Estimation of Foraging Grounds Based on Morphological and Molecular Analyses of Green Turtle Diets
Funding: JSPS KAKENHI Grant Number 19K06806—Ichimura Foundation for New Technology—Yanmar environmental sustainability support association.
ABSTRACT
Green turtles, Chelonia mydas, migrate to the Ogasawara Islands for breeding once every few years. However, the foraging grounds of these turtles prior to their visit to the Ogasawara Islands remain uncertain. In this study, we examined the genetic similarity between macroalgal specimens collected from various coastal regions and macroalgal fragments in the gut contents of green turtles visiting the Ogasawara Islands in order to estimate their foraging grounds. A total of 4 green, 13 brown, and 3 red algal species were identified from the gut contents of 62 adult green turtles. Macroalgal species composition differed by year of green turtle harvest, but not by sex of green turtle. Grateloupia angusta and Besa paradoxa were the most abundant temperate species obtained from the intestines, but they are rarely distributed around the Ogasawara Islands. Therefore, the cox1 gene and/or cox2-cox3 spacer region, which are useful to detect intraspecific genetic diversity, were analyzed for the two algal species obtained from the intestines and collected from various sea coasts. Four haplotypes of G. angusta recovered from the intestines were identical to specimens from some Japanese Pacific coasts and Korean islands. In contrast, a single haplotype of B. paradoxa retrieved from the intestines was identical to that of specimens from Enoshima, which is close to Tokyo. These data suggest that green turtles may have come to the Ogasawara Islands from different foraging grounds depending on the year. Determining the origin of green turtle diets is important to better understand the migratory connectivity between grazing and breeding grounds.
1 Introduction
Green turtles (Chelonia mydas) inhabit tropical, subtropical, and temperate waters, spending the majority of their lives on coastal foraging grounds (Seminoff et al. 2015). Adult green turtles typically migrate irregularly between their foraging grounds and distant breeding sites, with female turtles displaying a strong fidelity to these sites (Limpus et al. 1992; Broderick et al. 2007). It is therefore vital to understand the foraging grounds and migration routes of this endangered species if effective conservation and management strategies are to be developed (Webster et al. 2002; Boyle et al. 2009). However, it is challenging to determine the specific foraging grounds and migration routes due to the vast distances between breeding and foraging habitats, in addition to the protracted nature of these migrations (Hays and Hawkes 2018; Hays et al. 2020). Satellite transmitters, which are useful for tracking the migration route of green turtles (e.g., Kim et al. 2024), can extend for several years before falling off naturally, but the habit of green turtles rubbing their carapace against rocks causes detachment of transmitters. Consequently, the actual tracking period is only about 1 or 2 months (Hatase et al. 2006; Oki et al. 2019), which makes it challenging to examine their prenesting behavior.
Analysis of diet composition, which is important for understanding the feeding habitats and grounds of green turtles, has been performed using a variety of techniques, such as analysis of gut contents from wild-captured and stranded turtles (e.g., Reisser et al. 2013; Stokes et al. 2019), esophageal lavage and fecal examination (e.g., Seminoff et al. 2002), stable isotope analysis (Shimada et al. 2014; Pearson et al. 2017; Gama et al. 2021; Glen et al. 2024; Velasquez-Vacca et al. 2024), and DNA barcoding (Kim et al. 2021). It has been documented from multiple geographical locations that adult females exhibit greater size and accelerated growth rates in comparison to adult males (Caldwell 1962; Frazier 1971; Limpus 1993; Godley et al. 2002). Since females have higher fat and lower plasma uric acid levels than males (Bolten and Bjorndal 1992; Kwan 1994), their feeding habits may differ between the sexes. However, the majority of research on dietary preferences has been conducted on juveniles or nesting adult females (e.g., Carr et al. 1974; Mortimer 1982; Tucker and Read 2001; Fuentes et al. 2006), and there is a paucity of studies that have compared feeding habits between the sexes.
In Japan, green turtles foraging at the main island sites are predominantly of Ogasawara origin, with limited records from Taiwan and Micronesia (Hatase et al. 2006; Nishizawa et al. 2013; Seminoff et al. 2015), and the majority migrate to Ogasawara for breeding (Nishizawa et al. 2013). Green turtles that have been tagged at nesting sites on Ogasawara return to the same rookery after 3 or 4 years (Seminoff et al. 2015; ELNA unpublished data). The harvesting of green turtles for food has been a continuous process in the Ogasawara Islands and is currently strictly managed in accordance with the adjustment regulations of fisheries in Tokyo (Kondom et al. 2017). Gut contents obtained from these adult females and males can be used for gastrointestinal analysis.
Satellite tracking data have revealed that green turtles migrate to temperate neritic areas in Japan during postnesting periods from the Ogasawara Islands (Japan Fisheries Resource Conservation Association 1999; Hatase et al. 2006). Given that temperate macroalgae are frequently recovered from the gut contents of these green turtles (Kameda and Ishihara 2009), these macroalgae are likely to be grazed along temperate Japanese coasts prior to the migration to Ogasawara. However, the wide distribution of many temperate macroalgal species (Guiry and Guiry 2025) complicates the specification of foraging grounds based on the species composition of their diets.
Recent molecular analyses have revealed that many macroalgal species are genetically differentiated, and their haplotype composition differs among the Japanese coastal regions (e.g., Uwai et al. 2009; Hanyuda et al. 2016; Kurihara et al. 2016). Cytochrome c-oxidase (COX), the terminal enzyme of the mitochondrial respiratory chain, is encoded in the mitochondrial genome of all aerobic organisms, and the 5′ region of COX subunit I (COI-5P) and the spacer and its flanking region of COX subunit II and III genes (cox2-cox3 spacer) are frequently used for the study of inter/intraspecific diversity and phylogeography of macroalgal species, specifically for red algae (e.g., Saunders 2005; Sherwood et al. 2008; Kamiya and West 2014; Kurihara et al. 2016). The sequence comparison of these DNA regions between particular macroalgal species recovered from the gut contents and those collected from various Japanese coasts will provide useful information for identifying foraging grounds of prenesting green turtles.
The present study involved the examination of dietary components recovered from the esophagus, stomach, and intestine of harvested green turtles, as well as the analysis of differences in macroalgal species composition according to sex, size, and year of the green turtle harvest. Temperate red algae Grateloupia angusta and Besa paradoxa, which were recovered from the intestines of harvested green turtles, are rarely found around Ogasawara and are possibly grazed along temperate Japanese coasts before the green turtles migrate to Ogasawara. Therefore, the COI-5P and/or the spacer region of cox2 and cox3 were sequenced in G. angusta and B. paradoxa recovered from the gut contents and those specimens from various Japanese coasts, and the foraging grounds of prenesting green turtles were identified based on the sequence similarities.
2 Materials and Methods
2.1 Sampling and Gastrointestinal Content Analyses
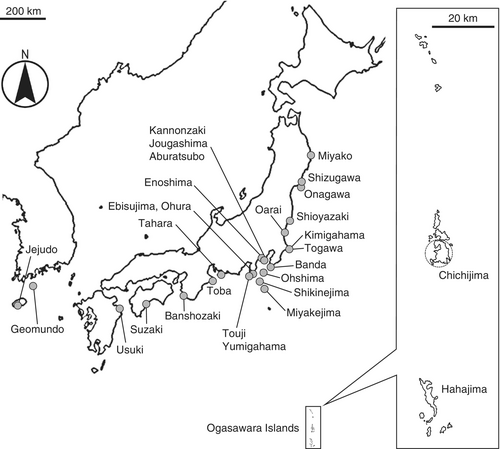
We used the green turtles harvested by fishermen with the permission of the governor of Tokyo. In March 2019 and March to May 2020, 62 adult green turtles (33 females and 29 males) harvested at Futami Bay in Chichijima, Ogasawara, Japan (27°02′ to 27°06′ N, 142°10′ to 142°14′ E (Figure 1)) were examined. Sex, body weight (BW), straight carapace length (SCL) and width (SCW) of each individual were recorded (Table S1). Macroalgal fragments obtained from the esophagus, stomach, and intestines of each turtle were washed with seawater and stored at −30°C until morphological and molecular analyses. Each macroalgal fragment was identified based on the gross morphology and internal structure of cross sections made by hand or cryomicrotome (Yamato Kohki Industrial Co., Saitama, Japan).
2.2 Statistical Analysis
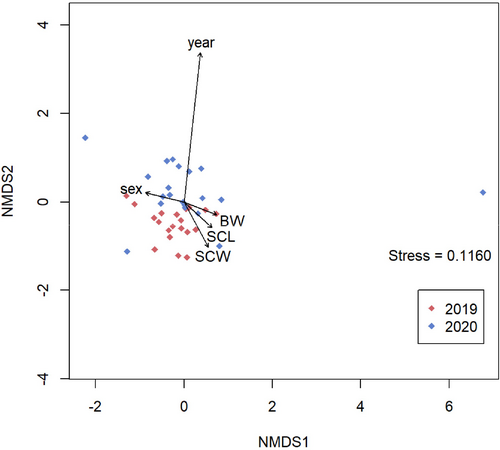
Two nonmetric multidimensional scaling (NMDS), based on the Jaccard distance (presence/absence data), was constructed to graphically assess the differences in macroalgal composition (others were excluded) among the year of harvest, sex, BW, SCL, and BW of green turtles. Macroalgal species (or genus) variables were plotted as vectors in the two NMDS. The influence of these factors on the macroalgal composition was evaluated by PERMANOVA (999 permutations) based on the Jaccard distance. All analyses were conducted using R software version 4.4.5 (R Core Team 2025).
2.3 Molecular Analysis
For molecular analysis, one to five macroalgal fragments of G. angusta and B. paradoxa were randomly selected from each turtle. In addition, living thalli collected from various Japanese coasts and dried specimens deposited in our laboratory or the National Museum of Nature and Science, Tokyo (Figure 1), were used for molecular analysis (Table 1). About a 1 cm long fragment was cut off from an apical part of a thallus, and epiphytes were carefully removed under a stereomicroscope (SZX12, Olympus, Tokyo, Japan). The DNA extraction method and the polymerase chain reaction (PCR) and sequencing procedures were based on Hayakawa et al. (2012). The fragment was cut into small pieces and placed into a 1.5 mL microtube with 20 μL (for frozen specimens) or 50 μL (for dried specimens) of 10 mM Tris (pH 8.0) solution containing 5% Chelex-100 resin (BioRad, Richmond, CA, USA) for 5 min or more. Then, the sample was finely ground using a Teflon grinder until few fragments were visible. After adding 230–260 μL of 5% Chelex-100 resin solution to bring the total volume to 280 μL, the extract was heated at 100°C for 10 min in a brock bath (MyBL-10, As One, Osaka, Japan) and immediately placed on ice. The extract was then centrifuged at 2000 g for 10 min, and 200 μL of the supernatant was transferred to a fresh microtube.
| Locality | Collecting date | Collector, herbarium no. or accession no. | State |
|---|---|---|---|
| Grateloupia angusta | |||
| Miyako, Miyako-shi, Iwate | 27 July 1988 | Unknown | Dried |
| Ohshima, Ooshima-cho, Tokyo | 30 July 2021 | Hashimoto, T. | Living |
| Miyakejima, Miyake-mura, Tokyo | 23 March 2019 | Unknown | Dried |
| Shikinejima, Niijima-mura, Tokyo | 28 June 2002 | Unknown | Dried |
| Aburatsubo, Misaki-cho, Kanagawa | 10 June 2021 | Kitano, M. | Living |
| Banda, Tateyama-shi, Chiba | 11 June 2021 | Kitano, M. | Living |
| Ebisujima, Shimoda-shi, Shizuoka | 24 July 2021 | Arai, T. | Living |
| Ohura, Shimoda-shi, Shizuoka | 2 October 2019 | Kamiya, M. | Living |
| Toba, Toba-shi, Mie | 14 July 2021 | Kurashima, A. | Dried |
| Banshozaki, Shirahama-cho, Wakayama | 22 April 2019 | Kamiya, M. | Living |
| Suzaki, Suzaki-shi, Kochi | 13 July 2021 | Tanaka, K. | Living |
| Usuki, Usuki-shi, Ooita | 22 August 2010 | TNS-AL190710 | Dried |
| Jejudo, South Korea | KJ48509, KJ48511, KJ48512, KC875853, KF475714, NC023094 | DNA | |
| Geomundo, South Korea | KJ648510 | DNA | |
| Besa paradoxa | |||
| Onagawa, Onagawa-cho, Miyagi | 5 August 2000 | TNS-AL179524 | Dried |
| Shizugawa, Minamisanriku-cho, Miyagi | 11 July 2021 | Kamiya, M. | Living |
| Shioyazaki, Iwaki-shi, Fukushima | 3 November 2017 | TNS-AL209140 | Dried |
| Oarai, Ooarai-cho, Ibaraki | 31 May 2021 | Hasui, S. | Living |
| Kimigahama, Choshi-shi, Chiba | 25 June 2020 | Kamiya, M. | Living |
| Togawa, Choshi-shi, Chiba | 25 June 2020 | Kamiya, M. | Living |
| Ohshima, Ooshima-cho, Tokyo | 30 July 2021 | Hashimoto, T. | Living |
| Kannonzaki, Yokosuka-shi, Kanagawa | 30 March 2021 | Kamiya, M. | Living |
| Jougashima, Miura-shi, Kanagawa | 14 July 2021 | Sadakane, K. | Living |
| Enoshima, Fujisawa-shi, Kanagawa | 17 April 2014 | TNS-AL186970 | Dried |
| 19 September 2021 | Oomori, T. | Living | |
| Touji, Shimoda-shi, Shizuoka | 14 May 2017 | TNS-AL208451 | Dried |
| Yumigahama, Minamiizu-cho, Shizuoka | 27 May 2021 | Fukuoka, M. | Living |
| Waji, Tahara-shi, Aichi | 16 May 2018 | TNS-AL209821 | Dried |
- Note: Accession no.; the registration number of sequence data in GenBank; Dried, dried specimen; Living, living specimen.
PCR amplification and sequencing of COI-5P and/or the cox2-cox3 spacer were carried out using primers listed in Table S2. PCR reaction was performed using a total volume of 10 μL containing 1 μL of template DNA, 0.5 μL of each 10 μM primer, 2 μL of 10 mM dNTP, 5 μL of 2xPCR buffer, and 0.2 units of KOD FX Neo DNA polymerase (Toyobo, Osaka, Japan). The thermal profile for PCR was as follows: initial denaturation at 94°C for 2 min, 30 cycles at 98°C for 30 s, 50°C for 30 s, and 68°C for 90 s, and final extension at 68°C for 7 min. PCR products were electrophoresed in 1.5% agarose gel to check PCR yield and specificity. If PCR yield was insufficient, the PCR product was used as template DNA for the second PCR, using the primer set used for the first PCR or a nested primer set. If more than one band was visible, the correct DNA band was excised and the DNA fragment was recovered from the gel using the QIAEX II Gel Extraction Kit (Qiagen, Gaithersburg, MD, USA). After purification of PCR products with ExoSAP-IT (GE Healthcare Japan, Tokyo, Japan), sequencing of amplicons was conducted by the DNA sequence service of Eurofins Genomics (Tokyo, Japan). The sequence data of COI-5P (603 bp long for G. angusta, accession number LC830510—LC830551, and 543 bp long for B. paradoxa, accession number LC830534–LC830533) and those of the cox2-cox3 spacer (377 or 378 bp long for G. angusta, accession number LC830552—LC830566, and 416 bp long for B. paradoxa, accession number LC830567–LC830570) were deposited in the DNA Data Bank of Japan. The COI-5P sequences were aligned with those of G. angusta or B. paradoxa deposited in GenBank using BioEdit software ver. 7.2.5 (Hall 1999). Haplotype network was constructed using TCS1.21 (Clement et al. 2000) and PopART software (Leigh and Bryant 2015).
3 Results
3.1 Species Composition of Green Turtle Diets
Three genera of green algae, eight genera of brown algae, and four genera of red algae were detected in the digestive organs of 55 green turtles (27 females and 28 males), and no macroalgae were found in any digestive organs of seven green turtles (Table 2). Among the green algae examined, Ulva species were found in five green turtles, all of which contained these species in more than one digestive organ, whereas Avrainvillea and Halimeda species were rarely detected. Among the brown algae examined, Lobophora species were most frequently detected (in 44 turtles), followed by Sargassum species (32 turtles) including S. horneri, S. micracanthum, and S. siliquastrum, Dictyopteris species (28 turtles), Homoeostrichus flabellatus (10 turtles), Distromium decumbens (eight turtles), Zonaria stipitata (six turtles), and Padina species (four turtles). Among the red algae examined, G. angusta was detected in the intestines of 14 turtles, whereas B. paradoxa was also detected in the intestines of four turtles and in both the esophagus and intestines of one turtle (20-29). Galaxaura falcate and Corallina species were found in one and three turtles, respectively.
| Year | 2019 | 2019 | 2020 | 2020 | Total Organs | Total Individuals | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex (no. of individuals) | Female (11) | Male (10) | Female (18) | Male (18) | ||||||||||
| Organ | Eso | Sto | Int | Eso | Sto | Int | Eso | Sto | Int | Eso | Sto | Int | ||
| Green algae | ||||||||||||||
| Ulva spp. | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 11 | 5 | ||
| Halimeda sp. | 1 | 1 | 1 | |||||||||||
| Avrainvillea sp. | 1 | 1 | 1 | 3 | 2 | |||||||||
| Brown algae | ||||||||||||||
| Dictyota sp. | 1 | 1 | 1 | 3 | 3 | |||||||||
| Dictyopteris spp. | 2 | 3 | 1 | 1 | 3 | 7 | 1 | 1 | 3 | 3 | 5 | 6 | 36 | 26 |
| Distromium decumbens | 3 | 1 | 1 | 3 | 8 | 8 | ||||||||
| Homoeostrichus flabellatus | 1 | 3 | 3 | 3 | 1 | 3 | 13 | 10 | ||||||
| Lobophora spp. | 7 | 5 | 6 | 4 | 4 | 8 | 7 | 9 | 8 | 8 | 8 | 12 | 86 | 44 |
| Padina spp. | 1 | 1 | 1 | 1 | 1 | 5 | 4 | |||||||
| Zonaria stipitata | 1 | 1 | 2 | 1 | 2 | 1 | 8 | 5 | ||||||
| Sargassum horneri | 1 | 2 | 3 | 3 | ||||||||||
| Sargassum micracanthum | 1 | 3 | 2 | 6 | 5 | |||||||||
| Sargassum siliquastrum | 1 | 1 | 1 | |||||||||||
| Sargassum spp. | 4 | 3 | 2 | 4 | 5 | 6 | 4 | 2 | 7 | 2 | 3 | 4 | 46 | 24 |
| Red algae | ||||||||||||||
| Dichotomaria falcata | 1 | 1 | 1 | |||||||||||
| Corallina sp. | 1 | 2 | 3 | 3 | ||||||||||
| Besa paradoxa | 1 | 1 | 1 | 1 | 2 | 6 | 5 | |||||||
| Grateloupia angusta | 3 | 4 | 2 | 5 | 14 | 14 | ||||||||
| Others | 7 | 7 | 7 | 7 | 6 | 10 | 10 | 7 | 9 | 5 | 4 | 12 | 91 | 48 |
| Total | 26 | 29 | 31 | 21 | 22 | 47 | 26 | 21 | 36 | 20 | 22 | 44 | ||
- Abbreviations: Eso, esophagus; Int, intestine; Sto, stomach.
The NMDS plots of macroalgal composition detected from the green turtles are shown in Figure 2. The macroalgal compositions of gut contents significantly differed by the year of harvest (p = 0.001) but not by sex, BW, SCL, and SCW (p > 0.05) (Table 3). Any interaction between the factors was not significant (data not shown).
| Df | SS | MS | F | p | |
|---|---|---|---|---|---|
| Year | 1 | 1.603 | 1.603 | 7.209 | 0.001 |
| Sex | 1 | 0.185 | 0.185 | 0.831 | 0.522 |
| BW | 1 | 0.234 | 0.234 | 1.054 | 0.415 |
| SCL | 1 | 0.125 | 0.125 | 0.564 | 0.697 |
| SCW | 1 | 0.175 | 0.175 | 0.787 | 0.544 |
| Residuals | 3 | 5.115 | 0.222 | ||
| Total | 4 | 11.36 |
- Note: Significant difference is indicated in bold (p < 0.05).
Although all macroalgal species except Avrainvillea and Halimeda species are distributed across temperate coasts, we considered that the algal thalli detected in the esophagi and/or stomachs were grazed around the Ogasawara Islands. Since most thallus fragments of G. angusta and B. paradoxa were detected only in the intestines, these algae were possibly grazed before the turtles migrated to Ogasawara and used for further molecular analysis.
3.2 Haplotype Analysis of G. angusta
Among the 21 fragments of G. angusta obtained from 14 turtles, the COI-5P region was successfully sequenced for 14 fragments from nine turtles (three females and six males) and six haplotypes were recognized. The same COI-5P site was also sequenced in 34 samples collected from Japanese coasts and two specimens deposited in the herbariums (Table 1), and six haplotypes were distinguished among these specimens. Figure 3 shows a haplotype network of G. angusta, including two haplotypes from two South Korean sites deposited in GenBank. Among the six haplotypes detected in the digestive organs, haplotypes G2, G5, and G7 were found at nine Japanese and two South Korean sites. Multiple fragments of G. angusta were examined in the turtles 20-02 (four fragments), 20-016 (two fragments), and 20-38 (two fragments), but more than one haplotype was not found from the same turtle.
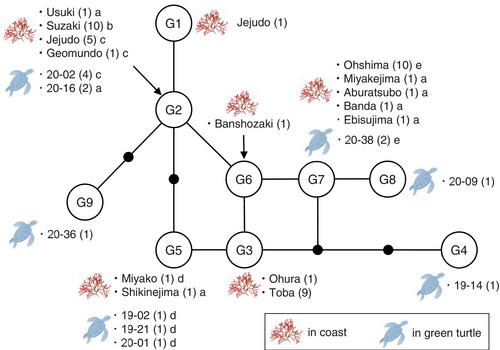
Many haplotypes were detected in more than one coastal site, causing difficulty in specifying the foraging grounds of each green turtle. For example, the haplotype G7 of G. angusta was detected in one green turtle and five Japanese sites, so this green turtle possibly grazed G. angusta at one of the five sites. Subsequently, we analyzed the cox2-cox3 spacer region of the algal fragments from six turtles as well as the specimens from nine Japanese sites; the cox2-cox3 spacer was 144 bp long, flanked by 63 bp long cox2 and 170 bp long cox3. Two sequences from South Korea (KC875853 and NC023094) deposited in GenBank were added to the haplotype analysis. A total of five haplotypes, a, b, c, d, and e, were distinguished (Figure 3); two substitutions were detected in both the spacer and cox2 regions, whereas five substitutions were found in the cox3 region. Three haplotypes (a, b, and c) were detected within the COI haplotype G2; haplotype G2c of the turtle 20-02 was identical to the specimens from Jejudo and Geomundo in South Korea, and haplotype G2a of the turtle 20-16 was identical to the specimen from Usuki. Two haplotypes (a and d) were detected within the COI haplotype G5; haplotype G5d of three turtles, 19-02, 19-21, and 20-01, was identical to the specimen from Miyako. Furthermore, two haplotypes (a and e) were detected within the COI haplotype G7; haplotype G7e of the turtle 20-38 was identical to the specimen from Ohshima.
3.3 Haplotype Analysis of B. paradoxa
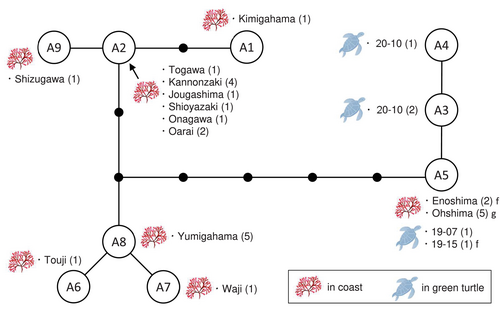
Among six fragments of B. angusta obtained from five turtles, we successfully sequenced the COI-5P region from five fragments in three turtles (one female and two males). COI-5P of 21 specimens collected from eight Japanese coasts and five specimens deposited in herbariums were also analyzed. Nine haplotypes were detected in green turtles and/or Japanese coasts (Figure 4); one to 11 bp differed between the haplotypes. Among the three haplotypes (A3, A4, and A5) detected in the green turtles, only one haplotype (A5) was identical to that detected in specimens from Ohshima and Enoshima, and the other two haplotypes did not match any haplotypes from the Japanese coasts examined in this study. The cox2-cox3 spacer region was successfully analyzed in one fragment from one turtle and the specimens from two Japanese sites; the cox2-cox3 spacer was 169 bp long, flanked by 63 bp long cox2 and 184 bp long cox3. Two haplotypes, differing in one bp, were detected within the COI haplotype A5, and haplotype A5f of the turtle 19-15 was identical to that of the specimen from Enoshima.
4 Discussion
Green turtles migrate to the Ogasawara Islands for the purpose of breeding from March to May (ELNA unpublished data) and the majority of them do not remain there for a long period; thus, a certain amount of macroalgal tissues grazed prior to migration may be retained in the digestive organs. In the present study, fan-shaped species belonging to the tribe Zonarieae, including Lobophora, Homoeostrichus, Zonaria, and Distromium, were frequently found in the digestive organs. The ingestion of these macroalgae by green turtles is likely to have occurred subsequent to their arrival, due to the distribution of these algae around Ogasawara (Kasaki and Oonishi 1973; Miyata 1991). Furthermore, these species were frequently detected in the esophagus and stomach, indicating their ingestion just prior to harvest by fishermen. In contrast, the temperate red algae G. angusta and B. paradoxa were predominantly detected in the intestines in both 2019 and 2020, suggesting ingestion prior to the migration to Ogasawara. It is noteworthy that both species are characterized by a relatively hard texture, which could result in their incomplete digestion in the intestines (Kameda and Ishihara 2009).
Genetic comparison of the gut contents of turtles and macroalgae collected from various coastal locations was useful in delineating the foraging grounds of green turtles. The analyses of the COI-5P and cox2-cox3 spacer regions revealed that the fragments of G. angusta obtained from six turtles exhibited the same haplotype as specimens collected from Usuki, Ohshima, Miyako, and South Korea (Geomundo and Jejudo). In the case of B. paradoxa, the fragment from one turtle was genetically identical to those from the two specimens from Enoshima. Living or dead green turtles have been reported from these locations (Hatase et al. 2006; Nishizawa et al. 2013; Fukuoka et al. 2015; Kim et al. 2021), which are regarded as potential foraging habitats.
Ohshima and Enoshima, located less than 1000 km from the Ogasawara Islands, have been identified as areas frequented by green turtles during their migratory journey towards Ogasawara; tagging experiments have suggested that green turtles visit the Pacific coast in the middle part of Honshu after laying eggs on Ogasawara (Hatase et al. 2006). In contrast, Usuki, Miyako, and South Korea are located at distances of 1120, 1300, and 1600 km from Ogasawara, respectively. The extent to which macroalgal fragments remain undigested in the intestine after more than 1000 km of migration remains unclear. The mean cruising swim speed of subadult green turtles is 0.27–0.47 m s−1 (Kinoshita et al. 2021); if the digestion time of green turtle diets is 31 days, the macroalgal fragments could remain in the turtle intestines after 723–1259 km of migration. The optimal swim speed of sea turtles is predicted to be proportional to their body mass (mb kg); the range is from mb0.05 to mb0.13 (Kinoshita et al. 2021). When the mean body mass of green turtles examined in this study, 121 kg, is applied in the above formula, the cruising swim speed can be calculated as 1.27–1.87 m s−1, and turtles can swim 3402–5009 km during 31 days. This suggests the possibility that some green turtles may undertake migratory movements to Ogasawara following grazing on G. angusta in Usuki, Miyako, or South Korea.
Amorocho and Reina (2008) examined the intake passage time of three different diets using green turtles from tropical coral reefs in Colombia; the turtles took a longer time to digest fresh leaves of land plants (31 days) than fish (20 days) or a combination of plants and fish (16–25 days), suggesting that a long intake passage time might enhance the digestibility of fiber. Green turtles, both subadults and adults, have a higher ratio of intestinal length to carapace length (12.6- to 13.9-fold) compared with the carnivorous Caretta (8.6-fold) or Dermochelys coriacea (9.5-fold), and the long intestines of green turtles may be important for digesting fibrous macroalgae and seagrasses (Bjorndal 1985). Temperature is a significant variable that limits digestive efficiency in poikilothermic animals, and the intake rate and digestive efficiency of turtles are affected by temperature (Amorocho and Reina 2008). The water temperature in Ogasawara during April and May ranges from 21°C to 25°C (Sea Temperature Info: https://seatemperature.info/june/ogasawara-water-temperature.html), which is colder than that observed in Colombian coral reefs where the intake passage time was examined (27.7°C–29.0°C) (Amorocho and Reina 2008). This suggests that green turtles may require a significantly longer time to fully digest macroalgae on the migratory route to Ogasawara.
In both G. angusta and B. paradoxa, a single haplotype has been identified in a single turtle, despite the examination of more than one macroalgal fragment in some green turtles. Different haplotypes were found from the same area (but different sites); for instance, haplotypes G3 and G7 of G. angusta were collected from Ohura and Ebisujima, respectively, only 3 km distant, and haplotypes A6 and A8 of B. paradoxa were found from Touji and Yumigahama, only 3.6 km distant. Previous studies on Sargassum horneri and Ulva australis demonstrated that several haplotypes were sympatrically distributed in the majority of the Japanese sites examined, and that the frequency of these haplotypes differed depending on the site (Uwai et al. 2009; Hanyuda et al. 2016). In order to ascertain the precise frequencies of these haplotypes, it is essential to analyze a greater number of macroalgal samples recovered from each turtle and collected from each location. The resulting data will facilitate the identification of foraging grounds.
Green turtles typically graze on macroalgae growing in the subtidal zone, at an average depth of 4.8 m (Fukuoka et al. 2019). However, B. paradoxa typically grows in a lower intertidal or upper subtidal zone. Of particular interest is the detection of COI haplotype A5 in both the intertidal zone (Enoshima) and the subtidal zone (3–8 m deep in Ohshima). However, the paucity of field surveys conducted in the subtidal zone along the Japanese coastline may be a reason why COI haplotypes A3 and A4 were not detected in the gut contents of green turtles. A similar phenomenon was observed in G. angusta, where several haplotypes (COI haplotypes G4, G8, and G9) detected in the gut contents did not correspond to any of the haplotypes in the specimens examined in this study. This alga is also distributed from the lower intertidal to subtidal zones and was reported from the gut contents of horned turbans (Turbo sazae) living at a depth of 17 m in Shikinejima, Japan (Iijima and Takase 2019).
Amplification of the required quantities of PCR products for subsequent sequencing was successfully achieved in the majority of field-collected samples. However, this was not the case for all of the samples from the pressed specimens and algal fragments in the gut contents. In instances where the initial PCR yielded an inadequate amount of product, subsequent nested PCRs, utilizing the initial PCR product as a template, occasionally resulted in a sufficient quantity for sequencing. It is notable that DNA from old specimens frequently fragments during long-term preservation (Nakahama 2021), and the DNA of the gut contents is degraded by digestion (Krehenwinkel et al. 2018), both of which reduce the efficiency of PCR amplification. To resolve these issues, it is recommended that DNA degradation should be minimized by quick freezing, prevention of repeated freeze–thaw cycles, and/or repair of DNA damage (Sproul and Maddison 2017).
Although Padina sp. and Sargassum horneri were detected in only a few females, no remarkable dietary preference was observed between the sexes. Differences in body size, growth rate, fat, and plasma uric acid levels between the sexes (Caldwell 1962; Frazier 1971; Bolten and Bjorndal 1992; Limpus 1993; Kwan 1994; Godley et al. 2002) may be due to factors other than dietary factors. According to Stokes et al. (2019), who assessed green turtle diets in the Western Indian Ocean, seagrass constituted 95% of the mean diet biomass for males and nonbreeding females and only 58% for gravid females. Although eight gravid females were included in the present study, the species composition of the gut contents was similar between the nonbreeding and gravid females (data not shown).
The species composition exhibited temporal variation; in 2019, Distromium decumbens and Homoeostrichus flabellatus were detected in eight and ten turtles, respectively, whereas neither species was detected in 2020. Conversely, Dictyota sp. was not observed in 2019 but was found in three turtles in 2020. The foraging grounds from which green turtles have come to the Ogasawara Islands may vary from 1 year to the next. Alternatively, the germination time and biomass of each macroalgal species may vary depending on the year (e.g., Lin et al. 2018; García-Gómez et al. 2021; Terada et al. 2021). For instance, the coverage of Dictyota sp. in the Mediterranean Sea exhibited substantial fluctuations over a 15-year period, potentially attributable to annual fluctuations in seawater temperature, nutrient concentrations, or storm activity (Medrano et al. 2020).
The dietary composition also varied among the green turtles, which may have been caused by differences in their arrival dates at the Ogasawara Islands. When green turtles remained on Ogasawara for several weeks, the majority of their gut contents were occupied by macroalgae growing around Ogasawara. Conversely, if green turtles have recently arrived at Ogasawara, macroalgae that they have grazed on their foraging grounds may remain undigested in the intestines. The foraging grounds for most of the green turtles migrating to Ogasawara are considered to be located in the vicinity of the Japanese coast (Nishizawa et al. 2013), implying that the macroalgal composition in the gut contents of these turtles is likely to vary depending on their foraging locations. Along the Japanese coast, the Oyashio and Liman cold currents flow from the north and the Kuroshio and Tsushima warm currents flow from the south, resulting in different macroalgal flora between regions (Tanaka 1997; Terada et al. 2021). Kameda and Ishihara (2009) reported the variations of macroalgal composition and relative volume of the gut contents among dead green turtles examined in western Japan. In the Yaeyama Islands, the gut contents were dominated by red algae, such as Gelidiella acerosa and Eucheuma spp., as well as seagrass. In the Sea of Japan coasts, the green alga Codium spp. and seagrass Phyllospadix japonicus were frequently detected. In Shikoku and the Kii Peninsula, green algae Codium spp., brown algae Sargassum spp., and various red algae, including Gelidium elegans, Ptilophora subcostata, Prionitis spp., and Grateloupia spp., were identified. However, given the distribution of these algal species across multiple areas, and the limitations of gut content analysis in accurately delineating foraging grounds, further research is necessary to fully understand the ecological dynamics of these ecosystems.
5 Conclusions
In the present study, various subtropical and temperate macroalgal species were recovered from the gut contents of green turtles, and the temperate red algae G. angusta and B. paradoxa were identified as having been grazed by the turtles shortly before their arrival at the Ogasawara Islands. The present study suggests that some green turtles migrating to the Ogasawara Islands graze on macroalgae around Miyako, Enoshima, Ohshima, Usuki, or South Korean Islands and that green turtles may have come to the Ogasawara Islands from different foraging grounds depending on the year. These results are useful for estimating the prenesting behavior and feeding strategies of green turtles, which are needed to conserve green turtles. If the timing of breeding migration is influenced by environmental conditions, such as water temperature and food availability (Enstipp et al. 2016), it is crucial to ascertain the origin of breeding turtles. To this end, further comprehensive data on factors such as haplotype composition and the frequency of macroalgal samples recovered from each green turtle and collected from each location are required to narrow the foraging grounds. Next-generation sequencing of pooled samples is an effective approach for outlining genetic variation and differentiation in populations (Hirao et al. 2017; Santos and Gaiotto 2020). Hence, we are currently collecting hundreds of samples from each habitat and applying this method to characterize the genetic structure of each habitat.
Author Contributions
S.R.: sampling, investigation, and writing of the draft. S.K. and C.K.: designing the field study, sampling, reviewing, and editing of the manuscript. H.S.: administration, supervision, reviewing, and editing of the manuscript. K.S.: statistical analyses. M.K.: conceptualization, methodology, supervision, and writing of the manuscript.
Acknowledgments
The authors sincerely appreciate the members of Ogasawara Marine Center, Ogasawara Fisheries Cooperative Association, and Sea Turtle Research Collegium, Tokyo University of Marine Science and Technology, for their invaluable assistance in the recovery of the gut contents from harvested green turtles. We would also like to acknowledge Drs. Akira Kurashima, Masanori Hiraoka, Kouki Tanaka, and Tomoaki Hashimoto for providing samples and Dr. Taiju Kitayama for the loan of specimens from the National Museum of Nature and Science, Tokyo. Additionally, we are indebted to Dr. Hiroto Murase for the improvement of the initial manuscript. Finally, we would like to thank Editage (www.editage.jp) for English language editing.
Ethics Statement
All green turtles used in this study were harvested by indigenous fishermen with the permission of the governor of Tokyo, and no turtles were killed specifically to provide samples for this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The datasets of this study may be available from the corresponding author on request.




