Mutational Analysis of Bile Cell-Free DNA in Primary Sclerosing Cholangitis: A Pilot Study
Handling Editor: Alessio Aghemo
Funding: This work was supported by grants PI22/00471 funded by Instituto de Salud Carlos III (ISCIII) and co-funded by the European Union, PI19/00613 funded by Instituto de Salud Carlos III (ISCIII) cofinanced by ‘Fondo Europeo de Desarrollo Regional’ (FEDER) ‘Una Manera de Hacer Europa’ and POSTD18014AREC and INVES223049AREC fellowship from AECC to MA; CIBERehd; European Union Horizon 2020 Transcan project [2022–784-024] to MA, MAÁ, MK, PS, KMS and CB; Thermo Fisher 2022 Oncomine Clinical Research Grant to MAÁ and CB; AECT Eurorregión Nueva Aquitania Euskadi Navarra “Innovación Eurorregional” [2020/101] and [2023/2] to MAÁ and CB; Rolf M Schwiete Foundation grant 2024–040 to MAÁ, CB and MK; Departamento de Salud Gobierno de Navarra [42/2021] to JU, Ramón y Cajal Program contract RYC-2018-024475-1 to MGFB. This study is based upon work from COST Action Precision-BTC-Network CA22125, supported by COST. The generous support of Mr. Eduardo Avila is acknowledged.
Matías A Ávila and Carmen Berasain share senior authorship.
ABSTRACT
Background
Primary sclerosing cholangitis (PSC) is a chronic liver disease characterised by inflammation and fibrosis of the bile ducts, conferring an increased risk of cholangiocarcinoma (CCA). However, detecting CCA early in PSC patients remains challenging due to the limited sensitivity of conventional diagnostic methods, including imaging or bile duct brush cytology during endoscopic retrograde cholangiopancreatography (ERCP). This study aims to evaluate the potential of bile cell-free DNA (cfDNA) mutational analysis, termed the Bilemut assay, as a tool for CCA detection in PSC patients.
Methods
Sixty-three PSC patients undergoing ERCP due to biliary strictures were prospectively recruited. Bile samples were collected, and cfDNA was extracted and analysed using the Oncomine Pan-Cancer Cell-Free assay. Twenty healthy liver donors were included for comparison. Samples with a mutant allele frequency (MAF) ≥ 0.1% were considered positive. Correlations between mutational status and clinical characteristics were assessed.
Results
cfDNA mutational analysis was successful in all bile samples. Mutations predominantly in KRAS, GNAS, and TP53 were detected in 36.5% (23/63) of PSC patients, compared to 10% (2/20) of healthy donors (p = 0.0269). The clinical characteristics of Bilemut-positive and -negative patients were comparable, though there was a trend towards a lower prevalence of inflammatory bowel disease in the Bilemut-positive group. Among PSC patients diagnosed with CCA during follow-up, 75% were Bilemut-positive, suggesting an association between mutational status and malignancy risk.
Conclusions
Mutational analysis of cfDNA obtained from bile collected from PSC patients undergoing ERCP is feasible. Implementing the Bilemut assay may help identify patients needing closer surveillance and further imaging studies.
Summary
- Patients with primary sclerosing cholangitis (PSC) suffer from frequent biliary strictures and have an increased risk of developing cholangiocarcinoma (CCA).
- However, early cancer detection remains challenging.
- Using the bile collected during diagnostic endoscopies, we have detected the presence of cell-free DNA mutations in a larger percentage of patients with CCA, suggesting the utility of bile DNA analysis for the management of PSC patients.
Abbreviations
-
- BD-IPMN
-
- branch duct intraductal papillary mucinous neoplasm
-
- CA-19-9
-
- carbohydrate antigen 19-9
-
- CCA
-
- cholangiocarcinoma
-
- cfDNA
-
- cell-free DNA
-
- eCCA
-
- extrahepatic cholangiocarcinoma
-
- ERCP
-
- endoscopic retrograde cholangiopancreatography
-
- IBD
-
- inflammatory bowel disease
-
- iCCA
-
- intrahepatic cholangiocarcinoma
-
- MAF
-
- mutant allele frequency
-
- MRCP
-
- magnetic resonance cholangiopancreatography
-
- MRI
-
- magnetic resonance imaging
-
- NGS
-
- next-generation sequencing
-
- pCCA
-
- perihilar cholangiocarcinoma
-
- PSC
-
- primary sclerosing cholangitis
-
- UDCA
-
- ursodeoxycholic acid
1 Introduction
Primary sclerosing cholangitis (PSC) is a chronic and progressive liver disease characterised by inflammation and fibrosis of the intrahepatic and extrahepatic bile ducts [1]. This condition leads to the formation of strictures, which impair bile flow and can eventually result in cirrhosis, liver failure, and confer a significantly increased risk of hepatobiliary malignancies. PSC is a rare disease, with an estimated prevalence of 0.5 to 1.3 per 100,000 individuals in North America and Northern Europe. It is particularly prevalent in countries such as Norway, Sweden, Poland, and the Netherlands [2].
PSC is closely associated with inflammatory bowel disease (IBD), particularly ulcerative colitis, which is present in up to 70% of PSC patients [3, 4]. PSC patients have a significantly increased risk of developing cholangiocarcinoma (CCA), with an incidence 160–1600 times that of the general population and a lifetime risk of 10%–20%, as well as an elevated risk of gallbladder cancer [5, 6]. It is known that more than 60% of PSC patients with CCA die within a year of diagnosis [7], and the 5-year survival is below 40% [5]. Despite being a clearly defined high-risk population, the diagnosis of CCA in patients with PSC remains challenging due to the lack of specific symptoms and the difficulty in distinguishing between benign and malignant strictures [6]. Many imaging modalities have been implemented for CCA surveillance and diagnosis in PSC patients, including ultrasound, magnetic resonance imaging (MRI) and magnetic resonance cholangiopancreatography (MRCP), and are recommended to be performed every 6–12 months, with or without testing for serum CA 19-9 levels, by clinical societies [6]. Unfortunately, when tumours are visible by imaging techniques, the disease is often already in an advanced stage, and thus with limited therapeutic options [5, 6].
In PSC patients presenting with severe/progressive bile duct changes or relevant strictures on MRI/MRCP, and thus with suspicion of CCA, endoscopic retrograde cholangiopancreatography (ERCP) is often performed [6, 8]. ERCP enables the collection of pathology specimens, such as bile duct brush cytology and forceps biopsy for laboratory diagnosis of malignancy [5, 6, 9]. However, the sensitivity of brush cytology and endobiliary biopsy performed during ERCP for the detection of CCA is often suboptimal, probably due to sampling errors [10, 11]. Therefore, additional tools are needed to improve CCA diagnosis and screening in PSC patients [5, 7, 8, 12]. Interestingly, the ERCP procedure also enables the collection of biliary fluid, in which molecules originating from cancer cells along the bile duct system may accumulate and can be assessed [13-19]. In patients with biliopancreatic tumours, cell-free DNA (cfDNA) present in bile was recently shown to contain DNA molecules from malignant cells, and the mutational analysis of this cfDNA demonstrated higher sensitivity for early cancer detection compared to the analysis of pathology specimens collected during ERCP [20]. In this prospective pilot study, we evaluated the feasibility of performing next-generation sequencing (NGS) using a commercially available cancer gene panel in cfDNA obtained from PSC patients who underwent ERCP. We also provide preliminary evidence supporting the potential of this approach as an additional surveillance tool for CCA malignancy in this high-risk population.
2 Methods
2.1 Patients
A cohort of 63 PSC patients who underwent ERCP because of a relevant bile duct stricture (any biliary stricture of the common bile duct or hepatic ducts associated with signs or symptoms of obstructive cholestasis and/or bacterial cholangitis according to the 2023 AASLD guidelines) was prospectively accrued for the study from 01.09.2021 to 31.05.2022 at the Department of Hepatology, Transplantology and Internal Medicine, Medical University of Warsaw, Warsaw, Poland. Their demographic and clinical characteristics are summarised in Table 1. All patients were older than 18 years and provided written informed consent for the examination of their samples and the use of their clinical data.
| Gender, n (%) | ||
| Female | 23 | (36.5%) |
| Male | 40 | (63.5%) |
| Median age years, n (range) | 38.4 | (18–67) |
| Location of biliary strictures, n (%) | ||
| Distal | 15 | (24%) |
| Perihilar | 6 | (9.5%) |
| Intrahepatic | 8 | (12.7%) |
| Distal/perihilar | 15 | (24%) |
| Distal/intrahepatic | 7 | (11%) |
| Perihilar/intrahepatic | 7 | (11%) |
| Distal/perihilar/intrahepatic | 5 | (8%) |
| Presence of cirrhosis, n (%) | ||
| Yes | 23 | (36.5%) |
| No | 40 | (63.5%) |
| Liver transplantation, n (%) | ||
| Yes | 14 | (22,2%) |
| No | 49 | (77.8%) |
| CA19-9, U/mL n (%) | ||
| < 44 | 47 | (74.6%) |
| ≥ 44 | 16 | (25.4%) |
| CCA, n (%) | ||
| Intrahepatic CCA (iCCA) | 1 | (1.5%) |
| Perihilar CCA (pCCA) | 3 | (4.7%) |
| Inflammatory bowel disease, n (%) | ||
| No IBD | 23 | (36.5%) |
| Ulcerative colitis | 33 | (52.5%) |
| Crohn's disease | 4 | (6.3%) |
| nd | 3 | (4.7%) |
2.2 Bile Collection
Patients were fasted overnight, and ERCPs were conducted by experienced endoscopists. Each patient received rectal diclofenac and antibiotic prophylaxis before ERCP. During the standard ERCP procedure, after cannulation of the bile duct over the wire, and in most cases before contrast injection (Iomeron, iomeprol), a bile sample of 1 to 5 mL from each patient was aspirated through the sphincterotome. Bile samples were taken from the common bile duct or common hepatic duct. The study was approved by the Institutional Ethics Committees (CEIm 2016/91 and #KB/112/2021).
A second group of patients (n = 20) included healthy living liver donors from whom gallbladder bile was collected at the time of surgery; six of them were already shown in [20]. These samples were collected at the Department of General, Transplant and Liver Surgery, Medical University of Warsaw, Warsaw, Poland, with the approval of the Institutional Ethics Committee (#KB/49/2015).
Informed consent was obtained from each patient, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki.
After collection, bile samples were maintained at 4°C, centrifuged for 10 min (4°C) at 3500 g, and the supernatant was stored in aliquots at −80°C. The time to freezing was less than 2 h.
2.3 cfDNA Extraction
Prior to cfDNA isolation, bile was slowly thawed at 4°C and centrifuged for 10 min (4°C) at 13000 g to ensure the removal of impurities in the supernatant. Bile cfDNA was extracted with the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. cfDNA was quantitated with the QuantiFluor dsDNA Sample Kit (Promega, Madison, WI, USA) and cfDNA size distributions were analysed by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
2.4 Next-Generation Sequencing DNA Analyses
Coded bile cfDNA samples were blindly tested with the Oncomine Pan-Cancer Cell-Free assay following the manufacturer's instructions (PanCancer, Thermo Fisher Scientific, Waltham, MA, USA). This panel includes 52 genes enabling hotspot single nucleotide variation (SNV) and short indel detection as well as copy number variation (CNV) detection in key genes frequently mutated in multiple cancer types, including pancreatobiliary cancers. Four PanCancer libraries were manually prepared prior to being sequenced within an Ion 540 Chip in the Ion S5 system. Using an input of 50 ng of cfDNA, tagging individual DNA fragments with short random oligonucleotides called unique molecular identifiers and defining 2 as the minimum number of variants supporting functional families, this assay allowed variant detection as low as 0.02%, given that on average ~8000–10 000 functional families were identified by the sequenced base. An MAF value ≥ 0.1% was considered suggestive of the presence of a mutation.
2.5 Statistical Analyses
Statistical analysis was performed using GraphPad Prism (version 8.0.1) software. Data were represented as mean ± SEM or median + range. Non-normally distributed data were analysed using the Mann–Whitney U test and normally distributed data using the t-test. Analysis of frequency distributions was performed using standard 2 × 2 contingency tables and Fisher or Chi-square tests, as appropriate. Statistical significance was labelled according to the following p values: *p < 0.05, **p < 0.01, ***p < 0.001.
3 Results
3.1 Patient Samples and Clinical Characteristics
Bile samples were obtained from 63 patients with PSC undergoing ERCP due to relevant bile duct strictures. The primary reasons for performing ERCP in these patients were therapeutic interventions (such as dilation of strictures or biliary stenting) or to collect histopathological/cytological material from relevant strictures where malignancy was suspected. The clinical characteristics of the patients are summarised in Table 1. The cohort consisted of 63% males with a median age at PSC diagnosis of 37 years (range: 18–67). All PSC patients received ursodeoxycholic acid (UDCA) as part of their treatment regimen. The median period from PSC diagnosis to bile collection was 58.45 months (range: 3.6–243 months). The median follow-up time for the cohort was 89.95 months (range: 31–272 months). The median follow-up time from bile collection was 24.38 months (range: 4.5–29 months). At the time of bile collection, 16 out of 63 patients (25.4%) had elevated CA-19-9 levels above 44 U/mL.
According to the literature [21], although heterogeneous, the location of the stenosis was mainly extrahepatic (87%), including perihilar and distal strictures (Table 1). Also, it is known that most PSC patients progress to cirrhosis [21]. In our cohort, 23 patients (36.5%) had been diagnosed with cirrhosis at the moment of bile collection.
In line with the association of PSC with IBD, 40 out of 63 patients (63.5%) had concurrent IBD: 33 (52.4%) had ulcerative colitis, four (6.3%) had Crohn's disease, and three (4.8%) had indeterminate colitis.
A total of 14 patients (22.2%) underwent liver transplantation during follow-up, with a median time of 96.12 months after PSC diagnosis (range: 25–214 months) and 13 months after bile sample collection (range: 1.3–24.4 months). All explanted livers were thoroughly checked for the presence of CCA.
Four patients were diagnosed with branch duct intraductal papillary mucinous neoplasm (BD-IPMN) during PSC follow-up. One of these patients was already diagnosed with perihilar CCA (pCCA).
Regarding the incidence of malignancies during follow-up, one patient was diagnosed with intestinal adenocarcinoma 127 months after PSC diagnosis, and 4 patients (6.3%) were diagnosed with CCA after a median follow-up of 163.8 months (range: 3.6–214.5 months): 1 patient with intrahepatic CCA (iCCA) and 3 patients with pCCA. In one of these patients, the pCCA tumour was detected during the examination of the explanted liver.
3.2 Mutational Analysis of Bile cfDNA From PSC Patients
We previously demonstrated the robust performance of the mutational analysis in bile cfDNA using the Oncomine Pan-Cancer Cell-Free assay (ThermoFisher) for the early diagnosis of malignancy in patients with biliary strictures [20]. From this point onwards, we will refer to the Pan-Cancer Cell-Free assay applied to bile cfDNA samples as the Bilemut assay, as in our previous study [20]. Here we isolated cfDNA from 1 mL of bile obtained from 63 PSC patients with biliary stenosis, with a median yield of 5.7 μg/mL (range 0.1–70.8 μg/mL) of DNA. The presence of mutations in these bile cfDNA samples was analysed using the Bilemut assay. Sufficient cfDNA was obtained from all patients to perform the analysis. As shown in Figure 1A, out of the 63 PSC patients, at least one mutation with a mutant allele frequency (MAF) ≥ 0.1% was detected in 23 individuals, representing 36.5% of the cohort. The amount of cfDNA between Bilemut-positive and Bilemut-negative patients was similar (Figure S1A). The time elapsed between PSC diagnosis and bile collection and between bile collection and the last follow-up was comparable between Bilemut-positive and Bilemut-negative patients (Figure 1B). We were able to detect between one and four mutations per patient; specifically, one mutation was detected in 12 patients, two mutations in 6 patients, three mutations in 3 patients, and four mutations in 2 patients (Figure S1B, Table S1). Among the patients with only one mutation, four of them had a MAF between 0.1% and 0.15% (Figure 1A).
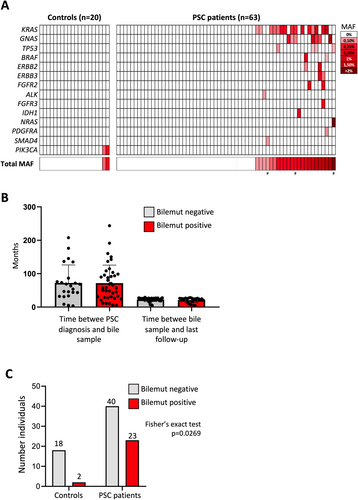
The most frequently mutated genes were KRAS (65% of cases), GNAS (30%), and TP53 (17%), followed by BRAF, ERBB2, ERBB3, and FGFR3, each in 9% of cases (Figure S1C). Detailed information on the specific mutations identified in bile cfDNA is provided in Table S1. This mutational distribution was similar to the landscape we found in patients with malignant biliary strictures in our previous report [20].
To assess whether a similar mutation profile could be found in bile from healthy controls, we performed the Bilemut analysis on 20 bile cfDNA samples collected from healthy living liver donors. In this cohort, a mutation in the PIK3CA gene was detected in two samples, resulting in a 10% positivity rate. The positivity rate in PSC patients (23/63; 36.5%) was significantly higher than that observed in healthy donor samples (Fisher's exact test, p = 0,0269; Figure 1C). Notably, the PIK3CA mutation detected in the control samples was not found in any of the PSC patients. Interestingly, in our previous study [20], among the 58 bile samples with detectable mutations, PIK3CA mutations were found in the bile of four patients and were always accompanied by other mutations, with patients carrying between three and seven mutated genes.
3.3 Clinical Characterisation of Bilemut-Positive and Bilemut-Negative PSC Patients
First, we evaluated where the Bilemut results depend on the location of the structures. As shown in Table S1, no clear association was detected as the location of the strictures was randomly distributed in both Bilemut-positive and negative PSC patients.
We then analysed various biochemical and clinical characteristics of PSC patients to determine whether Bilemut-positive patients differed from Bilemut-negative patients. As shown in Figure 2A, no differences were found in the levels of tumour and liver injury markers.

Our data do not show a possible contribution of a cirrhotic background to the mutational profile, as 8 out of the 23 (35%) PSC cirrhotic patients were Bilemut-positive and 15 out of the 40 (37.5%) PSC patients without cirrhosis at the moment of bile collection (Figure 2B, Table S1).
Although not statistically significant, the percentage of patients with IBD was higher in the Bilemut-negative group (Fisher's exact test, p = 0.0618; Figure 2C). Of the 23 Bilemut-positive patients, 11 (48%) had IBD, compared to 29 out of 40 Bilemut-negative patients (72.5%).
Additionally, 14 patients underwent liver transplantation: 5 out of 23 (22%) Bilemut-positive patients and 9 out of 40 (22%) Bilemut-negative patients (Figure 2D). Of these, 2 out of 5 (40%) Bilemut-positive liver transplants resulted in biliary complications, compared to 2 out of 9 (22%) Bilemut-negative transplants.
Among the four patients diagnosed with BD-IPMN, three of them, including the patient with CCA + BD-IPMN, were Bilemut positive (75%).
Among the four patients diagnosed with CCA, three tested positive for Bilemut, resulting in a 75% sensitivity for CCA diagnosis (Figure 3A). These three positive patients had pCCA, whereas the negative patients exhibited iCCA, suggesting that tumour location may influence the sensitivity of the analysis. The Bilemut-positive patients had between 1 and 2 mutations, with global MAFs of 0.1%, 0.35%, and 22% (Figure 1A, Table S1). Therefore, the Bilemut-positive patients with CCA did not exhibit the highest number of mutations nor the highest mutational load.
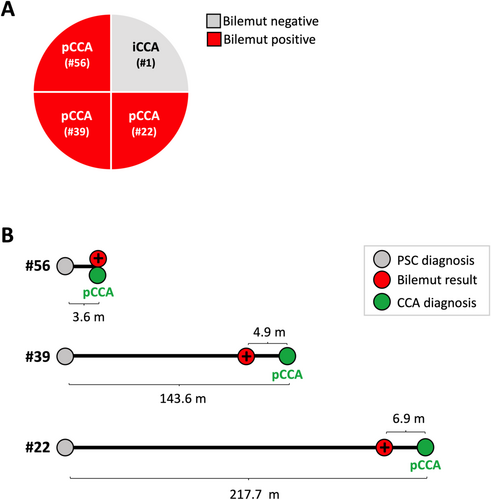
When we compared the time elapsed between PSC diagnosis and bile collection, we found it to be highly heterogeneous in both Bilemut-positive patients with CCA (median 136.7; range 3.06–207.7 months) as well as Bilemut-positive patients without CCA (median 58.6; range 5.06–176.1 months). Importantly, the Bilemut-positive patients with neoplasia were not necessarily those with the longest time from PSC diagnosis, further highlighting the complexity of the disease progression and its relationship with the detection of mutations.
Finally, we analysed the timeline of the final clinical diagnosis of the three Bilemut-positive PSC patients with CCA (Figure 3B). Patient #56 was diagnosed with pCCA based on a biopsy sample taken concurrently with bile collection during ERCP, only 3.6 months after the diagnosis of PSC. In this case, the Bilemut results confirm the presence of malignancy. However, for the other two patients, bile was collected before the CCA diagnosis was established: (1) In patient #39, a Bilemut-positive result was obtained in bile collected 4.9 months before the pCCA diagnosis, which occurred 143.6 months after the PSC diagnosis; (2) In patient #22, a Bilemut-positive result was obtained in bile collected 6.9 months prior to the diagnosis of pCCA in the explanted liver after the liver transplant, which took place 217.7 months after the PSC diagnosis.
4 Discussion
Despite recent advances in diagnostic tools and growing knowledge about the risks associated with PSC, the detection of malignancies in these patients often occurs late, limiting treatment options and negatively impacting prognosis [5, 6]. This situation highlights the urgent need for improving diagnostic approaches in this high-risk population. We had previously demonstrated the high sensitivity of mutational analysis in bile cfDNA, the Bilemut assay, for the early diagnosis of malignancy in patients with biliary strictures [20]. Here, we demonstrate that this assay also identifies mutations in cfDNA from bile samples obtained during an ERCP procedure performed in PSC patients, introducing bile analysis as a complementary tool for the management of this patient population at high risk of CCA development.
Currently, besides imaging approaches, sampling techniques such as endoscopic biopsy and biliary brush cytology remain the standard diagnostic tools for suspicious biliary strictures, although with limited sensitivity [11]. Additionally, cholangioscopy with biopsy is a valuable diagnostic tool for biliary strictures, offering high sensitivity and specificity. However, this procedure requires significant time, specialised equipment, and considerable expertise [22, 23]. These factors may limit its widespread implementation in clinical practice. Moreover, in a recent work, it was concluded that annual surveillance using CA19-9 and MRI/MRCP, followed by ERCP in those patients with severe/progressive bile duct changes, was not effective in detecting CCA early enough to significantly improve long-term survival [8]. In this regard, recent publications have demonstrated that applying NGS to biliary brush cytology samples significantly improves the sensitivity to detect neoplasia in PSC patients compared to conventional cytology [24-26], identifying mutations commonly reported in PSC-CCA patients [27-29]. During ERCP, bile can be aspirated without significantly increasing procedure time and the risk of complications for the patient. Thus, several studies, including ours, have demonstrated the feasibility and usefulness of detecting mutations in bile to confirm or improve the detection of malignancy in patients with biliary strictures [19, 20, 30-33]. However, to our knowledge, few reports have included patients with PSC. A study published in the early 2000s evaluated the presence of KRAS mutations in the bile pellet of 56 PSC patients, showing that, during follow-up, only those individuals with KRAS mutations developed CCA or dysplasia [34]. Aside from this, our study is the first to apply NGS to bile fluid samples from PSC patients. In our cohort of 63 patients, we successfully obtained sufficient cfDNA to be sequenced from all bile samples, highlighting the feasibility of using bile as a liquid biopsy matrix in this group of patients.
Across the entire cohort, 23 out of 63 patients tested Bilemut-positive, harbouring between one and four mutations. This elevated number of Bilemut-positive patients may be explained by the fact that in our cohort ERCP was performed and bile samples were collected exclusively from PSC patients with relevant strictures or clinical suspicion of malignancy. This specific selection likely skewed the results towards a higher prevalence of mutations, as these patients tend to be in more advanced stages of the disease, and importantly, this subgroup represents the target population where early detection of malignancy would have the most clinical relevance. Our results demonstrate no association between the detection of mutations and the location of the strictures. However, further studies are required to confirm this point. Moreover, given that most PSC-associated CCAs develop from a dominant stricture [21], efforts should be made to try to collect bile as close as possible to the location of the stricture.
Positive Bilemut results appear to be independent of the presence of cirrhosis. Interestingly, the development of CCA in PSC patients has also been shown to be independent of the presence of cirrhosis [21]. In our cohort, 3 patients without cirrhosis developed CCA, and 2 tested Bilemut-positive. Therefore, a predictive value for malignancy of a Bilemut-positive result in non-cirrhotic PSC patients warrants consistent evaluation.
Regarding the genes mutated, we found mutations in KRAS, GNAS, TP53, BRAF, ERBB2, ERBB3, and FGFR3. These mutations are consistent with those described in previous studies in CCA samples [27, 28] and ERCP-derived brushings [24-26] from PSC patients. In fact, the mutational landscape of PSC-associated CCA resembles eCCA more than iCCA, including the absence of gene translocations [28]. Although the mutated genes identified are similar across all studies, the prevalence of mutations differs depending on the type of samples. For instance, studies conducted on CCA samples commonly identify TP53 as the most frequently mutated gene. In contrast, studies performed on biliary brushings or, as in our case, bile samples consistently highlight KRAS as the most prevalent mutation. KRAS mutations are widely recognised as early events in the development of carcinoma [35]; therefore, the presence of these mutations does not necessarily indicate that a tumour is already present in these patients at the time of detection. However, as previously mentioned, it has been reported that during follow-up, only those PSC patients harbouring KRAS mutations in bile cfDNA developed CCA or dysplasia [34]. In our study, KRAS mutations were detected as the only mutation in seven patients, none of whom had signs of biliary neoplasia after a median follow-up of 94.5 months (range 60.3–203.3 months). Our findings confirm that KRAS mutation is an early event in patients with PSC, but not enough for tumorigenesis; therefore, its association with tumour development requires further studies.
Our study also describes the presence of single mutations in the gene PIK3CA in bile samples coming from two healthy liver-living donors. Importantly, although PIK3CA mutations have been described in PSC-CCA tumours [28], mutations in this gene were not detected in any of our PSC patients. Moreover, in our previous study, we did not detect isolated PIK3CA mutations in the bile of patients with malignant strictures, including CCA. We always detected PIK3CA mutations together with mutations in other genes. Interestingly, the presence of PIK3CA mutations has been described as a common event in normal tissues, which may indicate clonal expansions unrelated to tumour development [36].
Together with patients with mutations in a single gene, we were able to detect patients with mutations in four analysed genes. Determining the specific risk associated with individual mutations or combinations of mutations will require further studies. In fact, a notable limitation of our study is that we used a commercial panel analysing 52 genes, missing some genes frequently mutated in PSC-associated CCA such as CDKN2A [27, 28].
Within our cohort, four patients were diagnosed with CCA. Among these, three tested positive for Bilemut, yielding a sensitivity of 75% for CCA detection. Notably, all three positive cases were pCCA, while the negative case was an iCCA, suggesting that tumour location may influence the assay's sensitivity. More importantly, in two of these cases, the detection of mutations occurred 4.9 and 6.9 months prior to the diagnosis of CCA by current diagnostic strategies. This underscores the potential application of the Bilemut assay in providing earlier information for PSC patients, potentially offering a valuable window for early intervention.
Moreover, among the three Bilemut-positive patients diagnosed with CCA, two had only one mutation detected, while the other had two mutations. Our data, therefore, indicate that neither a higher number of mutated genes, a greater MAF, nor a longer follow-up time was consistently associated with positivity, highlighting the complexity and heterogeneity of disease progression.
Although the precise roles of specific bile cfDNA mutations might play in guiding clinical management is yet to be established, our data suggest that patients with a positive Bilemut assay may benefit from more frequent surveillance or expedited evaluation for liver transplantation. Additionally, the detection of targetable mutations could guide future therapeutic strategies, especially if malignancy is confirmed, allowing for more personalised and targeted treatments. Furthermore, if performed in a sequential manner, these analyses could reflect the chronology of mutations, distinguishing between early and late events in cancer development. This might also enhance our understanding of the progression of malignancy in these patients.
According to the British Society of Gastroenterology guidelines for the management of PSC [37], in patients undergoing ERCP for relevant strictures, it is mandatory to obtain pathological samples from suspicious strictures. Based on our findings, we propose that, in addition to standard brushings or biopsies, bile samples should also be collected for further molecular analyses. This additional sample could provide complementary information, enhancing the characterisation of the patient's condition and potentially leading to more tailored clinical decision-making.
Author Contributions
Conceived and designed the study: M.A., P.M., M.K., M.A.Á., C.B. Sample and clinical data collection: E.B., B.K., M.K., J.P., S.K., M.G., J.R.W., L.S.C., A.B., Ł.K., K.M.S. Sample processing: C.R., J.E., M.E. Data analysis: M.A., E.B., G.A.A., P.M., M.K., M.A.Á., C.B. Data interpretation: M.A., E.B., M.R., D.O., M.G.F.-B., Ł.K., J.U., P.S., C.T., P.M., M.K., M.A.Á., C.B. Funding acquisition: M.A., K.M.S., J.U., P.S., M.K., M.A.Á., C.B. Writing original draft: M.A., M.A.Á., C.B. Writing, reviewing, and editing: all authors. All authors read and approved the final manuscript.
Acknowledgements
We particularly acknowledge the patients for their participation. We also acknowledge the technical support of Sara Equiza, Teresa Imizcoz, Maria Isabel Mora, and Arancha Bielsa from CIMA LAB Diagnostics, University of Navarra.
Ethics Statement
The study was approved by the Institutional Ethics Committees (#KB/49/2015, #KB/112/2021 and CEIm 2016/91).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request.