express higher levels of total Agouti mRNA relative to wild-type mice (P = 0.0010, two-tailed t-test) (Fig. S3, Supporting information). In addition, like in adults, isoform 1C expression was significantly higher in wideband mice than in wild-type mice (P = 0.0003, two-tailed t-test), whereas there were no significant differences between the two strains in the expression of isoform 1D or 1E (P = 0.1575 and P = 0.3231, respectively, two-tailed t-tests) (Fig. S3, Supporting information). Together, the results of our measurements of isoform-specific Agouti mRNA levels in adults and pups indicate that the marked increase in Agouti expression seen in wideband mice, relative to wild-type mice, is primarily driven by upregulation of isoform 1C.
Agouti isoforms differ in luciferase production
To investigate functional variation associated with different Agouti isoforms, we measured their half-lives in vitro. After halting transcription from a vector expressing each isoform, we used qPCR to measure mRNA levels at different time points during the transcript decay that followed and did not detect any statistically significant differences between the half-lives of the three isoforms (P = 0.1230, one-way ancova) (Fig. S4, Supporting information). Thus, our experiment indicates that the three alternative exons simultaneously expressed in Peromyscus dorsal skin do not have measurable effects on the stability of the Agouti transcripts.
To determine whether the isoforms differed in their regulation of translation, we next examined whether each of the alternative exons affects the amount of protein produced from a luciferase transcript. Relative to a control vector expressing a luciferase coding sequence without a 5′UTR, a vector carrying exon 1C showed a marked increase in luciferase activity [P = 0.0390 (wideband sequence) and P = 0.0001 (wild-type sequence), two-tailed t-tests], whereas isoform 1D showed a marked decrease [P = 4.4 × 10−5 (wideband sequence) and P = 0.0002 (wild-type sequence), two-tailed t-tests]. Exon 1E from wideband mice was not significantly different from the control [P = 0.2577 (wideband sequence), two-tailed t-test], whereas exon 1E from wild-type mice showed increased luciferase activity compared with the control (P = 0.0082, two-tailed t-test) (Fig. 4A). Importantly, we found that mRNA levels did not differ between any of the vectors (P = 0.3790, anova), demonstrating that differences in translation, not transcription, are exclusively responsible for the differences in luminescence (Fig. 4B).
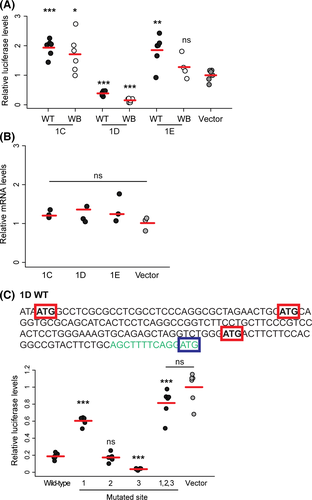
To further dissect the mechanisms underlying the differences in protein translation observed between the isoforms, we examined their sequences in more detail and found that exon 1D contains start codons (ATGs) upstream of the Agouti start codon (three in the wild-type 1D sequence and two in the wideband sequence) (Fig. 4C and Fig. S2, Supporting information). Upstream start codons have been shown to decrease translation efficiency by recruiting ribosomes away from the start codon (Kozak 2002; Rosenstiel et al. 2007; Song et al. 2007; Medenbach et al. 2011). To determine whether this can explain the reduced translation of isoform 1D, we mutated the ATG sites to ACG in the P. maniculatus wild-type sequence and quantified the relative amount of luciferase produced (Fig. 4C). When we mutated and tested each ATG site individually, we found that mutating site 1 led to a significant increase in luciferase production relative to the wild-type 1D sequence (P = 1.268 × 10−8, two-tailed t-test), mutating site 2 resulted in no significant difference (P = 0.5966, two-tailed t-test), and mutating site 3 led to a significant reduction (P = 1.6739 × 10−6, two-tailed t-test) (Fig. 4C). Thus, the different ATG sites in exon 1D differ in their ability to modulate luciferase production, possibly due to differences in their surrounding sequences (Kozak 1999, 2002). When we mutated the three ATG sites simultaneously, luciferase production from the mutant transcript was significantly higher than that from the wild-type 1D transcript (P = 0.000015, two-tailed t-test) and did not differ from the control (P = 0.0920, two-tailed t-test), indicating that the upstream start codons contained within exon 1D are responsible for the marked decrease in translation from this transcript. Together, these experiments demonstrate that the noncoding exons of the Agouti isoforms expressed in the dorsal skin of P. maniculatus differ in their regulation of protein translation, with noncoding exon 1C generating the highest level of luciferase, relative to the control, and noncoding exon 1D with its multiple ATG sites generating the lowest.
Genetic variation near exon 1C is associated with dorsal colour and Agouti expression in wild-caught mice
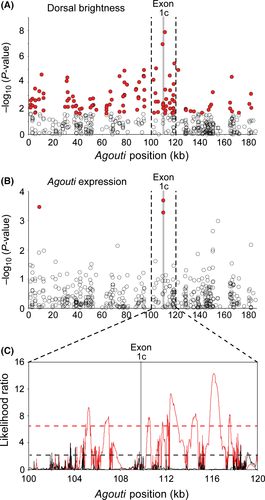
To evaluate evidence for exon 1C's contribution to adaptive coat colour variation in natural populations, we examined patterns of genotype–colour associations (from Linnen et al. 2013) and genotype–expression associations (this study) across the 180-kb Agouti locus in a phenotypically variable population of P. maniculatus. Here, we focus on dorsal brightness because we expect that this trait (i) has a large impact on substrate matching (hence, fitness) and (ii) has the potential to be strongly influenced by the hair cycle isoform 1C due to Agouti's direct impact on pigment deposition in hairs. For dorsal brightness, we previously identified a peak in association centred directly on exon 1C (Fig. 5A) (Linnen et al. 2013). Intriguingly, we also found a peak in association with expression at the same location (Fig. 5B). We note, however, that these expression data were generated using an assay that detects all Agouti isoforms. Nevertheless, when we evaluated the correlation between genotype and isoform 1C expression using 1C-specific probes, the exon 1C SNPs remained significantly associated (SNP 109 902, P = 0.0026; SNP 109 882, P = 0.0110). Although the lack of polymorphic positions in exon 1C that are derived in wideband mice, relative to the P. m. rufinus out-group (Linnen et al. 2009), suggests that the causal mutation is not in exon 1C itself, these association mapping data strongly suggest that there is a causal mutation somewhere in its immediate vicinity that simultaneously increases both expression of isoform 1C and, as a consequence, dorsal brightness.
Exon 1C has undergone strong positive selection
Although we have not yet identified the causal mutation, the results of our association mapping indicate that it should be in strong linkage disequilibrium with variants located near exon 1C (Fig. 5A, B). In this way, we can use the association mapping results to define light and dark haplotypes (i.e. those that contain the causal mutation and those that do not). If the light mutation has undergone positive selection in the light Sand Hills habitat, we expect to see signatures of selection on the light, but not dark, haplotypes. To evaluate evidence of selection on light and dark haplotypes, we previously (Linnen et al. 2013) used the composite-likelihood method implemented in Sweepfinder (Nielsen et al. 2005), a method that compares, for each location, the likelihood of the data under a selective sweep to the likelihood under no sweep. The significance of the CLR test statistic is then determined via neutral simulations. In our case, neutral simulations were conducted under a demographic model estimated from genomewide SNPs [as described in Linnen et al. (2013)]. Figure 5C depicts the resulting likelihood surfaces and significance thresholds for light and dark haplotypes across a ~20-kb window centred on exon 1C (Fig. 5A, B). This analysis indicates strong evidence of selection on the light, but not dark, haplotypes in this region of the Agouti locus. Specifically, within this window, the light haplotypes have a 3.6-fold increase in the number of sites rejecting neutrality and a 5.0-fold increase in the average selection coefficient. Together with the results of our association mapping, these analyses indicate that a mutation in the immediate vicinity of exon 1C contributes to dorsal colour and is currently undergoing strong positive selection [s = 0.14; Linnen et al. (2013)].
Evolutionary convergence of isoform regulation in Peromyscus
Our results show that Agouti isoform 1C is specifically upregulated in the light-coloured P. maniculatus wideband mice from the Nebraska Sand Hills relative to the dark-coloured ancestral population. We next investigated whether similar regulatory mechanisms are observed in another population of Peromyscus that independently underwent selection for light pigmentation. We examined patterns of isoform expression in light-coloured Santa Rosa Island beach mice (P. p. leucocephalus) and compared them to dark mainland P. p. subgriseus (Fig. 1). Quantitative PCR revealed that expression of Agouti was approximately threefold higher in beach mice compared with mainland mice (P = 0.0142, two-tailed t-test; Fig. 6A). Measurements of Agouti isoform-specific mRNA levels revealed that there were no significant differences in the expression of isoform 1D or 1E between beach mice and mainland mice (P = 0.5352 and P = 0.3826, respectively, two-tailed t-test; Fig. 6B). In contrast, we found that isoform 1C was significantly upregulated in beach mice compared with mainland mice (P = 0.0011, two-tailed t-test; Fig. 6B). Together, our measurements of mRNA levels demonstrate that the increase in Agouti expression seen in beach mice, relative to mainland mice, is produced primarily by a specific upregulation of isoform 1C, a pattern that matches what we observed in P. maniculatus (Fig. 3A).

Discussion
The Agouti locus, which contains multiple independently regulated transcription start sites and has been linked to pigment variation in Peromycus (Steiner et al. 2007; Mullen & Hoekstra 2008; Linnen et al. 2009, 2013) and other mammals (Rieder et al. 2001; Schmutz & Berryere 2007; Seo et al. 2007), represents an ideal study system to understand the importance of alternative transcript processing in adaptation to new environments. While different isoforms have been well studied in Mus musculus, in Peromyscus we identify both new dorsally expressed isoforms (1D and 1E) and the lack of expression of 1B across the body. These differences are consistent with genomewide surveys of isoform variation, which find rapid evolution of isoform usage between species (Barbosa-Morais et al. 2012; Merkin et al. 2012).
In this study, we also find that although the dorsal skin of Peromyscus mice expresses three different Agouti isoforms simultaneously, differing only in their first noncoding exon (1C, 1D and 1E), the marked differences in overall Agouti expression seen between P. maniculatus strains (wideband vs. wild-type) and between P. polionotus subspecies (P. p. leucocephalus vs. P. p. subgriseus) are exclusively driven by one of the isoforms (1C). Thus, populations of P. maniculatus and P. polionotus experiencing selection pressures for light dorsal pigmentation have independently converged not only on the same gene, but also on the specific upregulation of the same isoform. One explanation for this result is that exon 1C has inherent sequence properties that result in large amount of protein (relative to other Agouti isoforms), indicating that such convergence in isoform upregulation may be driven by selection for the molecular mechanism promoting the highest amount of Agouti protein production. In support of this, we find that in a admixed population in the Sand Hills, dorsal colour and Agouti gene expression is significantly associated with genetic variation around exon 1C and that this region shows a pattern of strong selection.
Given the patterns of phenotypic and gene expression association as well as the signatures of selection we have reported here and elsewhere (Linnen et al. 2013), it is likely that at least one causal polymorphism is located somewhere in the close vicinity of exon 1C (Fig. 5), as this population has low levels of linkage disequilibrium at the Agouti locus (Linnen et al. 2013). An important goal of future work is to characterize and functionally test the variants in this region, including indels and low-coverage SNPs that may have been absent in the capture-based genotype data. In the case of P. polionotus beach mice, despite the fact that selection on pigmentation is strong (s = 0.5; Vignieri et al. 2010) and Agouti is known to be a major contributor to pigment differences (Steiner et al. 2007; Mullen & Hoekstra 2008), any inferences of positive selection for regions in or around exon 1C are confounded by the unique demographic history of this species, which has experienced severe population bottlenecks associated with colonization events from the mainland to novel habitats in the Gulf Coast (Thornton et al. 2007; Domingues et al. 2012; Poh et al. 2014). Thus, it is not possible to evaluate with certainty whether specific regions in the Agouti locus show signatures of similar selective pressures between P. maniculatus and P. polionotus.
We find that the dorsal skin of Peromyscus expresses two isoforms that are not found in Mus or in other species (1D and 1E). However, it is unlikely that they have played a major role in the evolution of light coloration in Peromyscus because their expression patterns do not differ between the light- and dark-coloured strains examined here and do not follow the pulse of Agouti expression that occurs during hair growth (Vrieling et al. 1994). From a functional perspective, isoform 1D contains sequences that cause a marked repression of protein translation, so it is not surprising that this particular exon does not constitute a target for selection and does not show differences in expression between P. maniculatus strains or P. polionotus subspecies. In the case of isoform 1E, our functional experiments suggest that the sequence of exon 1E found in wild-type P. maniculatus increases protein translation, whereas the exon 1E sequence found in wideband P. maniculatus does not impact translation. The two strains’ sequences differ only at the end of the exon, where wild-type P. maniculatus has a six-base pair deletion (Fig S2, Supporting information). Importantly, however, the exon 1E sequence found in an out-group to the two strains, P. maniculatus rufinus, is identical to that found in wideband P. maniculatus, indicating that the deletion found in the wild-type 1E sequence is likely derived and arose after the split between wild-type and wideband populations. Thus, in the common ancestor of wild-type and wideband mice, exon 1C – and not exon 1E – would have been the only noncoding exon that promoted increased protein production. The driving forces underlying the evolution and maintenance of isoforms 1D and 1E in Peromyscus populations are yet unknown.
As Agouti's function is primarily linked to regulating pigment-type switching in melanocytes, changes affecting this gene are less likely to have negative pleiotropic consequences, and thus, selection pressures for eliminating particular transcripts from populations may be relaxed. Alternatively, isoforms 1D and 1E could be playing a role in other aspects of Agouti's function independent of its interaction with melanocytes that was not uncovered by our experiments, such as secretion and/or transport from the dermal papillae. Examining presence/absence and expression patterns of these isoforms in additional Peromyscus species and populations may shed light on some of these possibilities.
The findings presented here also bear on the molecular basis of convergent evolution. It has long been a topic of contention in evolutionary biology whether similar phenotypes that evolve independently tend to be generated by similar or different molecular changes (e.g. Manceau et al. 2010; Rosenblum et al. 2010; Stern 2013). Some studies of convergent phenotypes have found that they occur through independent mutations at different loci (e.g. Steiner et al. 2007; Weng et al. 2010; Kowalko et al. 2013), while others have found that similar evolutionary pressures on two populations can result in changes at the same gene (e.g. Woods et al. 2006), and in some cases, even the same amino acid substitutions (Zhen et al. 2012; van Ditmarsch et al. 2013). In the case of convergent pigmentation phenotypes in Peromyscus, not only is the same locus targeted, but the same pattern of isoform change has occurred – highlighting the various ways in which convergent evolution can occur at the molecular level.
Our results add an additional layer to the known mechanisms by which Agouti can play a role in the evolution of pigmentation phenotypes in Peromyscus. Cis-regulatory changes in Agouti are known to contribute to both the wideband phenotype in P. maniculatus (Linnen et al. 2009, 2013) and the beach mouse phenotype in P. polionotus (Steiner et al. 2007; Manceau et al. 2011). In addition, an amino acid change in the Agouti coding sequence of P. maniculatus wideband mice is strongly associated with light phenotypes and shows strong signatures of selection (Linnen et al. 2009, 2013). Here, we find that in addition to these changes, differences in Agouti isoform regulation have also been involved in the evolution of adaptive pigmentation variation in this genus. It is clear that alternative transcript processing provides a virtually limitless substrate for the generation of functional and structural transcriptomic and proteomic diversity, and genomic studies analysing rates of alternative splicing and alternative promoter usage have revealed the importance of this mechanism in originating diversity at large taxonomic scales (Barbosa-Morais et al. 2012; Merkin et al. 2012). Our study, by providing an example that links alternative mRNA processing with adaptation to a known selective pressure in different subspecies between sister species, highlights the importance of this mechanism as a driver of diversification and adaptation at smaller taxonomic scales as well.
Acknowledgements
We thank Judy Chupasko and Mark Omura for help with specimen preparation, Catalina Perdomo for advice and assistance with luciferase assays, Nicole Bedford for collecting samples for Santa Rosa Island Beach mice, and Nikki Hughes for providing logistical support. RM and his molecular work was supported by a Swiss National Science Foundation grant to HEH. TAL was supported by a Herchel Smith-Harvard Undergraduate Research Fellowship; CRL was supported by a Ruth Kirschstein National Research Service Award from NIH; HEH is an Investigator of the Howard Hughes Medical Institute. This work was supported by a grant from the Swiss National Science Foundation.
References
R.M., C.R.L. and H.E.H. conceived the project and designed the experiments. R.M., T.A.L. and C.R.L. performed experiments and analysed data. All authors wrote the manuscript.
Data accessibility
Data for coat colour measurements, soil reflectance and quantitative PCR have been deposited in Dryad doi: 10.5061/dryad.529p4.




