Pruning the Barcode Index Numbers tree: Morphological and genetic evidence clarifies species boundaries in the Eupithecia conterminata complex (Lepidoptera: Geometridae) in Europe
Contributing authors: Marco Infusino ([email protected]), Peter Huemer ([email protected]), Marko Mutanen ([email protected])
Abstract
enThe Barcode Index Numbers (BINs) are operational species units based on patterns of COI divergences that in most cases correspond to species. It has been repeatedly observed that more than one BIN can be found under the same species name particularly when large geographic scales are considered. One such case concerns Eupithecia conterminata, a species widespread in North European countries and restricted to mountainous regions in the rest of the continent, for which five BINs are found in Europe. In order to solve the question concerning the taxonomic status of these BINs and European populations, we employed an integrated approach by combining classical morphological traits (genitalia and wing markings) with those of molecular data, the latter involving both mitochondrial and nuclear genes. This approach allowed us to recognize two valid species in Europe, E. conterminata, currently known only in Fennoscandia, Baltic countries and Russia, and Eupithecia manniaria sp. rev., with distribution covering Central and South European countries. We furthermore synonymized Eupithecia pindosata syn. nov. from Greece with E. manniaria. The European range of these species and their mitochondrial diversity appear to be coherent with biogeographical histories of their foodplants Picea abies and Abies species.
Astratta
itI Barcode Index Number (BINs) sono unità tassonomiche operative basate sui pattern di divergenza del COI che in molti casi corrispondono ad una specie. È stato ripetutamente osservato che più di un BIN può essere trovato sotto uno stesso nome, soprattutto quando vengono considerate ampie scale geografiche. Uno di questi casi riguarda Eupithecia conterminata, diffusa nei paesi dell’Europa settentrionale e limitata alle regioni montuose nel resto del continente, per la quale in Europa sono stati trovati cinque BIN. Per chiarire lo status tassonomico di questi BIN e delle popolazioni europee, in questo lavoro abbiamo usato un approccio integrato che ha combinato caratteri morfologici classici (struttura dei genitali e pattern alare) con dati molecolari, gli ultimi riguardanti sia geni mitocondriali che nucleari. Questo approccio ci ha permesso di confermare la presenza di due specie in Europa, E. conterminata, attualmente conosciuta solo per Fennoscandia, paesi baltici e Russia, e E. manniaria sp. rev. nota per l’Europa centro-meridionale. Inoltre, abbiamo posto in sinonimia E. pindosata syn. nov. della Grecia con E. manniaria. La distribuzione europea di queste specie e la loro diversità mitocondriale sembra essere coerente con la storia biogeografica delle loro piante nutrici, Picea abies e diverse specie di Abies.
1 INTRODUCTION
In the recent years, it has been increasingly difficult to define univocally species boundaries due to the increasing trend of cryptic species discovery and incongruence between DNA and morphology in many cases (Platania et al., 2020). However, this cryptic portion of the biodiversity can provide useful models to understand mechanisms of speciation (Struck et al., 2018). The discoveries of cryptic species have accelerated greatly in well-studied areas along with the increased availability of molecular techniques enabling molecular screening of large numbers of specimens quickly and at a reasonable cost. The most used technique is DNA barcoding, which in animals is based on a relatively short sequence (658 bp) of the mitochondrial cytochrome c oxidase subunit I gene (COI). Its capacity to discriminate between species efficiently is well-documented (e.g. Hebert, Cywinska, et al., 2003; Hebert, Ratnasingham, et al., 2003). Barcode Index Numbers (BINs) are operational species units based on patterns of COI divergences (Ratnasingham & Hebert, 2013). Although good correspondence usually exists between BINs and species (Hausmann et al., 2013; Huemer & Hebert, 2015; Huemer et al., 2019), this technique alone can lead to wrong taxonomic conclusions (Kekkonen et al., 2015). More robust conclusions are obtained through integrating morphology, mitochondrial and nuclear DNA markers (Smith et al., 2008), or using efficient genomics approaches (e.g. Hundsdoerfer et al., 2019; Ivanov et al., 2018).
It has been observed in Lepidoptera that several BINs can frequently be found under the same species name particularly when large geographic scales are considered (Huemer et al., 2014), and when the degree of geographic isolation between the populations is high (Huemer & Karsholt, 2018). In Europe, many of these cases represent arctic-alpine species with isolated populations in different mountain regions and species with populations isolated in southern glacial refugia. Some of such isolated populations were proven to belong to the same taxonomic unit (Huemer et al., 2018), whereas others have been found to represent distinct lineages and are described as new to science (Huemer et al., 2013; Huemer & Hebert, 2011; Infusino et al., 2018; Ronkay & Huemer, 2018; Scalercio et al., 2016). They often likely reflect past glacial refugial history, and their study has contributed to highlight colonization/extinction dynamics of European territory during glaciations. In particular, these dynamics seem to have been produced many endemic but morphologically indistinguishable lineages in the Italian Peninsula (Menchetti et al., 2021).
In this study, we investigated the taxonomy and phylogeography of the European Eupithecia conterminata species complex including the recently described Eupithecia pindosata Weidlich, 2008 (Lepidoptera, Geometridae, Larentiinae) from Greece (Weidlich, 2008). We followed an integrated approach using classical morphological methods and molecular analyses involving both mitochondrial and nuclear genes.
The genus Eupithecia Curtis, 1825 is one of the most diverse and taxonomically difficult groups of Macroheterocera and covers 128 species in Europe (Mironov, 2003). Eupithecia conterminata (Lienig & Zeller, 1846) is an Eurasiatic species that is widespread in North European countries and restricted to mountainous regions in other parts of the continent. It belongs to the irriguata species group that is composed of five species. It is univoltine, flying from mid-March in the South to mid-June in the North of its range. The larva is monophagous, feeding on the young needles of Norwegian spruce (Picea abies (L.) H. Karst, 1881) (Mironov, 2003). Its westernmost range currently covers Central-East Europe (France, Switzerland, Austria, Germany, Italy, Slovenia, Czech Republic, Slovakia, Romania), Baltic countries (Poland, Lithuania, Latvia, Estonia, Russia, Belarus), Fennoscandia (Finland, Norway, Sweden), and Denmark (Karsholt & Nieukerken, 2013). In Italy, it was reported for the first time in a silver fir (Abies alba Mill., 1759) forest of the Central Apennines (Rotundo et al., 1999), but this record remains doubtful as the only collected specimen was not figured in the paper, is not present in the authors' collections, and as the food plant is not present in the area of collection. However, presumably the same taxon was discovered 16 years later in the Sila Massif, Calabria region, South Italy (Infusino & Scalercio, 2015), where some P. abies trees were planted a century ago. The authors postulated then that this population is allochthonous and postponed the clarification of its provenance to future research. One year later, a second and more abundant population was discovered in Calabria on the Serre Mountains by S. Scalercio and M. Infusino (unpublished data), but only A. alba woodlands are present in this area, while P. abies is absent. The discovery of this population prompted us to pose the following questions: Could this population belong to E. pindosata instead of to E. conterminata? Could two populations very isolated from any other populations and discovered only 95 km apart belong to different allopatric sister species? To clarify the taxonomic position of these populations, several specimens were initially barcoded. The results highlighted a situation much more complicated than expected, as the Calabrian populations belong to two clearly distinct BINs. Elsewhere in Europe, three more BINs exist, thus totaling the number of BINs to five, each under the name of E. conterminata.
2 MATERIALS AND METHODS
This study was based on specimens collected in the European countries corresponding to the known range of the species complex, with care taken to involve as many specimens as possible from the most isolated populations. A total of 99 specimens were studied, including some of them in more than one analysis. Precisely, 42 were used in the morphological study of genitalia, 46 in barcoding, and 34 in nuclear marker analyses for a total of 73 specimens (Appendix 1). In addition, 26 specimens were only used to compare wing patterns.
2.1 Genetic analyses
We analyzed the DNA barcode fragment of the mitochondrial DNA (mtDNA) COI gene for 46 specimens of the E. conterminata complex. We complemented this sampling by mining COI sequences of target species from GenBank and the Barcode of Life Data Systems (BOLD) repositories. Additionally, we sequenced seven nuclear markers altogether constituting 4237 bp for 34 specimens, each with barcode data available. The nuclear markers used were carbamoyl phosphate synthase domain protein (CAD), elongation factor 1 alpha (EF1α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), isocitrate dehydrogenase (IDH), cytosolic malate dehydrogenase (MDH), ribosomal protein S5 (RpS5), and wingless (wg). Dried legs and in some cases parts of the body were used for DNA extraction. For barcoding, the prescribed standards using the high-throughput protocol of deWaard et al. (2008) were used. The samples were processed at the Canadian Centre for DNA Barcoding (CCDB, Biodiversity Institute of Ontario, University of Guelph). For the sequencing of nuclear markers, and partially for DNA barcoding, genomic DNA was extracted with DNeasy Blood & Tissue Kit (Qiagen) at the University of Oulu. We mainly followed the protocols of Wahlberg and Wheat (2008) for the nuclear gene sequencing, except that PCR cleanup was carried out with ExoSAP-IT (Affymetrix) and Sephadex columns (Sigma-Aldrich; Table S2).
Sequencing was performed using an ABI 3730 DNA Analyzer (Applied Biosystems) at the University of Oulu. Sequences were manually aligned and edited using BioEdit (Hall, 1999). Nuclear sequence data were stored in the Voseq database (Peña & Malm, 2012), which was also used to generate datasets of the nuclear markers for various analyses and GenBank submissions.
Details of successfully sequenced voucher specimens (Appendix 1), including geographic data and images, can be accessed from BOLD (Ratnasingham & Hebert, 2007) in the public dataset EUPCONTE “E. conterminata species-group” https://doi.org/10.5883/DS-EUPCONTE. Accession numbers for nuclear markers are available in GenBank (BankIt2506175: OK413672–OK413704; BankIt2507032: OK413705–OK413736; BankIt2507117: OK413737–OK413763; BankIt2507178: OK413764–OK413788; BankIt2507192: OK413789–OK413818; BankIt2507197: OK413819–OK413841; BankIt2507201: OK413842–OK413868). Specimens were assigned to a given BIN according to the algorithms implemented in the BOLD Systems v4.
Degrees of intra- and interspecific variation in the DNA barcode fragments were calculated under p-distance model of nucleotide substitution using analytical tools of in BOLD Systems v. 4.0 (http://www.boldsystems.org). To visualize divergence patterns in COI, we constructed a neighbor-joining tree under p-distance model using Mega 10.0.5. For assessing node confidence, we used bootstrapping method (500 replicates). The tree was edited using CorelDraw v.22.0.0.412. We also run a maximum likelihood analysis of COI using IQ-TREE 1.6.1 (http://www.iqtree.org/) (Nguyen et al., 2015). IQ-TREE applies, by default, ModelFinder (Kalyaanamoorthy et al., 2017) to find the best-fit nucleotide substitution model and then reconstructs the tree using the model selected according to Bayesian information criterion (BIC). An ultrafast bootstrap (Hoang et al., 2018) with 1000 replicates to estimate robustness of tree topology was used. As COI analyses were not done to elucidate phylogenetic relationships between E. conterminata and other species of Eupithecia but to visualize genetic divergences, the tree was rooted to the midpoint without an outgroup. A tree of concatenated nuclear markers was constructed under with IQ-TREE 1.6.1 as for COI but partitioning the alignment by genes. The nuclear gene tree was rooted to Eupithecia indigata, a species evidently representing a close relative of E. conterminata (Lee et al., 2018). Inclusion of the outgroup here was to provide a phylogenetic framework within the ingroup specimens (E. conterminata s. l.). Furthermore, we constructed maximum likelihood trees gene by gene to investigate congruence between different nuclear markers. The trees shown in the main text were generated with IQ-TREE 1.6.1 and then edited with FigTree v.1.4.4 and CorelDraw v.22.0.0.412.
The data underlying the work are available partly as Supporting Information and from the above-mentioned public repositories GenBank and BOLD.
2.2 Morphometric analyses
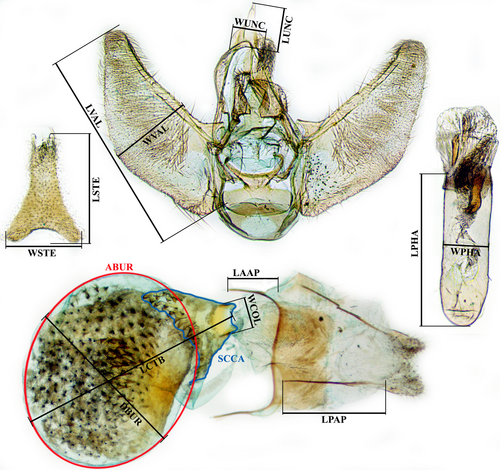
We compared genitalia of specimens belonging to different European populations. Morphometry of genitalia was carried out measuring eight traits for male and seven for female genitalia after mounting them on a microscope slide in Euparal. Optimally, these measurements are carried out before mounting (Yang et al., 2012), but Eupithecia genitalia are fragile and easily damaged during handling. To reduce effects of distortion on the measurements, we examined only traits less susceptible to deformation during slide preparation. In males, we measured length (LPHA) and width (WPHA) of phallus, length (LVAL) and width (WVAL) of valva, length (LUNC) and width (WUNC) of uncus, length (LSTE) and width (WSTE) of sternum A8 (Figure 1). In females, we measured the diameter of corpus bursae (DBUR), width of colliculum (WCOL), length of colliculum and bursa (LCTB), area of bursa (ABUR), area of the sclerotized part of colliculum (SCCA), length of anterior (LAAP) and posterior apophyses (LPAP; Figure 1). Because for some of these measures (LVAL and WVAL in males, DBUR and LCTB in females) it is not possible to provide clear landmarks, we specified that all of these measures were taken following the maximum distance, e.g., measuring the maximum length of valva and, perpendicularly to this, its maximum width in male genitalia, the largest diameter of corpus bursae computed perpendicularly to the maximum distance between the opposite extremes of colliculum and bursa in female genitalia. In total, we dissected 20 males and 22 females. Techniques for the preparation of microscope slides followed Parenti (2000), genitalia were stained in Chlorazol black and mounted in Euparal. Mounted genitalia were photographed with a biological microscope model BM 60 (Exacta + Optech Labcenter spa) equipped with a camera Invenio-10SIII (DeltaPix Co.), measurements and orthogonal lines were obtained using the software DeltaPix InSight. We performed principal component analysis (PCA) to summarize differences in the measurements for both sexes using the correlation matrix and the function Disregard Groups. Statistical analyses were performed using PAST version 2.17c (Hammer et al., 2001). Raw data resulting from morphometric analyses are available in Table S1.
3 RESULTS
3.1 Barcoding analysis
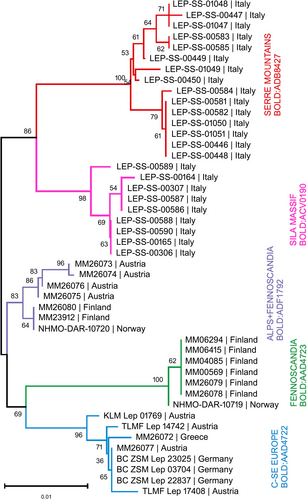
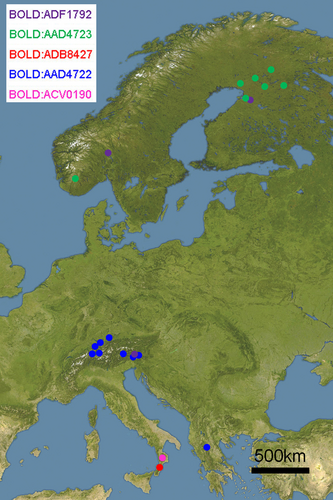
DNA barcoding resulted in a full 658 bp long barcode for 41 specimens, supplemented by five sequences longer than 534 bp. We recognized five BINs in the E. conterminata species complex (Figure 2; Table 1), most of them restricted to distinct geographic areas. Two BINs are endemic to different mountainous areas of Calabria, the Serre Mountains, and the Sila Massif, one includes specimens from Central-East Europe (Austria, Germany, and Greece), one includes Fennoscandian specimens, and one occurs in the Alps and in North Europe (Figure 3). The ML tree confirmed the grouping of specimens, showing a small difference concerning the BIN shared between the Alps and Fennoscandia (Figure S1).
| BIN | Geography | Member count | Average distance | Maximum distance | Distance to nearest neighbor | Nearest neighbor |
|---|---|---|---|---|---|---|
| BOLD:ACV0190 | Sila Massif (Italy) | 9 | 0.23% | 0.64% | 1.89% | BOLD:ADF1792 |
| BOLD:ADB8427 | Serre Mts (Italy) | 15 | 0.58% | 1.12% | 1.96% | BOLD:ADF1792 |
| BOLD:AAD4722 | C-SE Europe | 10 | 0.30% | 0.80% | 1.44% | BOLD:ADF1792 |
| BOLD:ADF1792 | Fennoscandia and Alps | 8 | 0.18% | 0.41% | 1.44% | BOLD:AAD4722 |
| BOLD:AAD4723 | Fennoscandia | 7 | 0.05% | 0.16% | 2.24% | BOLD:ADF1792 |
The BIN grouping exclusively Fennoscandian specimens (BOLD:AAD4723) includes only two haplotypes (n = 7), suggesting low regional genetic diversity of the COI gene. By comparison, other BINs were much more diversified: the Central-East European BIN (BOLD:AAD4722) included six haplotypes (n = 8) one of which belongs to E. pindosata from Greece; the Sila Massif BIN (BOLD:ACV0190) included four haplotypes (n = 9); the Serre Mountains BIN (BOLD:ADB8427) included nine haplotypes (n = 15); and the BIN shared by the Alps and Fennoscandia (BOLD:ADF1792) included five haplotypes (n = 7).
Minimum distance (p-distance) between BINs ranged from 1.44 to 2.24% (Table 1). The most differentiated BIN was found in Fennoscandia only. In addition, BINs endemic to southern Italy are clearly differentiated despite being only 95 km apart. The highest intra-BIN maximum distance was observed in the Serre Mountains (1.12%).
3.2 Nuclear markers
Sequencing of the nuclear markers was overall successful. Of the 34 included specimens, sequence data were recovered for 34 specimens for CAD, 32 specimens for EF-1α, 28 specimens for GAPDH, 26 specimens for IDH, 31 specimens for MDH, 24 specimens for RpS5, and 27 specimens for wg (Alignments S1–S7). For five specimens, less than four nuclear markers were successfully sequenced; however, a minimum of two nuclear sequences were obtained for all individuals.
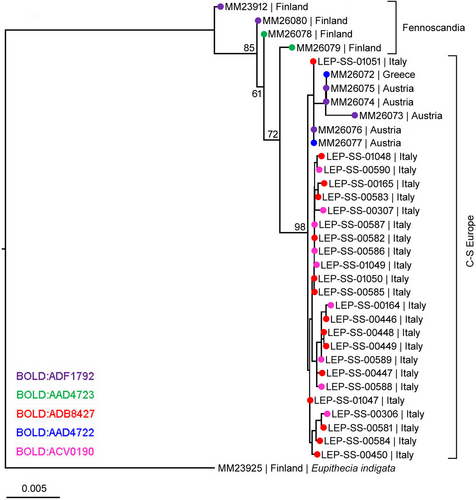
The maximum likelihood tree based on the seven nuclear genes revealed that the Fennoscandian specimens differed genetically from the Central and South European specimens (Figure 4, Alignment S8). The latter group, with E. pindosata included, formed a clade with strong support (BS = 98). The Fennoscandian specimens, in turn, showed greater genetic variability and formed a non-monophyletic grade. An examination of individual gene trees revealed that out of seven examined markers (and eight separate regions), the Finnish specimens formed a distinct and monophyletic cluster in four (CAD, EF-1α part 2, GAPDH, and IDH) (see Figures S2–S5). In addition, some less consistent indications for genetic differentiation of the Finnish specimens were observed in the three additional markers (MDH, RpS5 and wg). For example, in RpS5, the non-Fennoscandian specimens formed a monophylum while the Fennoscandian specimens formed a near-monophyletic grade.
3.3 Morphometric analysis
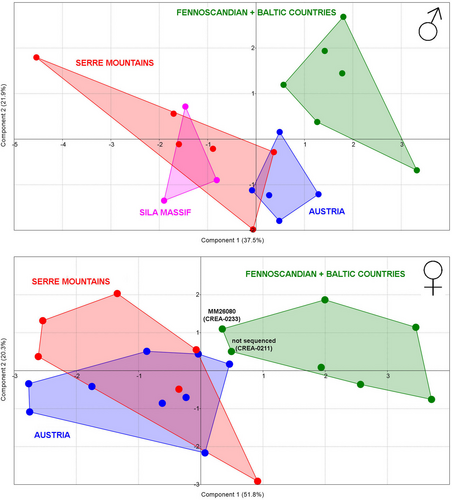
Morphometry of genitalia indicated two distinct groups for both sexes, the first including Fennoscandian and Baltic specimens and the second including specimens from the rest of Europe.
Fennoscandian and Baltic male genitalia are clearly separated from Alpine and Italian ones (Figure 5) by their different dimensions, as the highest loadings on the component 1 were composed by the length of phallus (LPHA) and the length of valva (LVAL; Table 2).
| Component 1 | Component 2 | Component 3 | |
|---|---|---|---|
| Eigenvalue | 3.000 | 1.752 | 1.180 |
| Cumulative eigenvalue | 3.000 | 4.752 | 5.932 |
| % variance | 37.504 | 21.905 | 14.756 |
| Cumulative % variance | 37.504 | 59.409 | 74.165 |
| Component loadings | |||
| LPHA | 0.371 | 0.394 | −0.172 |
| WPHA | 0.221 | 0.440 | 0.045 |
| LVAL | 0.494 | 0.122 | 0.257 |
| WVAL | 0.255 | −0.007 | 0.752 |
| LUNC | 0.373 | −0.216 | −0.526 |
| WUNC | 0.432 | −0.325 | −0.123 |
| LSTE | 0.034 | 0.650 | −0.210 |
| WSTE | −0.422 | 0.246 | 0.019 |
Note
- Length (LPHA) and width (WPHA) of phallus, length (LVAL) and width (WVAL) of valva, length (LUNC) and width (WUNC) of uncus, length (LSTE) and width (WSTE) of sternum A8.
Female genitalia of specimens collected in Austria and in Italy are intermixed on the Cartesian plan formed by the first two axis of the PCA, whereas the Fennoscandian and Baltic specimens grouped separately (Figure 5). The higher loadings on the component 1 are shown by features correlated with the dimensions of genitalia (ABUR, DBUR, and LCTB), meaning that dimensions of the northern specimens' female genitalia are significantly different to those from Austria and Italy (Table 3). However, the two smallest Fennoscandian female genitalia, corresponding to slides CREA-0233 and CREA-0211, are very close to Austrian and Italian females. Interestingly, one of the above-mentioned Fennoscandian females has a DNA barcode included in the BIN shared between Fennoscandia and Alps.
| Component 1 | Component 2 | Component 3 | |
|---|---|---|---|
| Eigenvalue | 3.625 | 1.422 | 0.858 |
| Cumulative eigenvalue | 3.625 | 5.047 | 5.905 |
| % variance | 51.780 | 20.317 | 12.258 |
| Cumulative % variance | 51.780 | 72.097 | 84.355 |
| Component loadings | |||
| ABUR | 0.503 | −0.021 | 0.176 |
| DBUR | 0.501 | −0.111 | −0.003 |
| LCTB | 0.483 | −0.033 | 0.299 |
| SCCA | 0.419 | 0.262 | −0.130 |
| WCOL | 0.150 | 0.594 | −0.733 |
| LPAP | 0.014 | 0.643 | 0.444 |
| LAAP | −0.255 | 0.460 | 0.343 |
- Abbreviations: ABUR, area of bursa; DBUR, diameter of corpus bursael; LAAP, length of anterior; LCTB, length of colliculum and bursa; LPAP, posterior apophyses; SCCA, area of the sclerotized part of colliculum; WCOL, width of colliculum.
3.4 Taxonomy of the Eupithecia conterminata complex in Europe
Morphometric and genetic evidence supports the co-presence of two distinct species, one in the Fennoscandian and Baltic countries, another covering Central and South European countries. As a consequence, we raise Eupithecia manniaria Herrich-Schäffer, 1848 sp. rev., type locality in Bohemia, from synonymy to bona species and establish E. pindosata Weidlich, 2008 syn. nov., described from Greece, as a junior synonym of E. manniaria Herrich-Schäffer, 1848.
Available descriptions of E. conterminata and E. manniaria suggest that the latter is only a variety of the former. Here, we re-describe both species in order to establish diagnostic features.
3.5 Eupithecia conterminata (Lienig & Zeller, 1846)
Larentia conterminata Lienig & Zeller, 1846: Isis 3: 197. Type locality: Latvia? Koknese (=Kokenhusen). Type: Natural History Museum, London, UK? [not traced]
- Finland – 5♀♀, 5♂♂: Revonkylä, 2.VI.1971, 1♀, A. Pyörnilä leg. (dissected, slide: CREA-0211); Pudasjärvi, 9-10.VI.2000, 1♂, M. Mutanen leg. (dissected, slide: CREA-0212); Rauma, 7.VI.1983, 1♀, J. Itämies leg. (dissected, slide: CREA-0213); Rovaniemi, 5.VI.1995, 1♂, M. Mutanen leg. (dissected, slide: CREA-0220); idem, 1♂ (dissected, slide: CREA-0222); idem, 1♂ (dissected, slide: CREA-0223); Kuusamo, 8.VI.2011, 1♀, M. Mutanen leg. (dissected, slide: CREA-0215); idem, 11.VI.1970, 1♂, J. Kyrki leg. (dissected, slide: CREA-0221); Kulju, 6.VI.1997, 1♀, A. Kallio leg. (dissected, slide: CREA-0216); Hollola, 22.V.1981, 1♀ (dissected, slide: CREA-0217) (Coll. Zoological Museum, University of Oulu)
- Norway – 1♀, 1♂: 1♂, Mo i Rana, 24.VI.1944, I. Feichtenberger leg. (dissected, slide: CREA-0159); 1♀, idem. (Coll. Bavarian State Collection of Zoology).
- Estonia – 1♀, 1♂: Dorpat, 24.VI.1928, 1♀, W. Petersen leg. (dissected, slide: CREA-0160); idem, 22.V.1926, 1♂ (Coll. Bavarian State Collection of Zoology).
3.5.1 Description
External characters (Figures 6a–e and 7a–e): Wingspan male 15–16 mm (n = 7), wingspan female 15–17 mm (n = 7). Forewings smoky gray, terminal area sometimes slightly darker. Transverse lines inconspicuous, slightly darker than ground color or absent, forming dark gray coastal spots. Medial and post-medial lines sometimes more distinct, sinuate; medial line curved around an ovate, medium-sized, black or dark gray discal spot. Hindwings concolor with forewings or slightly paler, with a small, sometimes inconspicuous, discal spot; transverse lines inconspicuous or absent. Fringe concolorous with wings, sometimes checkered light gray and gray. Underside of wings as upper side, but paler, with medial line often well-defined on the hindwings. Labial palpi gray, the rest of the head covered with grayish white scales. Females appear slightly lighter in color, and the wing shape appears a bit rounder.


Male genitalia (Figure 8a,b): Uncus short and narrow. Valva without ventral process, larger medially, with a curved ventral margin and rounded apex. Phallus about 3–4 times longer than medial width, bearing three dentate cornuti, of which one flat V-shaped at ductus ejaculatorius base and two smaller on the vesica covered by numerous very small spinules. Sternum A8 rectangular, with base broader than apex and with basal and apical hollows shallow, with two apical lobes.
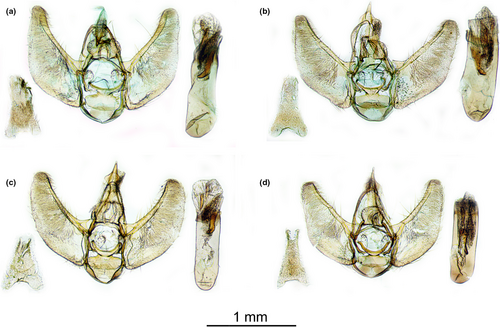
Female genitalia (Figure 9a,b): Bursa copulatrix pear-shaped, with a large area covered by spines, usually longer on the margin. Ductus bursae sclerotized, with two patches of spines at the margin of corpus bursae, where the narrow ductus seminalis is attached. Colliculum short, collar-like. Antrum long about half of the corpus bursae. Apophyses thin, posterior about twice longer than anterior ones. Tergum A8 rectangular. Papillae anales small and rounded.

Distribution: An Eurasiatic species ranging from northeastern Europe (Poland, Lithuania, Latvia, Estonia, Russia, Belarus), Fennoscandia (Finland, Norway, Sweden), and Denmark eastward to Ussuri region and Japan.
3.6 Eupithecia manniaria Herrich-Schäffer, 1848 sp. rev.
Eupithecia manniaria Herrich-Schäffer, 1848: Syst. Bearb. Schmett. Eur. 3 (32): 122–136. Type locality: Czech Republic, Bohemia. Type: lost.
- Austria – 11♀♀, 5♂♂: St. Johann im Walde, Steinbruch, 750m, 03.V.1998, 1♀, H. Deutsch leg. (dissected, slide: CREA-0191); Lengberg, 850m, 26.IV.2003, 1♀, H. Deutsch leg. (dissected, slide: CREA-0193); idem, 800m, 22.IV.2004, 1♀ (dissected, slide: CREA-0197); idem, 16.V.1985, 1♂ (dissected, slide: CREA-0192); idem, 1♀ (dissected, slide: CREA-0195); idem, 13.V.1986, 1♂ (dissected, slide: CREA-0198); idem, 1♀ (dissected, slide: CREA-0200); idem,7.V.1995, 1♂ (dissected, slide: CREA-0196); idem, 2.V.1990, 1♀ (dissected, slide: CREA-0199); idem, 26.IV.1992, 1♂, H. Mair leg. (dissected, slide: CREA-0194); Arriach, 1250 m, 30.IV.2013, 1♀ (barcoded, BOLD ID: MM26077); Irschen, 901m, 29.IV.2014, 1♂, C. Wieser leg. (barcoded, BOLD ID: KLM Lep 01769); Wimitz, 760m, 1♀, C. Wieser (dissected, slide: CREA-0229; barcoded, BOLD ID: MM26073); idem, 1♀ (dissected, slide: CREA-0230; barcoded, BOLD ID: MM26074); idem, 1♀ (dissected, slide: CREA-0231; barcoded, BOLD ID: MM26075); idem, 1♀ (barcoded, BOLD ID: MM26076); idem, 1♂ idem (dissected, slide: CREA-0228) (Colls. Tiroler Landesmuseum Ferdinandeum, Landesmuseum Kärnten).
- Italy – 49♀♀, 19♂♂: Santa Maria, Serra San Bruno, 925m, 6.IV.2016, 24♀♀, 2♂♂, S. Scalercio & M. Infusino leg.; idem, 1♀ (dissected, slide: CREA-0169); idem, 1♀ (dissected, slide: CREA-0168); idem, 1♀ (dissected, slide: CREA-0171); idem, 1♀ (dissected, slide: CREA-0163; barcoded, BOLD ID: LEP-SS-00446); idem, 1♀ (dissected, slide: CREA-0167; barcoded, BOLD ID: LEP-SS-00447); idem, 1♀ (barcoded, BOLD ID: LEP-SS-00583); idem, 1♂ (dissected, slide: CREA-0165); idem, 1♂ (dissected, slide: CREA-0172); idem, 1♂(barcoded, BOLD ID: LEP-SS-00584); idem, 883m, 6.IV.2016, 8♀♀, 1♂; idem, 1♂ (dissected, slide: CREA-0170); idem, 1♂ (dissected, slide: CREA-0161; barcoded, BOLD ID: LEP-SS-00449); idem, 860m, 2♀♀; idem, 1♀ (barcoded, BOLD ID: LEP-SS-00450); idem, 1♀ (barcoded, BOLD ID: LEP-SS-00448); idem, 1♀ (barcoded, BOLD ID: LEP-SS-00585); idem, 1♀ (barcoded, BOLD ID: LEP-SS-00581); idem, 1♀ (barcoded, BOLD ID: LEP-SS-00582); Il Palmento, Serra San Bruno, 840m, 6.IV.2016, 1♀, S. Scalercio & M. Infusino leg.; idem, 830m, 1♀; Rosarella, Serra San Bruno, 917m, 6.IV.2016, 1♀, S. Scalercio & M. Infusino leg.; idem, 1♀(dissected, slide: CREA-0158); idem, 1♂ (dissected, slide: CREA-0166); idem, 1♂ (dissected, slide: CREA-0157); Vivaio Sbanditi, Parco Nazionale della Sila, 1355 m, 1♂, S. Scalercio & M. Infusino leg. (dissected, slide: CREA-0162; barcoded, BOLD ID: LEP-SS-00164); idem, 1♂ (dissected, slide: CREA-0032; barcoded, BOLD ID: LEP-SS-00165); idem, 1350 m, 11.V.2015, 1♂ (barcoded, BOLD ID: LEP-SS-00307); idem, 4.V.2015, 1♂ (dissected, slide: CREA-0173; barcoded, BOLD ID: LEP-SS-00306); idem, 12.V.2017, 1♂, S. Scalercio, M. Infusino & A. Ienco leg. (barcoded, BOLD ID: LEP-SS-00590); idem, 1♂ (barcoded, BOLD ID: LEP-SS-00589); idem, 1♂ (barcoded, BOLD ID: LEP-SS-00588); idem, 3.V.2017, 1♂, S. Scalercio & S. Greco leg. (barcoded, BOLD ID: LEP-SS-00586); idem, 1♂ (barcoded, BOLD ID: LEP-SS-00587). (Coll. Research Centre for Forestry and Wood, Rende).
- Greece – 1ex: Ntoia Pindos, Dhokimi, Tria Potamia, 0.5 km NW FL 27, 890m (barcoded, BOLD ID: MM26072) (Coll. University of Oulu).
3.6.1 Description
External characters (Figures 6f–j and 7f–j): wingspan male 15–16 mm (n = 23), wingspan female 14–15 mm (n = 55). Forewings smoky gray, terminal area dark gray. Transverse lines slightly darker than ground color, sometimes inconspicuous or absent, forming dark gray costal spots. Medial and post-medial lines sometimes more distinct, sinuate, usually more evident in females; medial line curved around an ovate, large, black or dark gray discal spot. Hindwings paler, sometimes whitish gray, with a small discal spot; transverse lines inconspicuous or absent, more distinct in females; terminal area sometimes darker, especially toward termen. Fringe concolorous with wings, sometimes checkered light gray and gray. Underside of wings as upper side, but paler, with transversal lines often well-defined. Labial palpi gray, the rest of the head covered with grayish white scales.
Male genitalia (Figure 8c,d): Uncus short and narrow. Valva without ventral process, larger medially, with a curved ventral margin and rounded apex. Phallus about 3–4 times longer than medial width, bearing three dentate cornuti, of which one flat V-shaped at ductus ejaculatorius base and two smaller on the vesica covered by numerous very small spinules. Sternum A8 rectangular, with base broader than apex and with basal and apical hollows shallow, with two apical lobes.
Female genitalia (Figure 9c,d): Bursa copulatrix pear-shaped, with a large area covered by spines, usually longer on the margin. Ductus bursae sclerotized, with two patches of spines at the margin of corpus bursae, where the small ductus seminalis is attached. Colliculum short, collar-like. Antrum very short. Apophyses thin, posterior about twice longer than anterior ones. Tergum A8 rectangular. Papillae anales were small and rounded.
Distribution: European species with confirmed records from Czech Republic, Austria, Germany, Italy, and Greece. Likely present also in France, Switzerland, Slovenia, Slovakia, Poland, and Romania.
Diagnosis. Hindwings are concolorous or slightly paler than forewings in E. conterminata, whereas in E. manniaria they are markedly paler, sometimes whitish gray. Wing pattern is more contrasted in E. manniaria, especially in females, with terminal area of forewings always darker, and the wing shape appears a bit rounder. Discal spot of forewing is larger in E. manniaria. Male and female genitalia are smaller in E. manniaria (Table 4). Antrum is longer in E. conterminata than in E. manniaria.
| Male measurements | conterminata | manniaria | p(same) |
|---|---|---|---|
| N = 6 | N = 14 | ||
| LPHA | 1.07 ± 0.09 | 0.85 ± 0.07 | 0.0002 |
| WPHA | 0.31 ± 0.03 | 0.27 ± 0.02 | 0.0023 |
| LVAL | 1.13 ± 0.06 | 1.03 ± 0.05 | 0.0027 |
| WVAL | 0.45 ± 0.05 | 0.42 ± 0.04 | 0.1927 |
| LUNC | 0.24 ± 0.04 | 0.22 ± 0.03 | 0.5904 |
| WUNC | 0.19 ± 0.02 | 0.18 ± 0.03 | 0.4625 |
| LSTE | 0.63 ± 0.06 | 0.57 ± 0.04 | 0.0446 |
| WSTE | 0.35 ± 0.06 | 0.38 ± 0.03 | 0.1176 |
| Female measurements | N = 5 | N = 17 | |
|---|---|---|---|
| DBUR | 1.21 ± 0.12 | 0.90 ± 0.13 | 0.0003 |
| WCOL | 0.20 ± 0.02 | 0.19 ± 0.03 | 0.2052 |
| LCTB | 1.55 ± 0.14 | 1.21 ± 0.15 | 0.0005 |
| LAAP | 0.24 ± 0.02 | 0.26 ± 0.05 | 0.3219 |
| LPAP | 0.58 ± 0.08 | 0.56 ± 0.05 | 0.4487 |
| ABUR (mm2) | 1.24 ± 0.21 | 0.76 ± 0.17 | 0.0002 |
| SCCA (mm2) | 0.19 ± 0.03 | 0.11 ± 0.02 | 0.0002 |
Note
- Male genitalia—LPHA, length of phallus; LSTE, length of sternum A8; LUNC, length of uncus; LVAL, length of valva; WPHA, width of phallus; WSTE, width of sternum A8; WUNC, width of uncus; WVAL, width of valva. Female genitalia—ABUR, area of bursa; DBUR, diameter of corpus bursae; LAAP, length of anterior apophyses; LCTB, length of colliculum and bursa; LPAP, length of posterior apophyses; SCCA, area of the sclerotized part of colliculum; WCOL, width of colliculum. Significant differences between species of genitalia measurements tested with a Tukey test.
In the barcode sequences, we found seven diagnostic substitutions between species at nucleotide positions 103, 187, 385, 494, and 508, which in E. conterminata are cytosine (C) instead of thymine (T), and at positions 316 and 493 which E. conterminata are guanine (G) instead of adenine (A).
4 DISCUSSION
Using an integrative approach of genetic and morphological evidence, we demonstrated the presence of two sister species in the E. conterminata species complex in Europe. Eupithecia conterminata (Lienig & Zeller, 1846) is distributed from Fennoscandia and the Baltic countries to Japan, while the reestablished E. manniaria Herrich-Schäffer, 1848 is found in Central and South European countries. We found E. pindosata Weidlich, 2008 to be its junior synonym.
Several lines of evidence led us to separate E. conterminata from E. manniaria. These include constant differences in the wing pattern, the markings being more contrasted in E. manniaria, especially in the females, and with a discal spot of forewing being larger in E. manniaria. The two species can also be separated by characters in male and female genitalia, those being overall smaller in E. manniaria, and with the antrum being longer in E. conterminata. The validity of the two as separate species was further confirmed by nuclear markers, which showed slight differences between E. conterminata and E. manniaria but not between the latter and E. pindosata. These two taxa also share the same BIN and have the same wing pattern, which led us to conclude that E. pindosata is a junior synonym of E. manniaria.
It has recently been stressed that the recognition of taxa as diverging lineages or distinct, possibly paraphyletic species, mostly depends on the criteria adopted by different species concepts (Platania et al., 2020), which is due to the gradual nature of speciation process and the consequent difficulties of delimiting species in an unambiguous way. However, in our case we found congruent and diagnostic traits in wing pattern, genitalia, DNA barcodes, and nuclear genes. The similarity of species suggest either a relatively slow evolution rate or recent origin of the species.
Visual comparisons of genitalia shapes showed the presence of diagnostic features only in females, in which the antrum is relatively longer in E. conterminata. The fragility of genitalia and their possible distortions during slide preparations suggested us to exclude the use of geometric morphometric analysis that could have helped to recognize finer morphological differences (Mutanen & Pretorius, 2007). However, despite finding only one discriminating trait in the shape of genitalia, we found that the dimensions of genitalia are significantly different in both males and females, with those of E. conterminata being bigger than those of E. manniaria. It has been largely demonstrated in many invertebrate taxa that dimensions of genitalia are quite constant within species and typically show strong negative allometric relationships with the body size (Bernstein & Bernstein, 2002; Eberhard et al., 1998; Mutanen & Kaitala, 2006 and literature therein). In our case, we found small differences in wingspan between females (range in E. conterminata 15–17 mm, in E. manniaria 14–15 mm), while males have the same range (15–16 mm), suggesting significant differences in the dimensions of male and female genitalia being also of diagnostic value.
In mitochondrial DNA, we found five BINs in the European populations of the E. conterminata species complex, one exclusive for E. conterminata, three exclusive for E. manniaria, and one shared between them. The shared BIN suggests introgression, which likely has occurred in the recent past from E. manniaria to E. conterminata as suggested by the presence of only one haplotype of the shared BIN in northern countries. Introgression of mitochondrial DNA between well-diverged species is recognized as infrequent but not extremely rare in Lepidoptera and is regarded as the most reasonable explanation for observed mtDNA paraphyly (Mutanen et al., 2016; Zakharov et al., 2009). It is expected to be less rare between recently diverged species and has probably taken place in the couplet of E. manniaria–E. conterminata as well. However, in order to hypothesize when and where introgression occurred, larger genetic or genomic sampling covering a higher number of populations and individuals would be needed.
One E. manniaria BIN is widespread and covers Central and South–East Europe, whereas the two Italian BINs are endemic to two different refugia of the Calabria region. The presence of several distinct BINs is probably linked to the biogeographic history of the food plants and hence the prolonged isolation of populations of E. manniaria. The BIN unique to E. conterminata comprises only two haplotypes, each showing low genetic diversity.
The seven nuclear genes examined revealed strong mitonuclear discordance in both species. The variability of nuclear markers was rather low when compared with the presumably close E. indigata, which shows a remarkable genetic gap to the conterminata/manniaria couplet. The nuclear markers support our taxonomic conclusions by demonstrating the genetic integrity of mitogenetically variable E. manniaria, which in the nuclear tree forms a monophyletic and relatively compact clade. The E. pindosata type specimen is deeply nested within this clade and does not show genetic separation from several specimens of E. manniaria originating from Austria. In fact, together with mtDNA this result suggests that the Greek and Austrian populations of E. manniaria may have originated from a shared glacial refugium. Interestingly, the four Fennoscandian specimens of E. conterminata showed more genetic variability than E. manniaria and are not recovered as a monophylum in the ML analysis. We hypothesize that the North European E. conterminata originally had only a single main mitochondrial genotype and the second resulted from introgression from E. manniaria, although this could have happened in the opposite direction as well.
The distribution of E. manniaria in Central and South Europe perfectly matches that of A. alba (Konnert & Bergmann, 1995) that likely is its main food plant. As hypothesized by Weidlich (2008), another food plant could be Abies borisii-regis Mattf., the most common Abies species present in the locus typicus of E. pindosata. Available records concerning the feeding behavior of E. conterminata on P. abies are gathered from Finland (Seppänen, 1970; Weigt, 1993) and Russia (Draut, 1900), where only E. conterminata is confirmed to be present (Figure 10).
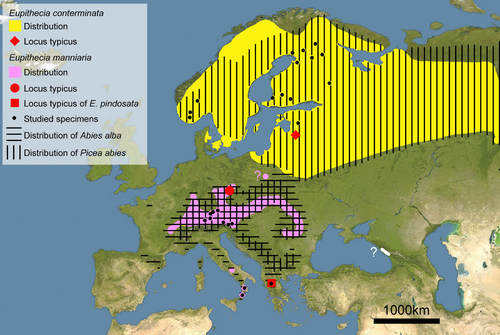
According to the available material, these species may be parapatric. They could possibly co-occur in Poland where E. conterminata has been recorded from the northeastern P. abies forests and from the south where A. alba is also present (Buszko & Nowacki, 2000). However, the taxonomic identity of the southern records must be ascertained.
The presence of E. manniaria only in Central and South European countries can be explained by the presence of its putative foodplant. However, the absence of E. conterminata in these areas is likely explained biogeographically as the range of P. abies overlaps with that of A. alba in several areas of Central and Eastern Europe. We hypothesize that glacial–interglacial cycles promoted isolated populations of the common ancestor of these species in southern glacial refugia to shift from P. abies to Abies species. This shift likely occurred during the bottleneck periods of P. abies during the Quaternary that resulted in the low genetic variability in European tree populations (Heuertz et al., 2006). The availability of Abies as an alternative and more abundant trophic resource allowed the southern population of the common ancestor to survive over unfavorable climatic periods. The higher tolerance of A. alba than P. abies to interglacial climate changes may further have promoted this shift.
To evaluate divergence time at this level and with the very erratic marker as mtDNA is difficult, but a rough estimation can be made. The divergence of 1.89% as found between Calabria populations (BOLD:ACV0190) can be dated as 1.89/1.5 (%/My) = 1.26 My following Quek et al. (2004) and 1.89/2.3 (%/My) = 0.82 My following Brower (1994). This is very interesting because these lineages are close to each other with no apparent barriers, and as this pattern echoes divergence time estimates of other species occurring in Calabria and diversified among mountain massifs, including Parnassius mnemosyne, Anthocharis damone, and Aglais urticae (Scalercio et al., 2020).
We further hypothesize that during glacial periods relict populations of E. conterminata survived only in the area of Moscow, recognized as the refuge area of the Baltic-Nordic genetic domain of the Norway spruce (Heuertz et al., 2006; Lagercrantz & Ryman, 1990). On the contrary, the silver fir survived in five refugia during the last glaciation, namely Central France, Balkan Peninsula, Central Italy, Pyrenees, and Calabria (Konnert & Bergmann, 1995), resulting in fragmentation of the E. manniaria range. During postglacial periods A. alba expanded to its current range only from the first three refugia, leaving the Pyrenean and Calabrian populations isolated. This scenario would explain the high COI diversity observed in E. manniaria, with the separation of BINs being further reinforced by the current fragmentation of its food plant's range in many southern mountain areas. Biogeographic isolation of the silver fir might also explain the great COI differentiation of Calabrian E. manniaria populations, likely the oldest populations of this species. In general, the fact that northern BINs are characterized by a lower genetic diversity is worth mentioning with regard to the well-known hypothesis of “southern richness and northern purity” (e.g., Hewitt, 1996), also recovered for European butterflies in a recent study (Dincă et al., 2021). Furthermore, our data are in line with repeated observations from recent years demonstrating that the taxonomic richness of southern areas is correlated with high genetic diversity (de Kort et al., 2021; Miraldo et al., 2016; Pelletier & Carstens, 2018; Yiming et al., 2021).
5 CONCLUSIONS
The distinctness of two cryptic species in the E. conterminata species complex remained undiscovered for a long time, until the recent discovery of relict populations in southern Europe where the only known food plant, P. abies, is absent. The discovery of a population in Greece and another in the southern Apennines, stimulated the present study, as only Abies species are present in these areas and this plant is not recognized as a food plant for E. conterminata. The integrated use of molecular and classical taxonomic methods provided concordant evidence for the presence of two distinct species and helped to depict the main traits of their evolutionary history, revealing an interesting adaptive history of plants and associated herbivores. Further studies are needed to refine the distributions of both species, especially in the eastern range of E. conterminata. The taxonomic identity of the population known from the West of Great Caucasus Mountains would deserve special attention as both A. alba and P. abies are absent in that area.
ACKNOWLEDGEMENTS
We thank Axel Hausmann (Bavarian State Collection of Zoology, Germany), Christian Wieser (Landesmuseum Kärnten, Austria) for the loan of specimens, Silvia Greco and Annamaria Ienco (Italy) for the help in the field collection of Italian specimens, and Tamara Hiltunen (Finland) for the English editing. This study was partly supported by the project “Progetto di monitoraggio dei Lepidotteri Notturni attraverso l'utilizzo di trappole luminose tipo Rothamsted” funded by the Sila National Park, and by the Project “ALForLab” (PON03PE_00024_1) co-funded by the National Operational Programme for Research and Competitiveness (PON R&C) 2007-2013, through the European Regional Development Fund (ERDF) and national resource (Revolving Fund—Cohesion Action Plan [CAP] MIUR). Furthermore, funding of the Promotion of Educational Policies, University and Research Department of the Autonomous Province of Bolzano—South Tyrol to the project “Genetische Artabgrenzung ausgewählter arktoalpiner und boreomontaner Tiere Südtirols” are acknowledged. Paul D.N. Hebert and the entire team at the Canadian Centre for DNA Barcoding (CCDB, Guelph, Canada) helped carrying out important parts of sequence analyses. The sequencing of nuclear markers was kindly conducted by Hannele Parkkinen and funded by the Academy of Finland through grant #314702 to Marko Mutanen. We thank Marko Prous for help in conducting IQ-TREE analyses. We thank also the five reviewers that strongly improved the manuscript, especially Leonardo Dapporto for several constructive suggestions. Finally, we thank the Sila National Park and the Serre Natural Regional Park that released us the permits for collecting.
APPENDIX 1: Specimens subjected to genetic and morphometric analyses
| Country | Geographic area | Sex | BOLD Sample ID | BOLD sequence ID | BIN | nDNA sequence data | Slide no. |
|---|---|---|---|---|---|---|---|
| Italy | Serre Mountains | f | LEP-SS-00446 | BIBSA1801-16 | BOLD:ADB8427 | Yes | CREA-0163 |
| Italy | Serre Mountains | f | LEP-SS-00447 | BIBSA1802-16 | BOLD:ADB8427 | Yes | CREA-0167 |
| Italy | Serre Mountains | f | LEP-SS-00448 | BIBSA1803-16 | BOLD:ADB8427 | Yes | N/A |
| Italy | Serre Mountains | m | LEP-SS-00449 | BIBSA1804-16 | BOLD:ADB8427 | Yes | CREA-0161 |
| Italy | Serre Mountains | f | LEP-SS-00450 | BIBSA1805-16 | BOLD:ADB8427 | Yes | N/A |
| Italy | Serre Mountains | f | LEP-SS-00581 | BCLEP130-17 | BOLD:ADB8427 | Yes | N/A |
| Italy | Serre Mountains | f | LEP-SS-00582 | BCLEP131-17 | BOLD:ADB8427 | Yes | N/A |
| Italy | Serre Mountains | f | LEP-SS-00583 | BCLEP132-17 | BOLD:ADB8427 | Yes | N/A |
| Italy | Serre Mountains | m | LEP-SS-00584 | BCLEP133-17 | BOLD:ADB8427 | Yes | N/A |
| Italy | Serre Mountains | f | LEP-SS-00585 | BCLEP134-17 | BOLD:ADB8427 | Yes | N/A |
| Italy | Serre Mountains | m | LEP-SS-01047 | BCLEP476-20 | BOLD:ADB8427 | Yes | CREA-0157 |
| Italy | Serre Mountains | f | LEP-SS-01048 | BCLEP477-20 | BOLD:ADB8427 | Yes | CREA-0158 |
| Italy | Serre Mountains | m | LEP-SS-01049 | BCLEP478-20 | BOLD:ADB8427 | Yes | CREA-0165 |
| Italy | Serre Mountains | f | LEP-SS-01050 | BCLEP479-20 | BOLD:ADB8427 | Yes | CREA-0169 |
| Italy | Serre Mountains | m | LEP-SS-01051 | BCLEP480-20 | BOLD:ADB8427 | Yes | CREA-0170 |
| Italy | Serre Mountains | m | N/A | N/A | N/A | No | CREA-0166 |
| Italy | Serre Mountains | f | N/A | N/A | N/A | No | CREA-0168 |
| Italy | Serre Mountains | f | N/A | N/A | N/A | No | CREA-0171 |
| Italy | Serre Mountains | m | N/A | N/A | N/A | No | CREA-0172 |
| Italy | Sila Massif | m | LEP-SS-00164 | BIBSA544-15 | BOLD:ACV0190 | Yes | CREA-0162 |
| Italy | Sila Massif | m | LEP-SS-00165 | BIBSA545-15 | BOLD:ACV0190 | Yes | CREA-0032 |
| Italy | Sila Massif | m | LEP-SS-00306 | BIBSA796-15 | BOLD:ACV0190 | Yes | CREA-0173 |
| Italy | Sila Massif | m | LEP-SS-00307 | BIBSA797-15 | BOLD:ACV0190 | Yes | N/A |
| Italy | Sila Massif | m | LEP-SS-00586 | BCLEP135-17 | BOLD:ACV0190 | Yes | N/A |
| Italy | Sila Massif | m | LEP-SS-00587 | BCLEP136-17 | BOLD:ACV0190 | Yes | N/A |
| Italy | Sila Massif | m | LEP-SS-00588 | BCLEP137-17 | BOLD:ACV0190 | Yes | N/A |
| Italy | Sila Massif | m | LEP-SS-00589 | BCLEP138-17 | BOLD:ACV0190 | Yes | N/A |
| Italy | Sila Massif | m | LEP-SS-00590 | BCLEP139-17 | BOLD:ACV0190 | Yes | N/A |
| Greece | N/A | MM26072 | BCLEP481-20 | BOLD:AAD4722 | Yes | N/A | |
| Germany | Bavaria | f | BC ZSM Lep 03704 | GWORH038-09 | BOLD:AAD4722 | No | N/A |
| Germany | Bavaria | m | BC ZSM Lep 23025 | FBLMS214-09 | BOLD:AAD4722 | No | N/A |
| Germany | Bavaria | f | BC ZSM Lep 22837 | GWORL555-09 | BOLD:AAD4722 | No | N/A |
| Austria | Nordtirol | N/A | TLMF Lep 17408 | LEATI023-15 | BOLD:AAD4722 | No | N/A |
| Austria | Kärnten | N/A | KLM Lep 01769 | ABOLA249-14 | BOLD:AAD4722 | No | N/A |
| Austria | Kärnten | f | MM26077 | BCLEP486-20 | BOLD:AAD4722 | Yes | N/A |
| Austria | Kärnten | f | MM26073 | BCLEP482-20 | BOLD:ADF1792 | Yes | CREA-0229 |
| Austria | Kärnten | f | MM26074 | BCLEP483-20 | BOLD:ADF1792 | Yes | CREA-0230 |
| Austria | Kärnten | f | MM26075 | BCLEP484-20 | BOLD:ADF1792 | Yes | CREA-0231 |
| Austria | Kärnten | f | MM26076 | BCLEP485-20 | BOLD:ADF1792 | Yes | N/A |
| Austria | Kärnten | m | N/A | N/A | N/A | No | CREA-0228 |
| Austria | Vorarlberg | N/A | TLMF Lep 14742 | LASTS290-14 | BOLD:AAD4722 | No | N/A |
| Austria | Oberösterreich | f | N/A | N/A | N/A | No | CREA-0191 |
| Austria | Osttirol | m | N/A | N/A | N/A | No | CREA-0192 |
| Austria | Osttirol | f | N/A | N/A | N/A | No | CREA-0193 |
| Austria | Osttirol | m | N/A | N/A | N/A | No | CREA-0194 |
| Austria | Osttirol | f | N/A | N/A | N/A | No | CREA-0195 |
| Austria | Osttirol | m | N/A | N/A | N/A | No | CREA-0196 |
| Austria | Osttirol | f | N/A | N/A | N/A | No | CREA-0197 |
| Austria | Osttirol | m | N/A | N/A | N/A | No | CREA-0198 |
| Austria | Osttirol | f | N/A | N/A | N/A | No | CREA-0199 |
| Austria | Osttirol | f | N/A | N/A | N/A | No | CREA-0200 |
| Estonia | Tartu | f | N/A | N/A | N/A | No | CREA-0160 |
| Norway | Nordre Land | N/A | NHMO-DAR-10720 | LON4755-16 | BOLD:ADF1792 | No | N/A |
| Norway | Bygland | N/A | NHMO-DAR-10719 | LON4754-16 | BOLD:AAD4723 | No | N/A |
| Norway | Nordland | m | N/A | N/A | N/A | No | CREA-0159 |
| Finland | Northern Ostrobothnia | f | MM26080 | BCLEP489-20 | BOLD:ADF1792 | Yes | CREA-0233 |
| Finland | Northern Ostrobothnia | N/A | MM23912 | LEFIJ7492-18 | BOLD:ADF1792 | Yes | N/A |
| Finland | Northern Ostrobothnia | N/A | MM00569 | LEFIB168-10 | BOLD:AAD4723 | No | N/A |
| Finland | Northern Ostrobothnia | f | MM06415 | LEFID473-10 | BOLD:AAD4723 | No | N/A |
| Finland | Northern Ostrobothnia | N/A | MM26079 | BCLEP488-20 | BOLD:AAD4723 | Yes | N/A |
| Finland | Northern Ostrobothnia | m | N/A | N/A | N/A | No | CREA-0212 |
| Finland | Northern Ostrobothnia | f | N/A | N/A | N/A | No | CREA-0215 |
| Finland | Northern Ostrobothnia | m | N/A | N/A | N/A | No | CREA-0221 |
| Finland | Eastern Finland | f | N/A | N/A | N/A | No | CREA-0211 |
| Finland | Ostrobottnia ultima | N/A | MM04085 | LEFIC487-10 | BOLD:AAD4723 | No | N/A |
| Finland | Lapland | f | MM06294 | LEFID383-10 | BOLD:AAD4723 | No | N/A |
| Finland | Lapland | N/A | MM26078 | BCLEP487-20 | BOLD:AAD4723 | Yes | N/A |
| Finland | Lapland | m | N/A | N/A | N/A | No | CREA-0220 |
| Finland | Lapland | m | N/A | N/A | N/A | No | CREA-0222 |
| Finland | Lapland | m | N/A | N/A | N/A | No | CREA-0223 |
| Finland | Satakunta | f | N/A | N/A | N/A | No | CREA-0213 |
| Finland | Pirkanmaa | f | N/A | N/A | N/A | No | CREA-0216 |
| Finland | Päijänne Tavastia | f | N/A | N/A | N/A | No | CREA-0217 |




