Multilocus genetic diversity and historical biogeography of the endemic wall lizard from Ibiza and Formentera, Podarcis pityusensis (Squamata: Lacertidae)
Abstract
Two monophyletic sister species of wall lizards inhabit the two main groups of Balearic Islands: Podarcis lilfordi from islets and small islands around Mallorca and Menorca and Podarcis pityusensis from Ibiza, Formentera and associated islets. Genetic diversity within the endangered P. lilfordi has been well characterized, but P. pityusensis has not been studied in depth. Here, 2430 bp of mtDNA and 15 microsatellite loci were analysed from P. pityusensis populations from across its natural range. Two main genetic groupings were identified, although geographical structuring differed slightly between the mtDNA and the nuclear loci. In general, individuals from islets/islands adjacent to the main island of Ibiza were genetically distinct from those from Formentera and the associated Freus islands for both mtDNA and the nuclear loci. However, most individuals from the island of Ibiza were grouped with neighbouring islets/islands for nuclear loci, but with Formentera and Freus islands for the mitochondrial locus. A time-calibrated Bayesian tree was constructed for the principal mitochondrial lineages within the Balearics, using the multispecies coalescent model, and provided statistical support for divergence of the two main P. pityusensis lineages 0.111–0.295 Ma. This suggests a mid-late Pleistocene intraspecific divergence, compared with an early Pleistocene divergence in P. lilfordi, and postdates some major increases in sea level between 0.4 and 0.6 Ma, which may have flooded Formentera. The program IMa2 provided a posterior divergence time of 0.089–0.221 Ma, which was similar to the multispecies coalescent tree estimate. More significantly, it indicated low but asymmetric effective gene copy migration rates, with higher migration from Formentera to Ibiza populations. Our findings suggest that much of the present-day diversity may have originated from a late Pleistocene colonization of one island group from the other, followed by allopatric divergence of these populations. Subsequent gene flow between these insular groups seems likely to be explained by recent human introductions. Two evolutionary significant units can be defined for P. pityusensis but these units would need to exclude the populations that have been the subjects of recent admixture.
Introduction
Coalescent theory has led to the development of statistical methods that enable key population parameters to be estimated. Application of these methods to sequence or length polymorphism data is popular because they can provide insights into population demographics and genetic structures, and help our understanding of causes of intraspecific diversity (Hey 2005; Rocha et al. 2011; Brown et al. 2012; Zieliński et al. 2013). They are also useful in conservation genetics, where population sizes and migration between populations with resultant changes in genetic structuring are key factors that could affect the long-term survival of conservation units (Paquette et al. 2007; Sly et al. 2010; Karl et al. 2011; Schoville et al. 2011; Reilly et al. 2012). The timing of intraspecific splitting events is also important, because the time periods over which populations have followed independent trajectories are informative when attempting to assess whether populations merit evolutionary significant- or management unit status.
Identification of suitable molecular markers is critical for informative coalescent-based inferences. Low substitution rates of nuclear sequences mean that they often contain relatively little information about recent population-level processes. In contrast, nuclear microsatellite markers have 100–1000 times higher evolutionary rates and are therefore more revealing about both recent historical and contemporary genetic patterns (Gilbert et al. 1990; Wan et al. 2004), but tend to be less suitable for examining earlier periods in a species' history. Despite some well-known drawbacks (Zhang & Hewitt 2003), the relatively high substitution rate of mtDNA (5–10 times that of nuclear sequences; Ballard & Whitlock 2004; Wan et al. 2004) makes it very useful for examining events that occurred during earlier periods of a typical species history. MtDNA is therefore quite a suitable marker for population and conservation genetics (Wan et al. 2004). This study employs microsatellite and mtDNA markers to examine populations of an island endemic lizard that appears to have originated just over 5 Ma, based on the rationale that they should be informative about very recent/current processes and older events in the species history, respectively.
Podarcis pityusensis (Boscá 1883) is found in the western Balearic Islands known as the Pityusic group. More specifically, it inhabits Ibiza and Formentera, along with 42 of their associated islets (Pérez-Mellado 2009; Salvador 2009). It is classified as ‘near threatened’ by the IUCN, unlike its sister species from the more eastern Balearic Islands (Gymnesic group), P. lilfordi, which is classified as ‘endangered’. Podarcis pityusensis shows considerable phenotypic variation among populations, in terms of size, sexual dimorphism, dorsal scales and coloration, which has led to the recognition of 23 subspecies (Salvador & Pleguezuelos 2002; Pérez-Mellado 2009). To date, there have been no detailed analyses of genetic diversity within this species. Isozyme markers were compared between subspecies by Guillaume & Cirer (1985), while Terrasa et al. (2004) examined mtDNA. Both studies detected low levels of diversity. Brown et al. (2008) included P. pityusensis from five sites in their mtDNA analysis of both Balearic Podarcis. Although this limited sampling captured the main mtDNA lineages, it was likely to represent only a small proportion of the total cross-archipelago diversity.
Some of the geographical events that might have impacted on the population history of Balearic Podarcis are well known. The Messinian Salinity Crisis (MSC) is a key event that includes the isolation and deep desiccation of the Mediterranean, and had an impact on many Mediterranean islands' taxa (Bover et al. 2008; Cano-Maqueda et al. 2008; Bauza-Ribot et al. 2011; Bidegaray-Batista & Arnedo 2011). After a long period of desiccation, 5.96–5.33 Ma ago (Krijgsman et al. 1999; Duggen et al. 2003), the Mediterranean Basin refilled within a very short time, possibly only a few months (Garcia-Castellanos et al. 2009). There is strong evidence to suggest that P. pityusensis became isolated from P. lilfordi at this time (Brown et al. 2008; Terrasa et al. 2009), so intraspecific lineages are expected to postdate the beginning of the Pliocene. MtDNA analysis of Balearic Podarcis dated the earliest sequence divergence among P. lilfordi at 1.45–3.99 Ma and the earliest divergence among P. pityusensis at 0.18–2.29 Ma (Brown et al. 2008). This predicts lower genetic diversity in P. pityusensis that could be explained by greater historical connections among populations: shallow channels (often <50 m) separate the main Pityusic islands of Formentera and Ibiza and their associated islets, and these will have been exposed during periods of low sea levels, as would have occurred during glacial maxima. Another crucial geological feature of the islands is that Formentera is mostly low and flat (although it does presently rise to a peak of 197 m), unlike Ibiza that reaches nearly 500 m. Major historical flooding during warmer interglacials, particularly Marine Isotope Stage 11 (~0.4 Ma ago), may also have led to the extinction of lineages confined to low-lying islands.
Knowledge of the genetic diversity within P. pityusensis is in part motivated by a political will to develop conservation strategies for the Balearic Islands endemic lizards. It will also contribute to our understanding of the historical biogeography of the Mediterranean. The species is subject to local (Balearic Islands Government) and international (CITES) levels of protection that do not distinguish among the different island populations. Any refinement of existing legislation and/or conservation strategies therefore requires more detailed knowledge of genetic structuring.
We aimed to provide the first detailed analysis of nuclear and mtDNA genetic diversity in P. pityusensis from across the Pityusic archipelago. The primary objectives were to establish the robustness of previously highlighted mtDNA lineages and establish whether these were also reflected in nuclear genealogies, determine the origin of these putative intraspecific units and assess the genetic evidence for their long-term persistence by examining effective population sizes and levels of migration between them.
Methods
Samples and DNA isolation
Seventy-four Podarcis pityusensis were captured by noose at 37 sites (1–5 individuals per site) (Fig. 1, Table 1), under license from the Balearic Islands Government (permit: CAP09/2010 from Conselleria de Medi Ambient). Islets with a documented history of introductions from populations from other islets were excluded (Böhme & Eisentraut 1981). The samples covered 22 of the 23 recognized subspecies. Tail tips were removed and stored in 100% ethanol. All individuals were released at the site of capture. Genomic DNA was extracted from the tissue samples using standard phenol–chloroform protocols.
| Id | n | Localities | Subspecies |
|---|---|---|---|
| Formentera Island | |||
| Ft | 5 | Punta Trocadors | P. p. formenterae |
| Fx | 4 | Sant Francesc Xavier | P. p. formenterae |
| Be | 1 | Cap Barbaria | P. p. formenterae |
| Sp | 1 | Sa Pujada | P. p. formenterae |
| Freus Islands | |||
| Er | 4 | Espalmador | P. p. formenterae |
| El | 3 | Espardell | P. p. formenterae |
| Al | 3 | Alga | P. p. formenterae |
| G | 3 | Castaví | P. p. gastabiensis |
| Nn | 4 | Negra Nord | P. p. negrae |
| P | 3 | Porcs | P. p. formenterae |
| Ip | 3 | Penjats | P. p. ahorcadosi |
| Cg | 3 | Caragoler | P. p. caragolensis |
| Tr | 3 | Torreta | P. p. torretensis |
| Ibiza Island | |||
| E | 5 | Sant Joan | P. p. pityusensis |
| At | 1 | Sant Josep de s'Atalaia | P. p. pityusensis |
| Sc | 3 | Sa Canal | P. p. pityusensis |
| Islands surrounding Ibiza | |||
| In | 3 | Illa Negra | P. p. pityusensis |
| Bc | 1 | Bosc | P. p. pityusensis |
| Cn | 1 | Conillera | P. p. carlkochi |
| Vd | 1 | Vedranell | P. p. vedrae |
| V | 1 | Vedrà | P. p. vedrae |
| Ee | 1 | Espardell de s'Espartar | P. p. frailensis |
| Ep | 2 | Espartar | P. p. kameriana |
| Ed | 1 | Escull d'Espartar | P. p. kameriana |
| Ev | 1 | Escull Vermell | P. p. maluquerorum |
| Bp | 1 | Bleda Plana | P. p. maluquerorum |
| Ng | 1 | Na Gorra | P. p. gorrae |
| Nb | 1 | Na Bosc | P. p. gorrae |
| Im | 2 | Illa Murada | P. p. muradae |
| Ma | 1 | Malvins Pla | P. p. schereitmuelleri |
| Cs | 1 | Cala Salada | P. p. calaesaladae |
| Se | 1 | Santa Eulària | P. p. redonae |
| Sh | 1 | S'Hort | P. p. hortae |
| Sr | 1 | Ses Rates | P. p. ratae |
| Ea | 1 | Es Canar | P. p. canensis |
| Ms | 1 | Ses Margalides | P. p. hedwigkamerae |
| Tg | 1 | Tagomago | P. p. tagomagensis |
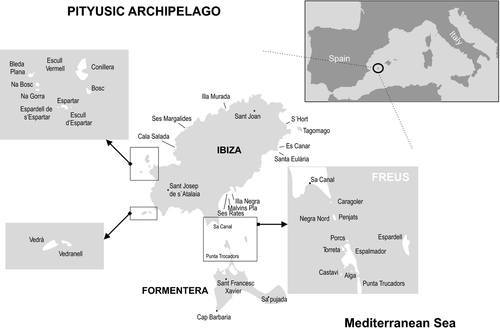
Microsatellite loci
Fifteen microsatellites were analysed, with all having been identified and tested in the sister species, Podarcis lilfordi (Bloor et al. 2011). PCRs were run in a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA, USA), using primers and conditions described in Bloor et al. (2011). Fluorescently labelled PCR products were run on an ABI 3130 DNA Sequencer (Applied Biosystems) with GeneScan-500 (LIZ) internal size standard. Fragment length was assigned using GeneMapper software v3.2 (Applied Biosystems).
General statistics of microsatellite diversity were obtained using Arlequin ver.3.1.1 (Excoffier et al. 2005). Analyses of molecular variance (amova) were also carried out using this program in order to test the statistical significance of genetic structure between the two main geographic groups (Formentera and adjacent Freus islands vs. Ibiza and associated islands, see Fig. 1). Genetic structure was inferred with a model-based clustering method implemented in the program STRUCTURE, version 2.3.3 (Pritchard et al. 2000), using an admixture model with correlated allele frequencies among populations. Twenty STRUCTURE runs (chain length = 200 000 steps, burn-in = 200 000 steps) were performed for all K between 1 and 20. The number of distinct genetic clusters was determined through assessment of ΔK (Evanno et al. 2005) using STRUCTURE HARVESTER (Earl & vonHoldt 2012). STRUCTURE and CLUMPP, version 1.1.2 (Jakobsson & Rosenberg 2007), were then used to assign individuals to clusters using the membership coefficient, Q. A threshold value of Q = 0.2 was used, because it is efficient and accurate at differentiating between purebreds and hybrids (Vähä & Primmer 2006), with Q values around 0.2–0.8 being indicative of hybridization between individuals from different clusters.
Mitochondrial DNA
Five mtDNA fragments were amplified to provide partial gene sequences from the following regions: 12S rRNA, cytochrome b (two separate regions), the control region, ND1 and ND2, and four tRNAs (tRNAThr,tRNAIle, tRNAGln and tRNAMet). Primers and amplification conditions are the same as those previously used for P. lilfordi (Terrasa et al. 2009). Both heavy and light strands were sequenced on an automated ABI 3130 sequencer (Applied Biosystems) using BigDye® Terminator v 3.1 Cycle sequencing kit (Applied Biosystems). The following homologous sequences were also obtained from previous studies: five P. pityusensis (GenBank Accessions nos: EF694768-69, EF694794-95, EU006725-26, EF694814-16, EF990552, EF694827-28) and ten P. lilfordi (GenBank Accession nos: EF694760-62, EF694764-66, EF694771, EF694773-75, EF694782, EF694785, EF694787-88, EF694799,EU006728, EU006730, EU006734, EU006738, EU006743, EU006745, EU006753, EU006756, EF694797, EF694799, EF694802, EF694805, EF694807, EF694809-10, EF990517, EF990522, EF990525, EF990531, EF990536, EF990540-41, EF990545-46, KC623944). Sequences were aligned using BioEdit Sequence Alignment Editor ver.7.0.5.2 (Hall 1999).
Estimation of genetic diversity and tests of neutrality were carried out using DnaSP ver.5.10.01 (Librado & Rozas 2009). Statistical significance of geographical structuring was examined in the same way as for the microsatellites, that is, using analysis of molecular variance (amova) on the same geographical groupings. The Bayesian method in BAPS, version 5.3 (Corander et al. 2003), was used to infer population structure. Genetic mixture analyses were performed with an upper bound of K = 40 (no priors on geographic location were used). The number of clusters was determined, and then the admixture analyses were executed to infer the ancestral source of the samples.
*BEAST, version 1.7.4, was used to infer the phylogenetic relationships and divergence times between the main Balearic Podarcis populations using the multispecies coalescent approach (Heled & Drummond 2010; Drummond et al. 2012). Nuclear microsatellites cannot be used in this analysis, and so the mitochondrial sequence alone was used. The method has the advantage of estimating divergence times while taking into account within-population ancestral polymorphism (of mtDNA in this case). In this application, we use genetically divergent potentially noninterbreeding Podarcis populations, rather than recognized species and so will use the term ‘population tree’, rather than the more widely applied ‘species tree’. The populations corresponded to two genetic groups of P. pityusensis that were detected here and five P. lilfordi lineages detected by previous studies (Brown et al. 2008; Terrasa et al. 2009). The age of the (P. lilfordi, P. pityusensis) node on the population tree was constrained using the normal distribution N(5.325, 0.0001), where 1 unit = 1 Ma. There is good evidence to suggest that rising sea levels separated the Gymnesic and Pityusic Islands groups at the end of the Messinian salinity crisis (Krijgsman et al. 1999; Duggen et al. 2003). Also, the degree of divergence in mitochondrial genes that have been well characterized in other reptiles strongly supports separation of these groups at this time (Brown et al. 2008).The time of the most recent common ancestor (MRCA) of all P. pityusensis was loosely constrained using the gamma distribution G(5.8, 0.2), and the MRCA of P. lilfordi was loosely constrained using G(3.48, 0.5). MtDNA sequence was assigned to eight partitions that are defined as: 12S rRNA, control region, all tRNAs, ND1/ND2 1st + 2nd codon position, ND1/ND2 3rd codon position and a single partition for each codon position within the cytochrome b sequence. Evolutionary models were tested using AICc (Akaike Information Criterion corrected for small samples) and for each partition, using MrAIC v 1.4.4 (Nylander 2004). If the selected model was not available in *BEAST, then the most similar more complex model was used instead.
The *BEAST MCMC sampler was run for 400 million generations, with one step per 10 000 being sampled. A strict clock model was specified (see Brown & Yang 2011), and a Yule speciation process used for the tree prior. Tracer, version 1.5 (Rambaut & Drummond 2007), was used to analyse the trace files generated by Bayesian MCMC runs, to check for convergence. Posterior trees were combined to obtain a tree with a maximum sum of posterior clade probabilities (for both population and gene trees).
Historical biogeography and demography
We applied the isolation-with-migration model (Nielsen & Wakeley 2001), implemented in IMa2 (Hey 2010a,b), to explore other aspects of population history using the entire mtDNA sequence and 15 microsatellite loci. This coalescent-based MCMC approach was applied to the two main islands groups (Ibiza and Formentera, each with their associated islands) in order to examine migration of gene copies between these groups after splitting. The Hasegawa–Kishino–Yano mutation model (HKY) (Hasegawa et al. 1985) was used for the mtDNA, while the stepwise-mutation model (SMM) (Kimura & Ohta 1978) was used for the microsatellites. We specified uniform priors for divergence time, population size and migration rates. A geometric heating model, with 40 independent heated chains, was found to provide good mixing, and high repeatability between the first and second halves of the run, and between analyses that began from different starting positions. After exploratory runs, we ran the MCMC chain for 5 × 106 steps, but discarded the first 5 × 105steps as burn-in. Conversion of parameters to real time was achieved using a mutation rate of 6.075 × 10−3 mutations per site per Ma, from previous estimates for this mtDNA fragment (Brown et al. 2008), and a generation time of 2.09 years (Galán 1999).
Results
Microsatellite variability and population structure
Sixty-nine Podarcis pityusensis were genotyped for all 15 microsatellites. Numbers of alleles per locus ranged from 9 (Pli22 or Pli24) to 25 (Pli4), with a mean of 15.7 (Table S1, Supporting Information). Observed heterozygosity was generally lower than expected heterozygosity for most of the loci, which is indicative of genetic structuring (Table S1, Supporting Information). Low values for the statistic in Garza & Williamson's test (M = 0.160–0.371) was indicative of recent size reductions (Garza & Williamson 2001).
Most variation was found among individuals within Formentera and Ibiza regions (97.6%) as opposed to between them (2.4%; FST = 0.024; P < 0.001). We also assessed an additional level of subdivision using amova, with the regions divided into groups, that is, Ibiza divided into the Ibiza main island plus associated islands and Formentera divided into the Formentera main island plus islets in the channel between Ibiza and Formentera (Freus islands). This indicated 1.5% variation between the Ibiza and Formentera regions, 1.5% between-groups within regions and 97% within populations.
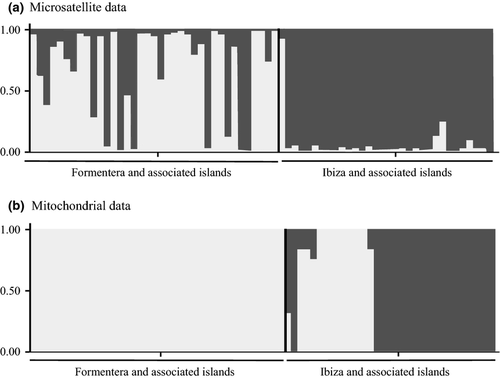
Analysis of ΔK (Evanno et al. 2005) estimated by STRUCTURE and STRUCTURE HARVESTER indicated two genetically distinct clusters (highest value of ΔK = 11.325) (Fig. 2a). K = 4 presented the second highest ΔK value (ΔK = 3.961). In general, groups are associated with geographic locations: one of them (cluster I) was found in Formentera and the associated Freus islands, with six individuals presenting slightly lower membership coefficients (ranging from 0.50 to 0.80). The second cluster (cluster II) contains populations from the main island of Ibiza and associated islets, but also included nine admixed individuals, with Q ranging from 0.61 to 0.98, some of them belonging to the same localities poorly assigned to cluster I. Individuals from Ibiza were unequivocally assigned to cluster II (Q > 0.9), with three exceptions: two individuals with slightly lower values of Q (Malvins Pla: Q = 0.76 and Illa Murada: Q = 0.87) and one of three individuals from Sa Canal (a peninsula sited in southern Ibiza, very close to Freus islands) (Q = 0.09) being assigned to cluster I.
MtDNA diversity and population structure
The five mitochondrial fragments were sequenced for 73 individuals (sequences have been deposited in GenBank: Accession nos JX852045-JX852108 and JX852118-JX852137) to provide a total concatenated fragment length of 2430 bp (cytochrome b, 831 bp; 12S rRNA, 373 bp; ND1, 59 bp; ND2, 415 bp; tRNAs, 271 bp; control region 481 bp). Sixty different haplotypes were detected within P. pityusensis, which differed by a mean (uncorrected) pairwise difference of 9.1, and contained 90 polymorphic sites. All diversity indices estimated in P. pityusensis were lower than corresponding values for P. lilfordi (Table 2).
| Species | No. | V | ti/tv | A+T (%) | No. hap | Nucleotide diversity | Mean pairwise differences | Haplotype diversity | Fu′s Fs (1997) | Fu & Li′s D (1993) | Fu & Li′s F (1993) | Fay & Wu′s H (2000) | Tajima's D (1989) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. lilfordi a(2382 bp) | 117 | 189 | 4.52 | 61.3 | 62 | 0.019 ± 0.005 | 45.067 ± 19.503 | 0.979 ± 0.005 | −1.129ns | 0.652ns | 0.860ns | −17.572ns | 0.770ns |
| P. pityusensis(2430 bp) | 73 | 90 | 3.20 | 61.1 | 60 | 0.004 ± 0.001 | 9.083 ± 4.17 | 0.992 ± 0.004 | −56.983*** | −2.432** | −2.589** | −25.341** | −1.745* |
- No., number of individuals sampled; V, variable position; No. hap, number of haplotypes; ns, not significant.
- *P < 0.1; **P < 0.05; ***P < 0.001.
- a Obtained from data in Terrasa et al. (2009).
The genetic structure was assessed by amova, which estimated the variance partitions as 39.4% between the Ibiza and Formentera regions and 60.6% within these regions (FST = 0.394; P < 0.001). Assessment of an additional subdivision of geographic populations (Ibiza: Ibiza island vs. associated islands; Formentera: Formentera island vs. Freus islands) partitioned the variance as 30.8% between Formentera and Ibiza regions, 14.5% among-groups within regions and 54.7% within populations.
The BAPS analysis of the mtDNA locus defined two genetic clusters (lnL = −1664.367) (Fig. 2b). The first cluster (A) included sites from Formentera, Freus islands, Ibiza main island, as well as from some of the Ibizan islands and islets (Es Canar, Cala Salada, Na Gorra, Vedranell and Vedrà). Ibiza main island showed considerable diversity, with two samples (from one site) assigned to the second cluster (B). This second cluster contained the majority of islands and islets associated with Ibiza, that is, Espartar and surrounding islets (Escull d'Espartar and Espardell de s'Espartar), Bosc, Na Bosc, Bleda Plana, Escull Vermell, Conillera, Illa Murada and Ses Margalides to the north of Ibiza; s'Hort and Tagomago in the east; and Ses Rates, Illa Negra and Malvins Pla in the south (see Fig. 1).
Species phylogeny and divergence times
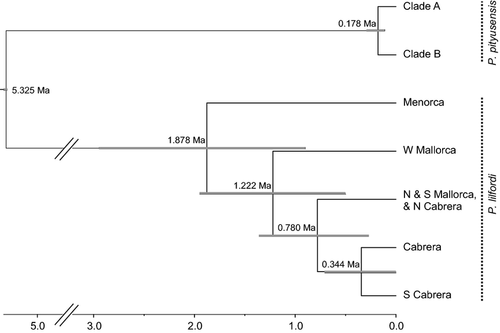
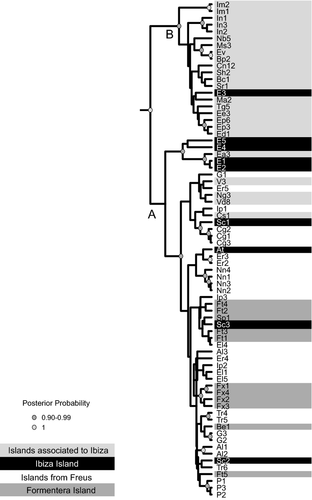
Strong posterior support (P = 1.00) was obtained for the basal Balearic Islands Podarcis nodes that corresponded to the MRCAs of the recognized species P. lilfordi and P. pityusensis. The mean posterior divergence time between the two P. pityusensis ‘species’ was 0.178 Ma (95% highest posterior density (HPD): 0.111–0.295 Ma; Fig. 3). The ancestral species node for P. lilfordi was dated at 1.878 Ma (95% HPD: 0.899–2.948 Ma; Fig. 3). The mitochondrial DNA tree for P. pityusensis is shown in Fig. 4.
Historical biogeography and demography
IMa2 provided informative posteriors for all parameters (Fig. 5). Replicated analyses that started from different positions converged on the same posterior. The posterior mean of t was 0.6697 (95% HPD: 0.3955–0.9765). Conversion into time in years suggests that Ibiza and associated islands diverged from the remaining Pityusic populations about 0.151 Ma (95% HPD: 0.089–0.221 Ma). Effective population sizes were approximately six times higher in Ibiza and associated islands (4.23 × 106; 95% HPD: 1.29 × 106–8.86 × 106) than in Formentera and Freus islands (6.47 × 105; 95% HPD: 3.31 × 105–9.93 × 105), with a size more similar to the ancestral population (4.36 × 105; 95% HPD: 2.50 × 105–6.28 × 105). Estimated effective number of migrant gene copies per generation (2NM) was higher from the Formentera group to the Ibizan group (7.19; 95% HPD: 0–22.33), than in the opposite direction (1.43; 95% HPD: 0–4.45).
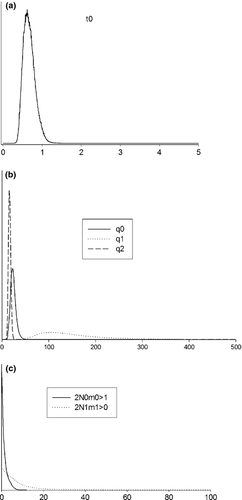
Discussion
We confirm the prediction of lower intraspecific diversity in Podarcis pityusensis from the Pityusic group of Balearic Islands, than that identified previously in Podarcis lilfordi from the Gymnesic group. This is largely due to the presence of two extant intraspecific lineages with a very recent origin in P. pityusensis, as opposed to five lineages (some of which date to the early Pleistocene or earlier) in P. lilfordi (Brown et al. 2008). Possible biogeographical scenarios that could explain the origin of the P. pityusensis lineages are discussed below. Our second major finding is that two major genetic groups are detected within both the microsatellite loci and mtDNA. While this could help recognition of units for conservation, this is complicated by evidence of some geographical discordance between markers (see below). Finally, our coalescent analyses detect nonzero migration as isolation between geographical regions that are currently isolated by sea channels.
The finding of relatively high migration between the Ibiza and Formentera regions could be explained by recent introductions. There are well-documented records of translocations of lizards between islets (Böhme & Eisentraut 1981), and although we did not use specimens from these populations, it would seem likely that there have been additional introductions that we are unaware of. For example, the pattern found in Formentera and associated island populations appears to evidence recent introductions. Specimens from this region are largely homogenous in terms of mtDNA, but several Formentera specimens show high affinities with Ibizan populations in terms of nuclear microsatellites. The presence of admixed individuals provides evidence of hybridization and intermediate membership coefficients (of around 0.5 in some cases) even suggest first generation hybrids. A second example of possible introductions is found in the Ibizan populations. They form a clear genetic cluster in terms of nuclear microsatellites. However, the mtDNA of the majority samples from the main island of Ibiza, as well as some associated islands (Es Canar, Vedrà, Vedranell, Na Gorra and Cala Salada), corresponded to the Formentera group. This could be explained by mitochondrial capture after introductions from Formentera that could arise under a selective advantage for Formentera mtDNA, a relatively common explanation of mitonuclear discordance (Toews & Brelsford 2012). We detected greater effective migration of gene copies from Formentera to Ibiza, which supports this hypothesis. Some Ibiza specimens were clearly distinct from Formentera in terms of both nuclear and mtDNA (two specimens from the main island and those from the offshore islets of Espartar, Escull d'Espartar, Espardell de s'Espartar, Bosc, Na Bosc, Bleda Plana, Escull Vermell, Conillera, Illa Murada, Ses Margalides, s'Hort, Tagomago, Ses Rates, Illa Negra and Malvins Pla). This appears to be of considerable significance for the conservation genetics of this species. The offshore populations in particular will have been isolated from the effects of mitochondrial colonization of the main island populations and so represent the genetic composition of the original Ibiza lineage prior to putative introductions. These populations therefore merit special conservation measures because they represent the Ibizan lineage prior to introductions. Of course, we cannot be certain that mitonuclear discordance is not due to incomplete lineage sorting. However, the observed biogeographical patterns do not generally show the random characteristics that might be expected if this was the case (Funk & Omland 2003; Toews & Brelsford 2012). Also, the populations are highly fragmented and have probably been this way for some time, which seems to rule out causes that involve high levels of natural gene flow between divergent lineages, such as sex-biased differences in hybridization.
Podarcis pityusensis is an endemic island vertebrate that makes it more prone to extinction (Frankham 2003). It exists mostly in small fragmented populations with reduced gene flow. Small fragmented populations are expected to suffer both immediate effects of genetic diversity loss (inbreeding and fixation of deleterious alleles) as well as longer-term effects such as decreased long-term ability to adapt to environmental change (Frankham 2003). This means that descriptions of patterns of genetic diversity and identification of management units are essential for conservation strategies.
Recent introductions preclude easy definition of evolutionary significant (ESU) or other management units. ESUs are a common output from conservation genetics studies and of considerable use in ensuring that the evolutionary heritage that has led to high genetic and ecological distinctiveness is recognized and protected (Funk et al. 2012; Reilly et al. 2012). The criteria frequently used for recognition of ESUs based on mtDNA and microsatellite data are reciprocal monophyly and significant divergence of alleles frequencies, respectively (Moritz 1994; Fraser & Bernatchez 2001). These criteria are only met in P. pityusensis if we do not consider islands/islets where lizards appear to have been introduced. This would mean that many of the populations adjacent to Ibiza (but excluding Ibiza itself) would represent one ESU, while most of the Formentera and Freus populations (with the exception of those showing Ibizan microsatellite alleles) would represent a different ESU.
There was no genetic support for the ~23 morphological subspecies of P. pityusensis, a finding that is not unusual in vertebrates (Zink 2004). Small genetic differences may explain the morphological characteristics on which some of these subspecies are based. For example, three of the populations are melanic, and it has been shown that small genetic substitutions in certain genes can cause dark/light variation in dorsal colour within some species (Rosenblum et al. 2010)(note that a recent study rules out an association between the mc1r gene and melanism in P. pityusensis: Buades et al. 2013). It would seem inappropriate to base an intraspecific taxonomy on discrete genetic differences that do not closely reflect the population tree. However, the recognition of distinct taxa could help promote conservation of these unique morphological forms. They may represent adaptive variations that could be important for the future existence of the species by providing a reservoir of genetic variation for the species. Another way to conserve morphological diversity, besides the maintenance of distinct taxa, is to identify managements units (MUs). MUs are defined as demographically independent populations, whose populations dynamics depend largely on local birth and death, rather than on immigration (Palsbøll et al. 2007; Funk et al. 2012). Nongenetic measures, such as the degree of similarity of morphological traits, could be used to define MUs (Palsbøll et al. 2007). In P. pityusensis, some phenotypes are associated with specific small islands or islets, which could be delineated as MUs. For example, specimens from Bleda Plana and Escull Vermell islands are melanic and have been assigned to P. p. maluquerorum (Mertens 1921). It exists as two demographically independent populations and could be defined as a MU, which would help conserve this component of the morphological diversity.
It is useful to speculate on the biogeographical scenario that underpins the origin of the two main lineages in P. pityusensis. Lizard diversity across and within islands is often explained by ancient dispersal between distant islands and population vicariance due to geophysical events within islands (Brown & Pestano 1998; Malhotra & Thorpe 2000; Stenson et al. 2004; Cox et al. 2010). Smaller sizes and closer proximities of islands within the Ibiza and Formentera group could reduce the potential for lineage divergence in P. pityusensis compared with its sister taxon P. lilfordi. However, a more reasonable explanation is that ancient P. pityusensis lineages have been lost through extinctions. It seems unlikely that gene flow was so high between these highly fragmented populations that cladogenesis was impeded from the time of the origin of the species (5.33 Ma) until just 0.1–0.3 Ma (our estimate of the time of the most basal divergence within P. pityusensis). Instead, it seems more credible that lineages appeared and subsequently became extinct. Ibiza is relatively high (487 m), as are some of its associated islets (e.g. Vedrà, 381 m and Tagomago, 115 m), but most of Formentera (and many offshore islets) is below 50 m (although it does contain two peaks of 105 and 197 m). As a result, Formentera would have been largely flooded by temporary Plio- and Pleistocene rises in sea level, providing a potential extinction mechanism. There is some evidence to support a major rise in the Mediterranean sea level some 0.6 Ma (Emig & Geistdoerfer 2004), which fits this pattern because it considerably pre-dates the divergence of the Ibiza and Formentera groups. This was followed by significant drops in sea level ~0.5 Ma, around 0.15 Ma and again during the last glacial maximum marine isotopic stages (MIS 2, 0.02 Ma, with decreases of 120 m) (Emig & Geistdoerfer 2004). These falls in sea level must have joined Ibiza and Formentera, which are separated by a channel of only 50 m. These dates do not seem to be incontrovertible. In Mallorca, sea level increases of 55, 35 and 8 m have been proposed for MIS, corresponding to interglacial periods, 11 (0.4 Ma), nine (0.33 Ma) and seven (0.2 Ma) (Butzer 1975; Cuerda 1975), while during the last interglacial (MIS 5; 0.085–0.135 Ma), sea level decreases of 15 and 20 m have also been proposed (Dorale et al. 2010). Despite these differences, there does seem to be a consensus that there was a sea level rise prior to the estimated divergence time that could have led to the extinctions of ancient populations/lineages in low-lying areas across Formentera and many of the islets. Recolonization of previously flooded islands would be most likely when between-island channels were exposed by drops in sea level, and subsequent rises in sea level would in turn isolate these populations. The divergence times that we estimated between the two P. pityusensis populations could therefore be explained by isolation of Formentera populations that had previously colonized the island from Ibiza.
Finally, it is important to consider the population divergence times using *BEAST and IMa2 with reference to estimates of sequence divergence times obtained by a previous analysis (Brown et al. 2008). Divergence time estimates are considerably more recent than those estimated previously, despite a similar tight calibration on the (P. lilfordi, P. pityusensis) node. For example, the oldest sequence divergence within P. pityusensis was previously estimated as 0.94 Ma ago, while here, population divergence is estimated at around 0.2 Ma ago. This is largely expected because these new analyses account for the ancestral polymorphism within populations. This approach has also been found to considerably reduce estimated timing of shallow population divergence in other species (Brown et al. 2012). Bayesian dating of shallow phylogenies requires considerable care (Brown & Yang 2010). Here, we aimed to overcome the main sources of error by using a strict clock and a substantial amount of informative sequence (Brown & Yang 2011). Nevertheless, the problem of unexpected prior distributions is still potentially problematical (Heled & Drummond 2012). The similarity of the mtDNA-based *BEAST estimate and the mtDNA and microsatellite-based IMa2 estimates of population divergence time provided confidence in their robustness, although important differences need to be outlined. First, the *BEAST analysis estimated timing between the two major mtDNA population units. This appeared sensible because we solely aimed to date the population split that led to these two mtDNA lineages. In contrast, the IMa2 analysis was used to estimate time since population splitting and migration of gene copies between the two geographical populations, using both microsatellites and mtDNA (*BEAST does not incorporate migration). Second, time calibrations within the two approaches were related because the mutation rate in IMa2 and the species node calibration in *BEAST were based on the post-Messinian divergence of the two Balearic Podarcis. However, divergence time in years in IMa2 requires an estimate of generation time adding uncertainty to the estimates. In sum, the fact that both methods provide results that fit with our biogeographical scenario, despite the existence of significant differences between them, provides considerable confidence in the estimated divergence time.
To summarize, we have applied coalescent analyses to microsatellite and mtDNA data to provide insights into the evolutionary history of this island endemic. We find strong evidence for two genetic groupings that we believe may have been initiated by changes in sea level, as well as finding evidence that the current phylogeographical distribution is quite likely to have been influenced by man-mediated introductions. The genetic integrity of ancient lineages is compromised by these introductions, although our study allows us to identify genetically ‘uncontaminated’ populations that may be important when formulating conservation policy.
Acknowledgements
We are grateful to the IFISC (Institute for Cross-Disciplinary Physics and Complex Systems), we used the Nuredduna Mosix cluster to run IMa2 analyses and we thank J.A. Jurado for computational support. This work was financed by the Research Grants CGL2009-12926-C02 and CGL2012-39850-C02 of the Ministerio Español de Economía y Competitividad and European Regional Development Fund (ERDF). V.R. was granted with an FPI fellowship from Conselleria d'Educació, Cultura i Universitats (Govern de les Illes Balears), cofinanced by European Social Fund.
References
Conceived and designed the experiments: M.M.R., J.A.C., R.P.B.; Sampling: V.P.M.; Performed the experiments and analyzed the data: V.R.; Contributed reagents/materials/analysis tools: V.P.M., A.P., B.T.; Wrote the paper: V.R., R.P.B., M.M.R.
Data accessibility
DNA sequences: GenBank Accessions nos: JX852045-JX852108 and JX852118-JX852137. IMa2 input file: Dryad Digital Repository. doi: 10.5061/dryad.r1538.




