Diversification and secondary contact in the magpie-jays (Calocitta) throughout the pacific lowlands of Mesoamerica
Contributing authors: Luis A. Sánchez-González ([email protected]), Vicente J. Castillo-Chora ([email protected]), Enrique Arbeláez-Cortés ([email protected])
Luis A. Sánchez-González and Vicente J. Castillo-Chora contributed equally.
Abstract
enThe Mesoamerican dry forests (MDF), rich in species and in endemic taxa, are distributed nearly continuously nearly continuously along the American Pacific slope from Mexico to Costa Rica; however, several of the bird species inhabiting the MDF show clear phenotypic differentiation recognized through the description of subspecies. There are two currently recognized species of magpie-jays of the genus Calocitta (Corvidae) distributed throughout the MDF: the monotypic black-throated magpie-jay (C. colliei) and the polytypic white-throated magpie-jay (C. formosa). These two species have sometimes been considered conspecific and have been reported to hybridize in sympatric areas, where birds with intermediate plumage characters are recorded. Using mitochondrial and nuclear DNA markers from individuals of the two species, we analyze the phylogeographic structure and the genetic diversity within Calocitta under an isolation with migration (IM) model. The results showed strong genetic structure, in which the two currently recognized species and some of the C. formosa subspecies grouped into four well-supported and reciprocally monophyletic clades. IM analyses suggested divergence dates for the split between C. colliei and C. formosa that were congruent with geological factors, as well as with the deep divergence of the three lineages within C. formosa. These factors likely led to a dynamic demographic history in all lineages. We also found strongly limited gene flow, null or near null migration values, and large genetic fixation and genetic distance values. We suggest that the strong genetic differentiation between lineages is the result of allopatric differentiation with later secondary contact, further supporting a highly dynamic biotic history in MDF.
Resumen
esLos bosques secos Mesoamericanos (MDF) son ricos en especies y están distribuidos casi de manera continua a lo largo de la vertiente pacífica de México hasta Costa Rica; sin embargo, varias de las especies de aves que los habitan muestran clara diferenciación fenotípica, lo que se ha reconocido a través de la descripción de subespecies. Existen dos especies actualmente reconocidas de urracas del género Calocitta (Corvidae) que se distribuyen en los MDF: la monotípica Urraca Hermosa Carinegra (C. colliei) y la politípica Urraca Hermosa Cariblanca (C. formosa). Estas dos especies a veces han sido consideradas conespecificas y se ha reportado que hibridizan en áreas de simpatría, donde se han registrados individuos con caracteres intermedios de plumaje. Usando marcadores de DNA mitocondrial y nuclear para individuos de ambas especies, analizamos la estructura filogeográfica y la diversidad genética dentro de Calocitta bajo un modelo de Aislamiento con Migración (IM). Los resultados muestran una fuerte estructura genética, en la cual las dos especies reconocidas y algunas de las subespecies de C. formosa se agrupan en cuatro clados bien soportados y recíprocamente monofiléticos. Los análisis de IM sugieren fechas de divergencia para la división entre C. colliei y C. formosa que son congruentes con factores geológicos, al igual que para las tres divergencias profundas entre los linajes dentro de C. formosa. Esos factores posiblemente llevaron a una historia demográfica dinámica en todos los linajes. También encontramos flujo génico muy limitado, valores de migración nulos o casi nulos, y valores altos de fijación y distancia genéticas. Sugerimos que esa fuerte diferenciación genética es el resultado de diferenciación alopátrica con posterior contacto secundario, apoyando aún más una historia biótica altamente dinámica en los MDF.
1 INTRODUCTION
Although the Neotropical dry forests of the American continent are distributed over a large area, that distribution is discontinuous, which has likely promoted the evolution of very high species richness and endemism among many different biotic groups (Banda et al., 2016; Bullock et al., 1995; Ceballos et al., 2010; Dirzo et al., 2011; Pennington et al., 2006; Prieto-Torres et al., 2018). Biogeographic patterns show strong structure, which has led to the recognition of different biotic ecoregions that largely match the main dry forest patches (see Olson et al., 2001). While most of these distributional patterns have mainly been defined based on plants (e.g., Banda et al., 2016; Pennington et al., 2006; Werneck et al., 2011), congruent patterns in a number of taxa support the distinctiveness of different regions, one of which is the Mesoamerican dry forests (hereafter MDF; Montaño-Arias et al., 2018; Prieto-Torres et al., 2018).
The distribution of the MDF is nearly continuous along the Pacific slope of Mesoamerica, from northwestern Mexico (southern Sonora) to northwestern Costa Rica. On the Atlantic slope, these forests are limited to only sparse patches in eastern Mexico, the northern Yucatan Peninsula, and in eastern Guatemala (Ceballos et al., 2010). MDF, particularly on the Pacific slope of Mexico, have been considered the most biodiverse forests in the world (Ceballos et al., 2010). An estimated 190–255 species of birds are found in the MDF, of which up to 20% are endemic (Escalante et al., 1998; Navarro-Sigüenza et al., 2014). These species richness and endemism estimates vary depending on the application of different species concepts (e.g., Navarro-Sigüenza & Peterson, 2004; Peterson & Navarro, 2000) and changing understanding of the genetic variation in different taxa (e.g., Arbeláez-Cortés et al., 2014; Arbeláez-Cortés & Navarro-Sigüenza, 2013; Mason et al., 2018; Montaño-Rendón et al., 2015). Several bird species in the region show marked phenotypic differentiation, which has been recognized through the description of subspecies in many taxa (Dickinson et al., 2003; Navarro-Sigüenza & Peterson, 2004). In many cases, this differentiation is not associated with any clear ecological or geographical barriers, making it difficult to recognize both intraspecific and interspecific boundaries. For example, within the Ortalis chachalacas and the Calocitta magpie-jays, taxa have been considered single polytypic species or several species-level taxa (AOU, 1998; dos Anjos et al., 2009; Banks, 1990; Phillips, 1986). Decisions on species limits have traditionally been subject to the significance and frequency of phenotypically intermediate individuals or putative hybrids.
Magpie-jays of the genus Calocitta (Passeriformes: Corvidae) are medium-sized, conspicuous birds with a mostly blue, black, and white plumage, characterized by a long tail and a noticeable crest (Figure 1a). The two currently recognized species are distributed along the Pacific slope (dos Anjos et al., 2009; Howell & Webb, 1995). The monotypic black-throated magpie-jay (C. colliei) is endemic to northwestern Mexico, from southern Sonora and southwestern Chihuahua through western Jalisco and Colima. The polytypic white-throated magpie-jay (C. formosa) includes three recognized subspecies: C. f. formosa, distributed from western Jalisco to central Oaxaca and the Balsas Basin; C. f. azurea, distributed from southeastern Oaxaca and Chiapas in Mexico to western Guatemala; and C. f. pompata, found from interior eastern Oaxaca and the Central Chiapas Depression in Mexico to northwestern Costa Rica (Figure 1b; Gill & Donsker, 2016).
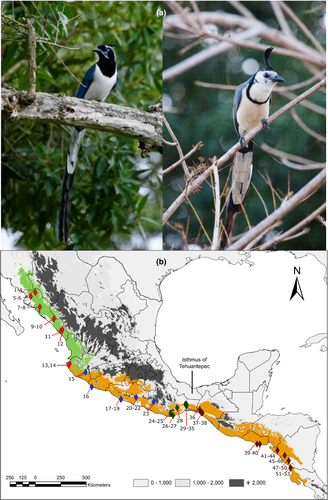
Black-throated and white-throated Magpie-jays have sometimes been considered conspecific (see AOU, 1998; dos Anjos et al., 2009; Phillips, 1986). This has been largely based on suspected hybridization in western Jalisco and Colima evidenced by a locally high incidence of birds with intermediate plumage characters (dos Anjos et al., 2009; Pitelka et al., 1956). Phillips (1986), however, suggested that current hybridization may not be as common as the plumage characters would seem to suggest, proposing instead that there may be individual phenotypic variation due to limited past interbreeding in the zone where the two taxa are sympatric in western Jalisco. This casts doubt on the significance of hybridization or phenotypically intermediate in terms of species recognition.
Within C. formosa, subspecies have been recognized based on plumage features such as the amount of black on the face and breast and the intensity of the blue coloration of the upperparts. Intergradation has been reported between the azurea and formosa subspecies in coastal Oaxaca (Miller et al., 1957). This region also includes the distribution of a described but no longer recognized fourth subspecies: C. f. impudens, which was described from southern Oaxaca and southeastern Chiapas in Mexico to the interior arid valleys of Honduras and disjunct from southeastern Guatemala and El Salvador. However, its range is interrupted by C. f. azurea in southern Chiapas and S Guatemala (Phillips, 1986). Although impudens is not currently recognized, its description suggests high variability in plumage patterns in Calocitta and emphasizes the difficulty of using these characters to establish limits for taxonomic differentiation. In addition, interspecific hybridization has been also reported between C. formosa and the closely related Brown jay (Psilorhinus morio) in western Chiapas (Pitelka et al., 1956).
Calocitta magpie-jays are a well-suited model to test diversification and differentiation in birds of the Pacific MDF; the two currently recognized sister species probably diverged relatively recently in the middle to Early Pleistocene (Fernando et al., 2017), and subspecific differentiation is observed in C. formosa along the apparently continuous dry forests of the region. In addition, individuals with intermediate plumage have been recorded in areas where species or subspecies currently met, which allows us to hypothesize that these individuals are the result of secondary contact. Also, following current systematic schemes, we expect higher and more recent gene flow within subspecies of C. formosa than between C. colliei and C. formosa, which may be reflected in higher similarity of phenotypic traits within C. formosa.
The shared evolutionary patterns revealed by research on different biological groups have allowed more accurate estimations on how biotic communities in different regions are assembled (Byrne et al., 2008; Crisp, 2006; Pennington et al., 2004). In this sense, the study of polytypic taxa may be highly informative, especially in habitats that are continuous in the present, such as the Pacific MDF. Here, under an isolation with migration (IM) model framework and using DNA data from both mitochondrial and nuclear markers from individuals of the two species of Calocitta throughout their geographic range, we: (a) describe the genetic diversity and phylogeographic structure within the two Calocitta species; (b) estimate divergence times among the recognized species and subspecies and compare them with patterns in other studies in search of common causes for differentiation; (c) explore demographic changes at the species and subspecies levels; (d) assess whether currently recognized phenotypic differentiation corresponds with genetic differentiation among the recognized subspecies of C. formosa; and (e) attempt to clarify the taxonomic status of the C. formosa populations of the Isthmus of Tehuantepec, where up to three subspecies have been recorded.
2 METHODS
2.1 Sampling and laboratory procedures
To assess patterns of genetic variation, we obtained samples of 54 individuals from 25 localities of C. colliei and all three currently recognized subspecies of C. formosa (Table 1, Figure 1). We used taxa from some of the major New World corvid clades: Aphelocoma coerulescens, Cyanocorax yncas, Cyanocitta cristata, we also included Psilorhinus morio, the putative sister taxon to Calocitta (Bonaccorso & Peterson, 2007; Bonaccorso et al., 2010; Fernando et al., 2017).
| Species | Catalog | ND2 | COI | Locality | Longitude | Latitude | Name used |
|---|---|---|---|---|---|---|---|
| C. colliei | UWBM 88909 | MW401854 | MW401951 | Mexico: Sinaloa | −108.80833 | 26.305 | Sin_12 |
| C. colliei | UWBM 88935 | MW401861 | MW401947 | Mexico: Sinaloa | −108.8089 | 26.3054 | Sin_03 |
| C. colliei | UWBM 88941 | MW401864 | MW401948 | Mexico: Sinaloa | −108.39 | 26.654 | Sin_04 |
| C. colliei | MZFC 19681 | MW401857 | MW401952 | Mexico: Sinaloa | −106.76333 | 24.303333 | Sin_15 |
| C. colliei | MZFC 23182 | MW401858 | MW401953 | Mexico: Sinaloa | −108.39 | 26.65333 | Sin_14 |
| C. colliei | MZFC 24015 | MW401860 | MW401945 | Mexico: Sinaloa | −107.99272 | 25.364556 | Sin_08 |
| C. colliei | MZFC 24016 | MW401856 | MW401943 | Mexico: Sinaloa | −107.99272 | 25.364556 | Sin_09 |
| C. colliei | MZFC 24017 | MW401852 | MW401956 | Mexico: Sinaloa | −106.13611 | 23.255369 | Sin_10 |
| C. colliei | UWBM 81146 | MW401862 | MW401950 | Mexico: Sinaloa | −106.764 | 24.304 | Sin_07 |
| C. colliei | MZFC 15312 | MW401853 | MW401955 | Mexico: Sinaloa | −105.97444 | 23.367778 | Sin_11 |
| C. colliei | MZFC 25311 | MW401855 | MW401949 | Mexico: Jalisco | −105.3575 | 20.34523 | Jal_01 |
| C. colliei/formosa? | MZFC 25313 | MW401871 | MW401954 | Mexico: Jalisco | −105.42156 | 20.259389 | Jal_03 |
| C. colliei | UWBM 82704 | MW401859 | MW401944 | Mexico: Sinaloa | −108.39 | 26.654 | Sin_05 |
| C. colliei | FMNH_343602 | DQ912591.1 | NA | Mexico: Sinaloa | −107.785 | 25.4533333 | Sin_13 |
| C. formosa | KUNHM_9352 | DQ912602.1 | MW401916 | El Salvador: Usulután | −88.508833 | 13.224 | Sal_01 |
| C. formosa | KUNMH_9368 | MW401895 | MW401917 | El Salvador: Usulután | −88.761667 | 13.238333 | Sal_02 |
| C. formosa | MZFC ALTIBAL_99 | MW401900 | MW401934 | Mexico: Guerrero | −99.470826 | 17.358891 | Gue_03 |
| C. formosa | MZFC 20751 | MW401877 | MW401930 | Mexico: Chiapas | −93.72381 | 15.995455 | Chi_03 |
| C. formosa | MZFC 19422 | MW401879 | MW401931 | Mexico: Chiapas | −93.874628 | 16.303057 | Chi_02 |
| C. formosa | MZFC 23580 | MW401878 | MW401933 | Mexico: Chiapas | −93.806667 | 16.158056 | Chi_01 |
| C. formosa | MZFC 18738 | MW401865 | MW401904 | Mexico: Oaxaca | −95.829638 | 16.44958 | Oax_01 |
| C. formosa | MZFC 22066 | MW401881 | MW401906 | Mexico: Oaxaca | −96.427361 | 15.934003 | Oax_06 |
| C. formosa | MZFC 16362 | MW401873 | MW401942 | Mexico: Michoacán | −103.40459 | 18.300109 | Mic_01 |
| C. formosa | MZFC 16363 | MW401874 | MW401938 | Mexico: Guerrero | −100.79667 | 17.203333 | Gue_05 |
| C. formosa | MZFC 16638 | MW401876 | MW401941 | Mexico: Guerrero | −100.79667 | 17.203333 | Gue_06 |
| C. formosa | MZFC 16881 | MW401866 | MW401910 | Mexico: Oaxaca | −95.0425 | 16.7925 | Oax_12 |
| C. formosa | MZFC 16882 | MW401867 | MW401912 | Mexico: Oaxaca | −95.011622 | 16.670619 | Oax_11 |
| C. formosa | MZFC 16880 | MW401868 | MW401914 | Mexico: Oaxaca | −95.115 | 16.745278 | Oax_10 |
| C. formosa | MZFC 16849 | MW401869 | MW401911 | Mexico: Oaxaca | −95.0425 | 16.7925 | Oax_09 |
| C. formosa | MZFC 16848 | MW401870 | MW401915 | Mexico: Oaxaca | −94.990109 | 16.632563 | Oax_08 |
| C. formosa | MZFC 28061 | MW401883 | MW401908 | Mexico: Oaxaca | −96.1945 | 15.76953 | Oax_07 |
| C. formosa | UWBM 69251 | MW401893 | MW401922 | Nicaragua: Chinandega | −86.95 | 12.683333 | Nic_01 |
| C. formosa | UWBM 69008 | MW401894 | MW401926 | Nicaragua: Chinandega | −86.95 | 12.683333 | Nic_02 |
| C. formosa | UWBM 69010 | MW401889 | MW401927 | Nicaragua: Chinandega | −86.95 | 12.683333 | Nic_03 |
| C. formosa | UWBM 69011 | MW401887 | MW401929 | Nicaragua: Chinandega | −86.95 | 12.683333 | Nic_04 |
| C. formosa | UWBM 69312 | MW401891 | MW401918 | Nicaragua: Rivas | −85.795 | 11.145 | Nic_09 |
| C. formosa | UWBM 69068 | MW401890 | MW401919 | Nicaragua: Rivas | −85.795 | 11.145 | Nic_10 |
| C. formosa | UWBM 69069 | MW401892 | MW401920 | Nicaragua: Rivas | −85.795 | 11.145 | Nic_11 |
| C. formosa | UWBM 69172 | MW401896 | MW401924 | Nicaragua: Granada | −85.9583 | 11.76 | Nic_05 |
| C. formosa | UWBM 69428 | MW401898 | MW401928 | Nicaragua: Granada | −85.9583 | 11.76 | Nic_06 |
| C. formosa | UWBM 69178 | MW401899 | MW401921 | Nicaragua: Granada | −85.9583 | 11.76 | Nic_07 |
| C. formosa | UWBM 69435 | MW401897 | MW401932 | Nicaragua: Granada | −85.9583 | 11.76 | Nic_08 |
| C. formosa | UWBM 70235 | MW401888 | MW401925 | Nicaragua: Managua | −86.314054 | 12.228059 | Nic_12 |
| C. formosa | UWBM 56228 | MW401886 | MW401923 | Nicaragua: Managua | −86.314054 | 12.228059 | Nic_13 |
| C. formosa | MZFC HJ6_022 | MW401875 | MW401936 | Mexico: Guerrero | −100.82156 | 17.194778 | Gue_04 |
| C. formosa | MZFC MAF_300 | MW401884 | MW401909 | Mexico: Oaxaca | −96.1945 | 15.76953 | Oax_13 |
| C. formosa | MZFC 26437 | MW401902 | MW401935 | Mexico: Guerrero | −99.470826 | 17.358891 | Gue_01 |
| C. formosa | MZFC 28902 | MW401901 | MW401937 | Mexico: Guerrero | −99.470826 | 17.358891 | Gue_02 |
| C. formosa | MZFC 17261 | MW401885 | MW401913 | Mexico: Oaxaca | −95.02317 | 16.677953 | Oax_03 |
| C. formosa | MZFC 17256 | MW401880 | MW401905 | Mexico: Oaxaca | −95.0425 | 16.7925 | Oax_05 |
| C. formosa | MZFC 14115 | MW401903 | MW401939 | Mexico: Oaxaca | −97.906667 | 16.963333 | Oax_02 |
| C. formosa | MZFC 21313 | MW401882 | MW401907 | Mexico: Oaxaca | −96.420892 | 15.92515 | Oax_04 |
| C. formosa x colliei | MZFC 25312 | MW401872 | MW401940 | Mexico: Jalisco | −103.94241 | 19.59591 | Jal_02 |
| C. formosa | FMNH 343605 | HQ123802.1 | NA | Mexico: Guerrero | −100.225 | 17.2616667 | Gue_07 |
| Cyanocitta cristata a | KUNHM 4390/SPP1513-23733 | DQ912604.1 | DQ434558.1 | NA | NA | NA | C_cristata |
| Aphelocoma coerulescens a | FMNH 396259/FMNH 396261 | DQ912601.1 | DQ432737.1 | NA | NA | NA | A_coerulescens |
| Cyanocorax yncas a | MZFC 15927/2021 | GU144847.1 | DQ433558.1 | NA | NA | NA | C_yncas |
| Psilorhinus morio a | KUNHM B−1896/USNM:Birds:614081 | DQ912607.1 | JQ174603.1 | NA | NA | NA | P_morio |
- Abbreviation: NA, data not available.
- Museum acronyms: UWBM, University of Washington Burke Museum; MZFC, Museo de Zoología Facultad de Ciencias–UNAM; FMNH, Field Museum of Natural History; KUNHM, University of Kansas Natural History Museum).
- a Used as out-group.
DNA sequences were obtained from samples of muscle, heart, or liver tissue stored in ornithological collections (Table 1; see Acknowledgements). DNA extraction was performed using a phenol–chloroform method (da Silva et al., 2012) or using the Qiagen DNeasyTM Blood and Tissue Kit (Qiagen Inc.) following manufacturer's protocol. We amplified two mitochondrial DNA loci (mtDNA): the mitochondrial encoded NADH dehydrogenase 2 (ND2), using the primers H6313 and L5219 or L5216 (Sorenson et al., 2004) and mitochondrially encoded cytochrome c oxidase I (COI), using primers COIBirdF1-COIBirdR2 (Hebert et al., 2004). We also amplified two nuclear DNA (nuDNA) loci: transforming growth factor beta 2 intron 5 (TGFB2), using the primers TGFB2.5F and TGFB2.6R (Sorenson et al., 2004), and glyceraldehyde-3-phosphate dehydrogenase intron 11 (GAPDH), using the primers G3P13b and G3P14b (Fjeldså et al., 2003). Primer sequences are included in Table S1.
The mtDNA and nuDNA loci were amplified by PCRs containing PCR buffer 1X, 1.5 mM MgCl2, 0.2 mM of dNTP, 0.4 to 0.5 µM of ePach primer, 1–2 µl of DNA polymerase, and 2 µl of the DNA extract. All PCR procedures included negative controls to rule out contamination, and products were visualized on agarose gels stained with ethidium bromide. Sequencing was performed in two different laboratories, depending on the original of the tissue samples. PCR products of tissues housed at the MZFC were sequenced at the High Throughput Genomics Center of the University of Washington (USA). PCR products from tissues from other collections were prepared for sequencing using BigDyeTM, v3.1 terminator chemistry (Qiagen Inc.), using the same primers as in the PCR, purified with ethanol precipitation, and sequenced using an ABI 3730XL automated sequencer at the Biodiversity Institute of the University of Kansas (USA). The sequences were edited and aligned manually using Sequencher 4.8 (GeneCodes, An Arbor, MI). The nuDNA chromatograms were inspected for double peaks, which were coded using standard IUPAC codes for ambiguities. The allelic phase of the nuclear loci was determined using PHASE (Stephens & Donnelly, 2003; Stephens et al., 2001), a Bayesian method based on coalescence with default parameters as implemented in DnaSP v5 (Librado & Rozas, 2009). For all subsequent analyses, we used only individuals for which all four study loci could be successfully sequenced.
2.1.1 Phylogenetic analyses and divergence time estimation
All of our phylogenetic analyses were run using a concatenated matrix (mtDNA + nuDNA, 2272 bp in the final alignment; Alignment S1). We reconstructed phylogenetic relationships using Bayesian inference (BI). The appropriate DNA substitution models for each locus were estimated using jModeltest v2.1.4 (Darriba et al., 2012; Guindon & Gascuel, 2003), and the best models were selected via the Akaike information criterion. The BI tree was obtained using MrBayes v3.1 (Ronquist & Huelsenbeck, 2003), in which we carried out two parallel runs for 1 × 107 generations, sampling every 1000 steps. We used Tracer v1.5 (Rambaut et al., 2014) to evaluate the convergence of the chains, which was verified by inspection of the effective sufficient sample size values (ESS >200; Drummond et al., 2006). After convergence, the first 2000 trees were discarded as burn-in, and a majority-rule consensus tree was calculated. We considered nodes with a posterior probability ≥0.95 to be well supported (Larget & Simon, 1999). Additionally, to compare the relationships among haplotypes, we constructed a haplotype network using Network v4.6 (Bandelt et al., 1999) using the median joining method on the mtDNA dataset.
We constructed a species tree using the *BEAST 2 template (Heled & Drummond, 2010) as implemented in BEAST 2 v2.6.3 (Bouckaert et al., 2014). *BEAST 2 implements a multispecies coalescent model and allows the use of a different number of sequences for different genes per taxon in a single, not-concatenated analysis (Heled & Drummond, 2010). Substitution rates used for mtDNA were 0.029 (0.024—0.033) substitutions/site/ lineage/Million years (s/s/l/My) for ND2, and 0.016 (0.014—0.019) s/s/l/My for COI; while for nuDNA were 0.0012 (0.0007–0.0017) s/s/l/My for GAPDH, and 0.0017 (0.0013—0.0022)-s/s/l/My for TGFB2 (Lerner et al., 2011; Lim & Sheldon, 2011). Priors for the analysis included a strict molecular clock for all of the loci, an initial UPGMA tree, a Yule speciation process, linear growth, and a constant root population size model. The analysis was run for 200 million generations, sampling every 10,000 generations, discarding the initial 20% as burn-in. Chain convergence and ESS (>200 in all of the parameters) were inspected in Tracer v1.5 (Rambaut et al., 2014). We then used TreeAnnotator v1.7.5 (Drummond et al., 2006) to generate a consensus tree with median node heights in order to summarize information on topology and divergence times. We visualized the species tree in DensiTree 2 (Bouckaert & Heled, 2014).
2.1.2 Genetic diversity and structure
To analyze the mtDNA under a population genetics framework, we clustered Calocitta individuals into four groups following the clades obtained in the phylogenetic analyses (see Results) as follows: (i) northwestern Mexico (NWM, n = 15), which included all samples of C. colliei; (ii) Mexican Central Pacific (MCP, n = 9); (iii) Oaxaca (OAX, n = 12); and (iv) Middle Central America (MCA, n = 18). The NWM, MCP, and OAX groups included all of the samples of C. formosa. We assessed the genetic diversity within each lineage by estimating the number of haplotypes (h), haplotype diversity (Hd), and nucleotide diversity (π) using DnaSP (Librado & Rozas, 2009). We also explored genetic structure using a three-way AMOVA with the samples grouped as above. Two groups (C. formosa-C. colliei) and three subgroups corresponding to the clades within C. formosa were used in the analysis. The significance of the AMOVA results was assessed via 10,000 non-parametric permutations. We also estimated the genetic differentiation FST parameter and assessed its significance using 10,000 permutations. These statistics were calculated in Arlequin ver. 3.5.1.3 (Excoffier & Lischer, 2010). Interpretation of the FST values followed Hartl and Clark (1997).
Due to the patterns of genetic structure detected within C. formosa, we decided to run tests for genetic divergence among the different lineages within this taxon. We also included C. colliei for comparison. We estimated genetic divergence among the three groups within C. formosa, as well as between each group and C. colliei using Nei's genetic distance values (Dxy) with a Jukes-Cantor correction (Nei, 1987), as implemented in DNAsp v5 (Librado & Rozas, 2009).
2.1.3 Demographic history analysis
We estimated gene flow among the different clades using the coalescent-based MCMC method in IMa2 (Hey, 2010), which fits an IM model of genetic data from closely related populations or species. Gene flow estimates included effective population size (Ne), directional migration rates, and population divergence times, all of which are calculated simultaneously using both the mtDNA and the nuDNA datasets. We used our *BEAST species tree for IM model estimations and assumed a 2-year generation time, which is based on the age at which males disperse from the natal territory in C. formosa (Langen, 1996a; Skutch, 1953).
We conducted four independent MCMC runs of 50 million of generations with a burn-in of 500,000 and an HKY substitution model using the following geometric heating scheme: -hfg -hn20 -ha0.96 -hb0.9. All demographic parameters were scaled by the overall mutation rate given by the geometric mean of each locus-specific mutation rate (Hey & Nielsen, 2004). Demographic parameters represent 95% of the highest posterior density (HPD) intervals, as well as the peak height value in the IM model (Nielsen & Wakeley, 2001). We applied likelihood ratio tests to assess the significance of the IM model migration rate estimates using an α = 0.001. We determined convergence of parameter estimates and chain mixing using two methods: (a) inspecting for clear trend lines in each independent run and (b) inspecting values for effective sample sizes (ESS >50; Hey, 2005).
2.1.4 Population clustering
We inferred population differentiation using GENELAND v. 1.0.7 (Guilliot et al., 2005) considering the mtDNA genetic data as well as the location of samples to determine the optimal number of genetic groups (K) and the geographic position of each (Guilliot et al., 2008). The algorithm identifies genetic discontinuities while estimating both the number and locations of populations without any a priori knowledge on the population units and limits (Zepeda et al., 2019). We conducted 10 independent runs with 1×106 MCMC iterations, sampling every 100 generations of the Markov Chain. We used K values ranging from 1 to 4 assuming an admixture model and correlated haplotype frequencies to estimate the posterior probability of the data. From the posterior distribution, we drew a map of probability isoclines of population membership, one for each population or cluster inferred by the model.
3 RESULTS
3.1 Phylogenetic analyses
We obtained mtDNA sequences for 38 samples of C. formosa and 14 of C. colliei. These sequences varied in length for the ND2 gene; 22 sequences had 646 base pairs (bp) and 30 sequences had 1041 bp. The COI gene sequences had 494 bp. We concatenated the mtDNA genes into a single alignment of 1535 bp analyses. For the GAPDH gene, we obtained 38 sequences of C. formosa and 14 of C. colliei, with a length of 305 bp; for TGFB2, we obtained 35 sequences for C. formosa and eight sequences for C. colliei, with a length of 432 bp. All sequences are deposited in GenBank (Table 1).
The Bayesian phylogenetic tree showed two well-supported major clades (Figure 2). One clade included all the C. formosa individuals, and the other included all C. colliei individuals. The C. formosa clade was further divided into three well-supported lineages, indicating clear phylogeographic structure (Figure 2), although the relationships among them were unclear due to low support values. The first sub-clade grouped samples from the southern Mexican Pacific coast including Oaxaca and the Isthmus of Tehuantepec regions (OAX). A second sub-clade included samples from the Mexican Central Pacific Coast (MCP), mainly from the state of Guerrero, but also including samples from Jalisco, Michoacán, and Oaxaca. The third sub-clade grouped the samples from southeastern Mexico (Chiapas) and the Pacific coast of Central America (Nicaragua and Salvador; MCA) (Figure 2). Interestingly, although we recovered three highly supported lineages, the distribution of the samples in these lineages did not correspond to the delimitation of the three currently recognized subspecies.
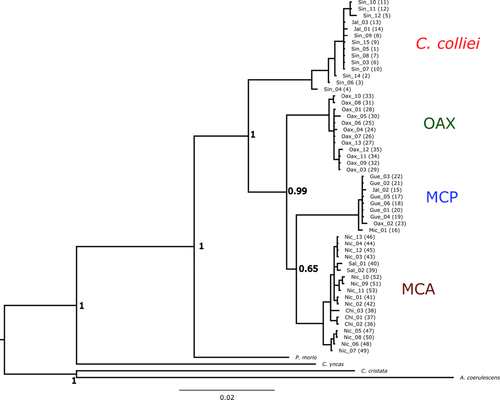
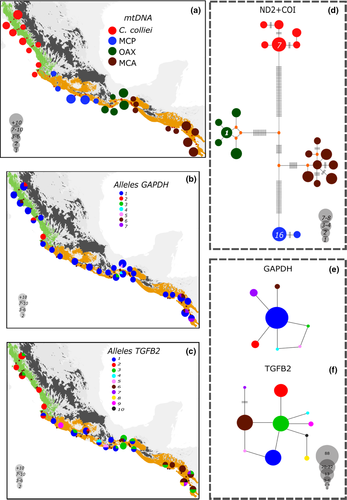
The mtDNA haplotype network showed 45 mutations between C. colliei and the OAX population of C. formosa. There were another 24 mutations between the haplogroups conformed by the OAX and MCA populations of C. formosa, and 29 mutations between the MCA and MCP populations (Figure 3d), indicating a marked divergence among the lineages within C. formosa. Using the nuDNA markers, GAPDH showed seven alleles, with a one private allele for C. colliei, three private alleles in the OAX and one in the MCA populations of C. formosa. The MCP population of C. formosa was monomorphic for the most common allele at this locus (Figure 3e).
3.2 Genetic diversity and structure
Patterns of genetic diversity for mtDNA and nuDNA in each of the main lineages are summarized in Table 2. For both datasets, MCA presented the highest number of haplotypes and alleles, but this population did not exhibit high levels of nucleotide diversity, which varied according to the locus analyzed. For the mtDNA, OAX had the highest nucleotide diversity (π = 0.0017) and MCP had the lowest values (π = 0.0004) and low haplotype diversity. In the case of nuDNA, C. colliei also had high values of nucleotide diversity (π = 0.001 and π = 0.0017 for GADPH and TGFB2, respectively.
| n | bp | S | h | Hd | π | |
|---|---|---|---|---|---|---|
| mtDNA | ||||||
| C. colliei | 14 | 1130 | 8 | 5 | 0.593 | 0.00289 |
| MCP | 10 | 1130 | 21 | 3 | 0.378 | 0.00741 |
| OAX | 12 | 1130 | 6 | 5 | 0.803 | 0.00169 |
| MCA | 18 | 1130 | 14 | 8 | 0.889 | 0.00264 |
| GADPH | ||||||
| C. colliei | 14 | 305 | 2 | 3 | 0.36 | 0.001 |
| MCP | 9 | 305 | 1 | 2 | 0.2 | 0.0007 |
| OAX | 11 | 305 | 3 | 5 | 0.34 | 0.0014 |
| MCA | 18 | 305 | 2 | 3 | 0.41 | 0.0016 |
| TGFB2 | ||||||
| C. colliei | 8 | 432 | 3 | 4 | 0.44 | 0.0017 |
| MCP | 9 | 432 | 4 | 6 | 0.63 | 0.0023 |
| OAX | 11 | 432 | 3 | 4 | 0.62 | 0.0016 |
| MCA | 15 | 432 | 7 | 7 | 0.65 | 0.0026 |
Note
- Populations were defined according to phylogenetic analysis.
- Abbreviations: A, alleles number; Ad, allelic diversity; bp, base pairs; h, haplotypes; Hd, haplotype diversity; n, sample size; S, segregating sites; π, nucleotide diversity.
The AMOVA revealed that 39.2% of the variation was in the two major groups defined for this analysis (C. formosa and C. colliei) and the three subgroups corresponding to the clades within C. formosa that were used in the analysis (Table 3). Most of the variance (56.45%) occurred within the three subclades of C. formosa. The FST values for the mtDNA dataset were higher than 0.91, indicating remarkably strong genetic structure among all of the populations of Mesoamerican Calocitta (Table 4). Genetic differentiation distances (Dxy) among C. colliei and the lineages within C. formosa were about 4.3% (range: 4.1– 4.7%), while among the subclades of C. formosa the average genetic differentiation was about 2.9% (range 2.5–3.5%; Table 4).
| Source of variation | d.f. | Sum of squares | Variance components | % of variation |
|---|---|---|---|---|
| Among groups | 1 | 417.660 | 11.45862 Va | 39.20 |
| Among populations within groups | 2 | 413.825 | 16.50190 Vb | 56.45 |
| Within populations | 47 | 59.850 | 1.27341 Vc | 4.36 |
| Total | 50 | 891.335 | 29.23393 |
Note
- Fixation Indexes—FSC: 0.92836; FST: 0.95644; FCT: 0.39196.
- Boldface values are significant at α < 0.05.
| Dxy/FST | C. colliei | MCP | MCA | OAX |
|---|---|---|---|---|
| C. colliei | — | 0.060/0.062 | 0.050/0.053 | 0.039/0.041 |
| MCP | 0.95663 | — | 0.027/0.029 | 0.035/0.036 |
| MCA | 0.95062 | 0.948 | — | 0.024/0.026 |
| OAX | 0.95663 | 0.97118 | 0.91935 | — |
Note
- Probability values were obtained after permutation tests with 1000 replicates. Uncorrected pairwise distances/Dxy values (above the diagonal).
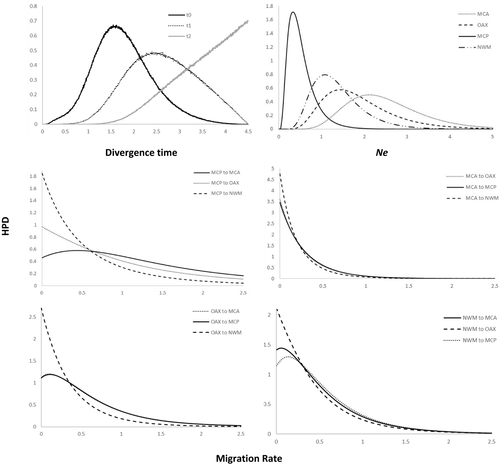
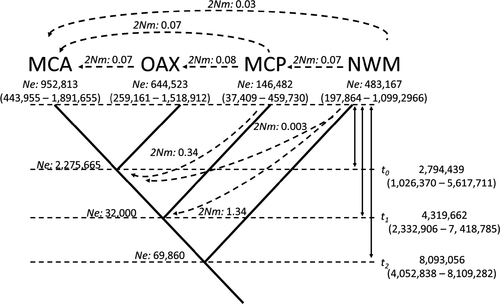
Based on the species tree (Figure S1), gene flow estimates using the IM model showed a dynamic demographic history within Calocitta (Figures 4 and 5). Based on the peak HPD values of Ne, the initial divergence in Calocitta occurred during the Middle Miocene (HPD 8.09 mya) leading to a population increase in NWM, while the rest of the lineages, currently grouped within C. formosa, decreased in population size. These three lineages then increased in Ne toward the Early Pliocene (HPD 4.3 mya), during which MCP diverged from OAX and MCA. The most recent divergence, the split between OAX and MCA, occurred during the Late Pliocene (HPD 2.79 mya). In general, our estimated migration rates showed non-significant values slightly above zero (0.03–0.08, i.e., less than one migrant every fourth generation) for geographically adjacent groups (Figure 5). Gene flow was highly limited (i.e., less than one migrant every fourth generation) in a north to south direction (from NWM to MCP to OAX to MCA), but not in the opposite direction (Figure 5). In addition, we detected limited ancestral gene flow from NWM to the ancestor of all of the C. formosa lineages (1.34; at least one migrant every generation), as well as from MCP to the ancestor of OAX and MCA (0.34), but no gene flow in the opposite direction (Figure 5).
3.3 Population clustering
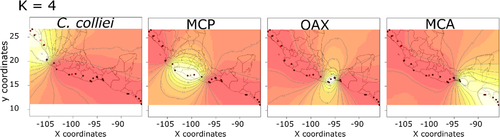
Population clustering analyses from the mtDNA and nuDNA suggested an optimal value of K = 4. These four genetic clusters are geographically well-delimited (Figure 6) and correspond to the same clades recovered by the phylogenetic analyses.
4 DISCUSSION
4.1 Divergence in Calocitta
Recent studies on different vertebrate groups in the dry forests of western Mexico (Arbeláez-Cortés et al., 2014; Arbeláez-Cortés & Navarro-Sigüenza, 2013; Arcangeli et al., 2018; Montaño-Rendón et al., 2015; Reyes-Velasco et al., 2013; Zarza et al., 2008) have revealed unexpected geographic structure in spite of current habitat continuity and the apparent absence of contemporary geographic or ecological barriers in Mesoamerica since at least the Holocene (Castillo-Chora et al., 2021). This pattern suggests that past climate and geological changes have deeply impacted the biotic history of this tropical lowland region. It has been proposed that these changes were most favorable during the Middle Miocene, which allowed for the development, coalescence, and radiation of seasonally drier communities and lineages (Graham, 2011).
This dynamic history is also reflected in the evolutionary history of Calocitta Magpie-jays. The clear phylogeographic structure in our analyses showed that the two currently recognized species (C. colliei and C. formosa) grouped into well-supported and reciprocally monophyletic clades. According to the estimated divergence dates, this initial split may have occurred during the Middle Miocene, coinciding with the genesis of the Trans-Mexican Volcanic Belt and a period of high vulcanism (from 5 to 3 mya) near the border between Jalisco and Nayarit (Frey et al., 2007). Furthermore, strong phylogeographic structure was recovered within C. formosa, in which three well-supported and reciprocally monophyletic groups are also apparent (Figure 2). The estimated divergence dates for these splits (Figure 5) are consistent with the transition between the Late Pliocene and the onset of the Pleistocene, suggesting a significant role of climatic change. Divergence in these three clades may have been maintained through periods in which favorable environmental conditions were highly localized in isolated climatic stable areas (Castillo-Chora et al., 2021).
In terms of demographic history, after the first split, the population in NWM experienced an increase in the effective population size, while the ancestor of the three groups within C. formosa apparently decreased in effective population size, probably due to a reduction in the dry forest area throughout western Mesoamerica during moister periods. Later climatic changes promoted the expansion and reconnection of previously isolated forest patches, allowing populations to established contact zones and gene flow, at least between adjacent populations (Castillo-Chora et al., 2021). However, several of our results support the idea of genetic divergence. Our results using the IM model suggest that gene flow is highly limited and not significant (Figure 4), with less than one individual every fourth generation in modern lineages. Furthermore, the large FST and Dxy values for all Calocitta lineages and the star-like haplotype network (Figure 3) suggest population expansion. Further studies incorporating data from more nuclear loci would provide additional support for our conclusions.
Our analyses suggest that the current distribution of the four lineages detected in the Calocitta magpie-jays has resulted from secondary contact, allowing for recent but highly limited and non-significant gene flow among formerly separated populations. Both intra- and interspecific gene flow and hybridization (sensu Moore, 1995; Phillips, 1986) have been reported as locally common in Calocitta (dos Anjos et al., 2009; Pitelka et al., 1956). However, observations by Phillips (1986) of phenotypic differences in individuals of flocks including both C. colliei and C. formosa in western Jalisco led the author to suggest that hybridization may not be as common as previously thought, and that phenotypically intermediate individuals may rather represent individual variation or past interbreeding, and that the reported variation in plumage patterns may be restricted to young individuals that do not yet express their definitive adult plumage. Our results also seem to support this, since gene flow is highly limited in recent times but was higher at the time of the initial split. Therefore, areas of present sympatry among the different lineages seem likely to be the product of secondary contact. Further studies using novel sequencing and analysis techniques are ongoing and will be published elsewhere (Castillo-Chora et al., unpublished data).
The strong genetic differentiation suggesting population isolation was less evident from the analysis of the nuclear dataset, which was expected given the different coalescent times for nuDNA versus mtDNA because of their population sizes (Hung et al., 2016; Zink & Barrowclough, 2008), Nonetheless, the geographic distribution of the nuclear alleles showed several similarities to the distributions of the mtDNA clades. When the nuDNA is grouped according to the previously identified mtDNA clades, the fact that the distribution of only a few alleles in these nuDNA groups shows geographically sorted private haplotypes suggests gene flow (Figure 2). The nuDNA allele frequencies among populations suggest incomplete lineage sorting, probably due to the slow mutational rates, but probably also to processes such as migration and introgression between the two taxa. However, IM coalescent analyses suggest that gene flow is highly limited in modern lineages but was somewhat higher in past lineages (NWM to OAX-MCA-MCP; Figure 5), suggesting a lag on the path to reciprocal monophyly (Omland et al., 2006). In addition, we detected differences in the divergence dates in this study compared with those estimated by Castillo-Chora et al. (2021), which may be due to the ability of IMa2 (Hey, 2010) to accommodate gene flow into a single coalescent analysis.
Factors may account for maintenance of the observed genetic divergence in Calocitta: (1) ecological barriers and (2) behavioral traits and natural history. No clear modern geographic barriers are associated with the distribution of the lineages in Calocitta. Therefore, ecological barriers may have acted first as promoters of diversification, and later contributed to maintaining the differentiation among adjacent lineages in more recent times (Castillo-Chora et al., 2021). Thus, after habitat modification due to orogenic and volcanic activity, diversification between colliei and formosa may have evolved due to habitat differences between them. These two species can form mixed flocks in western Jalisco, where oak forests mix with tropical dry forest vegetation (Phillips, 1986), suggesting secondary contact. Populations of OAX would have been prevented from expanding toward the range of the MCA by the humid lowland rainforests of southern Chiapas (the Soconusco region), as well as by a low-altitude mountain range near Rizo de Oro, which may have limited dispersal to the Central Depression in Chiapas, as suggested by Campbell (1999). In the case of MCP and OAX, no contemporary geographic barrier is apparent, placing a stronger emphasis on the significance of ecological barriers for genetic differentiation in these populations.
Sampling individuals from presumed suture zones would be highly informative, as they could reveal novel population dynamics (e.g., Harrison & Larson, 2016) between the different genetic groups in our study group. In addition, sampling is also needed to cover geographic gaps within some of the recovered clades, such as in Nayarit (for C. colliei) and from southern Chiapas to western El Salvador (for C. [f.] pompata). More complete sampling in these areas may reveal further details on the demographic history, as well as on the significance of past interbreeding that resulted in phenotypic intermediates or hybridization. On the other hand, behavioral differences and natural history may have also played a role in differentiation (Barber & Klicka, 2010); however, studies on these aspects throughout the distribution of Calocitta are virtually completely lacking and are presently limited to populations that are presumably outside of the secondary contact zones (e.g., Innes & Johnston, 1996; Langen, 1996b).
4.2 Taxonomic implications
Our research revealed unexpected levels of phylogenetic structure, historical demography, and genetic distinctiveness among the lineages found within Calocitta which may have taxonomic implications. High levels of genetic differentiation and the strongly reduced gene flow detected among the three C. formosa lineages are comparable to the levels of genetic divergence between the two currently recognized species in Calocitta (Table 4) and are also consistent with deep divergence times. The unmatched geographic distribution of the recognized subspecies in C. formosa as compared to the genetic lineages found in our study is especially complex in coastal Oaxaca and the Isthmus of Tehuantepec, where up to three recognized subspecies (plus one unrecognized taxon) meet, and for which our analyses seem to provide a clearer picture. Following our results, the MCP lineage may be assigned to C. [f.] formosa, which ranges from Jalisco to Guerrero and the Balsas Basin in southern Mexico; the MCA lineage includes two currently accepted subspecies: C. [f.] pompata, distributed from southern and central (the Central Depression) Chiapas in Mexico south to northwestern Costa Rica, and C. [f.] azurea from the SE Oaxaca to western Guatemala. This lineage should be recognized as azurea, which has priority over pompata since it was assigned to birds described from SE Chiapas in 1897, while the second was assigned to birds described from Costa Rica in 1914. This situation leaves the OAX lineage without a name, because although the unrecognized name impudens may be available, the type locality for birds under this name is Conchagua, El Salvador, which is within the range of azurea, therefore invalidating the use of impudens. The formal description of this putative new taxon from Oaxaca will be published elsewhere (Sánchez-González et al., unpublished data).
4.3 Evolutionary differentiation in the dry forests of Mesoamerica
Our results add to a growing body of evidence suggesting that the dry forests of the Mexican Pacific slope have had a highly dynamic history (Graham, 2011; Prieto-Torres et al., 2018, 2019) that has influenced the diversification and differentiation of a large number of plant taxa (e.g., De-Nova et al., 2012), and animal taxa, including frogs (Zaldı́var-Riverón et al., 2004), reptiles (Bryson et al., 2011; Suárez-Atilano et al., 2014; Zarza et al., 2008), birds (Arbeláez Cortés et al., 2014; Vázquez-Miranda et al., 2009), and mammals (Arcangeli et al., 2018), all of which exhibit clearly structured monophyletic lineages. These patterns highlight a very dynamic region, which was first isolated by tectonic processes, such as the rise of the Sierra Madre Occidental and the Trans-Mexican Volcanic Belt (Becerra, 2005; Sosa et al., 2018), allowing for the diversification of several plant and animal taxa. These patterns, however, differ widely in the age of diversification for different groups, which suggest that taxa in the region have also experienced further diversification events due to climatic changes since at least the Miocene (De-Nova et al., 2012; Navarro-Sigüenza et al., 2017). These climatic shifts seem to be the most likely cause of divergence and differentiation in other dry forest birds in western Mexico (see Arbeláez-Cortés et al., 2014; Castillo-Chora et al., 2021; Montaño-Rendón et al., 2015; Navarro-Sigüenza et al., 2017) as well as in the Calocitta magpie-jays explored here.
ACKNOWLEDGEMENTS
We thank the following curators and scientific collections for access to tissue loans: University of Kansas Biodiversity Institute and Natural History Museum (M. B. Robbins and A. T. Peterson), Museo de Zoología Facultad de Ciencias UNAM, University of Washington Burke Museum (J. Klicka and S. Birks), and the Field Museum of Natural History (S. Hackett and J. Bates). We express particular acknowledgment and gratitude to all of the collectors of specimens and tissues used in this study. We thank D. Roldán-Piña for logistical support in laboratory procedures and Alexander Llanes-Quevedo for assistance with demographic analyses. Financial support was obtained from the Dirección General de Asuntos del Personal Académico UNAM (PAPIIT IN 215518), and the Consejo Nacional de Ciencia y Tecnología (CONACyT) grant 152060 (to AGN-S), and through a master’s student scholarship (Posgrado en Ciencias Biológicas, UNAM) and a SNI assistantship grant, both to VJC-C.




