Conflicting relationships of Vipera walser inferred from nuclear genes sequences and mitochondrial DNA
Contributing authors: Sylvain Ursenbacher ([email protected]), Konrad Mebert ([email protected]), Samuele Ghielmi ([email protected]), Lorenzo Laddaga ([email protected]), Patricia Sourrouille ([email protected]), Mert Kariş ([email protected]), Pierre-André Crochet ([email protected])
Abstract
enThe description of Vipera walser from the Northern Italian Alps as a new species (Ghielmi et al., 2016, Journal of Zoological Systematics and Evolutionary Research, 54, 161) was one of the most unexpected surprises of European herpetology in the 21st century. In mitochondrial (mt) DNA, it is closely related to a group of vipers only present in the Caucasus region and Northeastern Anatolia. However, its morphology is similar to the V. berus populations that inhabit nearby mountains in the Swiss-Italian Alps, which raises questions on its relationships and status. We thus sequenced five nuclear (nu) genes to determine the position of V. walser relative to V. berus and to the Caucasian/Northeastern Anatolian vipers in nuDNA. We also reanalyzed five previously sequenced mtDNA fragments. NuDNA markers recovered V. walser as closely related to Italian populations of V. berus and not to the Caucasian/Anatolian species, thus contradicting the mtDNA phylogeny. We checked that each of the five mtDNA fragments independently amplified by Ghielmi et al. (2016, Journal of Zoological Systematics and Evolutionary Research, 54, 161) produced individual gene trees compatible with the concatenated mtDNA phylogeny, thus excluding the hypothesis that NUMTs sequencing generated the mtDNA relationships reported by Ghielmi et al. (2016, Journal of Zoological Systematics and Evolutionary Research, 54, 161). Given the low level of nuclear differentiation between V. walser and the Italian population of V. berus, we argue that ancient admixture between V. berus and the ancestral population of V. walser is the most likely explanation for this case of cyto-nuclear discordance and we discuss the consequences of these results on the systematic status of V. walser.
Résumé
frLa description de Vipera walser comme une nouvelle espèce des Alpes italiennes du Nord (Ghielmi et al., 2016, Journal of Zoological Systematics and Evolutionary Research, 54, 161) a été l'une des surprises les plus inattendues de l'herpétologie européenne au 21e siècle. Son ADN mitochondrial est étroitement apparenté à celui d’un groupe de vipères uniquement présent autour du Caucase et de l'Anatolie orientale. Cependant, sa morphologie est similaire aux populations de V. berus qui vivent dans la même région des Alpes italiennes, ce qui soulève des questions sur ses relations de parenté et son statut systématique. Nous avons donc séquencé cinq gènes nucléaires pour déterminer la position de V. walser par rapport à V. berus et aux vipères du Caucase/Anatolie orientale pour l'ADN nucléaire. Nous avons également réanalysé les cinq fragments d'ADN mitochondrial précédemment séquencés. Les marqueurs nucléaires ont identifié V. walser comme étroitement apparenté à la population italienne de V. berus et non aux espèces caucasiennes/anatoliennes orientales, contredisant ainsi la phylogénie de l'ADN mitochondrial. Nous avons vérifié que chacun des cinq fragments d'ADN mitochondrial amplifiés indépendamment par Ghielmi et al. (2016, Journal of Zoological Systematics and Evolutionary Research, 54, 161) ont produit des arbres de gènes individuels compatibles avec la phylogénie mitochondriale concaténée, excluant ainsi l'hypothèse selon laquelle le séquençage de NUMTs a généré les relations mitochondriales identifiées par Ghielmi et al. (2016, Journal of Zoological Systematics and Evolutionary Research, 54, 161). Compte tenu du faible niveau de différenciation nucléaire entre V. walser et la population italienne de V. berus, nous pensons qu’une introgression ancienne entre V. berus et la population ancestrale de V. walser est l'explication la plus probable de cette discordance cyto-nucléaire; nous discutons en conclusion des conséquences de ces résultats sur le statut systématique de V. walser.
1 INTRODUCTION
The description of a new species of viper from the Italian Alps, Vipera walser Ghielmi et al., 2016, was one of the most unexpected discoveries of European herpetology in the 21st century. This small and isolated population is only known from several valleys of the north-western Italian Alps about 60 km west from the closest known viper populations of the Adder Vipera berus. Vipera walser is morphologically close to the neighboring populations of Vipera berus, which belong to a distinct lineage (the Italian clade in Ursenbacher et al., 2006) for which the name Vipera berus marasso applies (Schmidtler & Hansbauer, 2020). All the morphological characters investigated overlap with Vipera berus marasso, even if a higher fragmentation of cephalic scales results in significant differences between these two groups in several characters, allowing correct classification of 94% and 88% of females and males, respectively, by a discriminant analysis based on six meristic variables (Ghielmi et al., 2016). Cranial osteology suggests one diagnostic feature as well (Seghetti et al., 2021), but this will need to be confirmed on larger sample sizes as four specimens of V. walser and five V. berus were examined, three of which only were from the Italian subspecies. The main argument to describe this population as a new species was its highly divergent mtDNA, which is not even sister to the mtDNA of Vipera berus but groups within a distinct clade of Vipera including the Meadow vipers group (V. ursinii, V. graeca, V. eriwanensis) and the Caucasian and Anatolian species V. kaznakovi, V. anatolica, V. darevskii, and V. dinniki (Ghielmi et al., 2016). Two nuclear markers were sequenced as well by Ghielmi et al. (2016) for a single specimen of four Western European species of Vipera. One marker revealed strongly divergent haplotypes between V. berus and V. walser (V. walser being even closer to V. aspis), but the second identified berus alleles as the closest relative of walser alleles with only one mutation (versus four mutations to V. aspis). However, the berus specimen sequenced for the two nuclear loci by Ghielmi et al. (2016) comes from France (SU unpublished data), which makes the interpretation of the nuclear data difficult. While the validity of V. walser was accepted by Freitas et al. (2020), who gave much weight to mtDNA data, the lack of sufficient nuclear data supporting the unexpected mtDNA relationships was one of the main arguments used by Speybroeck et al. (2020) to reject its validity pending further studies.
The unexpected biogeographical scenario suggested by the mtDNA data and the close morphological resemblance of V. walser with V. berus raise concern about the reliability of the relationships suggested by the mtDNA data. The aim of this study was thus to use independent nuclear markers to test whether the relationships of the V. walser population inferred from several nuclear loci agree with the mtDNA gene tree or not. For this purpose, we sequenced representative samples of most of the European species of Vipera including the newly described V. walser for five nuclear introns. We also repeated the mtDNA analyses of Ghielmi et al. (2016), using the same dataset as these authors did, as they are important to evaluate the various hypotheses that could explain our results, and provide single-fragment trees that were not presented in the main text in Ghielmi et al. (2016, see “Sampling” below). Our main aim was to assess whether nuDNA markers group V. walser with the Caucasian taxa (as suggested by mtDNA) or with the Italian populations of V. berus (as suggested by morphology and biogeography). Consequently, we did not include all taxa of the genus Vipera, but we selected samples from the Caucasian taxa, from several subspecies of V. berus, and from a few other European species. The first step was to verify that every well-supported species of viper formed a distinct cluster with our nuDNA data. Once this was done, we used these data to assess the position of V. walser in relation to the species we sequenced.
2 MATERIALS AND METHODS
2.1 Sampling
Most tissue samples used for the nuclear genes sequencing originated from the collection of the “Biogéographie et Écologie des Vertébrés” team (CNRS & École Pratique des Hautes Études, BEV-EPHE) housed in the UMR 5175-CEFE in Montpellier, France. Other samples were obtained from the private collections of SU and KM while V. walser samples were collected by SG and LL. Most samples were muscle samples obtained from road-killed specimens, or tail tips, ventral scales, or buccal swabs obtained from live specimens. All samples were stored in >95% ethanol prior to extraction. Our dataset included 35 individuals from most of the species of the genus Vipera: two samples of Vipera kaznakovi, four samples of Vipera darevskii, three samples of Vipera eriwanensis, four samples of Vipera ursinii (subspecies ursinii and rakosiensis), five samples of Vipera aspis (subspecies aspis and zinnikeri), two samples of Vipera seoanei, 11 samples of Vipera berus (subspecies bosniensis and berus and five individuals from the subspecies marasso from the Italian Alps), and five samples of Vipera walser. The complete list of all samples and their origin is given in Table 1. Non-invasive tissue sampling does not require ethical approval in French or Italian institutions. For fieldwork in Turkey, we received ethical permission (Ege University Animal Experiments Local Ethics Committee, 2013#050) and special permission (2018#101792) for field studies from the Republic of Turkey, Ministry of Agriculture and Forestry, General Directorate of Nature Conservation and National Parks. No permit was requested for sampling V. berus or V. walser in Italy.
| Taxon | Locality | Latitude | Longitude | Precision (m) | Voucher and/or tissue number | RAG1 | NT3 | R35 | BDNF | OD |
|---|---|---|---|---|---|---|---|---|---|---|
| Vipera kaznakovi | E of Kiyicik, Turkey | 41.3056 | 41.2530 | 10 | Vika7 | MW248848 | MT969159 | MW248820 | MT912385 | MW292429 |
| Vipera kaznakovi | Esenkıyı, Turkey | 41.4324 | 41.4580 | 10 | Vika17 | MW248849 | MT969160 | MW248821 | MT912386 | - |
| Vipera darevskii | Oguzyolu, Turkey | 41.2486 | 42.9923 | 10 | Vida9 | MW248846 | MT969157 | MW248818 | MT912383 | MW292427 |
| Vipera darevskii | Zekeriyaköy, Turkey | 40.9875 | 42.1367 | 10 | Vida11 | MW248847 | MT969158 | MW248819 | MT912384 | MW292428 |
| Vipera darevskii | 2 km E. Zekeriya, Turkey | 40.9941 | 42.1655 | 500 | BEV.8369/T741 | MW248845 | MT969155 | MW248812 | MT912381 | MW292425 |
| Vipera darevskii | 2 km E. Zekeriya, Turkey | 40.9941 | 42.1655 | 500 | BEV.8855/T742 | - | MT969156 | MW248813 | MT912382 | MW292426 |
| Vipera eriwanensis | 6 km ENE Arpaçay, Turkey | 40.8898 | 43.3530 | 500 | BEV.8856/T743 | MW248852 | MT969163 | MW248816 | MT912389 | MW292432 |
| Vipera eriwanensis | 8 km E of Gndevaz, Armenia | 39.7650 | 45.7105 | 20 | BEV.14134/T10554 | MW248850 | MT969161 | MW248814 | MT912387 | MW292430 |
| Vipera eriwanensis | Kechut, Armenia | 39.7950 | 45.6681 | 20 | BEV.14524/T11130 | MW248851 | MT969162 | MW248815 | MT912388 | MW292431 |
| Vipera walser | Valle Strona, Italy | 45.9394 | 8.2253 | 2000 | 2788 | MW248836 | MT969145 | MW248803 | MT912371 | - |
| Vipera walser | Valle Strona, Italy | 45.9447 | 8.2276 | 2000 | 2791 | MW248837 | MT969146 | MW248804 | MT912372 | - |
| Vipera walser | Valle Elvo, Italy | 45.6046 | 7.9044 | 2000 | 2792 | MW248834 | T969142 | MW248800 | MT912368 | MW292414 |
| Vipera walser | Upper Valle Sesia, Italy | 45.7283 | 7.9346 | 2000 | 2794 | MW248853 | MT969143 | MW248801 | MT912369 | MW292415 |
| Vipera walser | Valle Mastallone, Italy | 45.9324 | 8.0986 | 2000 | It5 | MW248835 | MT969144 | MW248802 | MT912370 | MW292416 |
| Vipera berus marasso | Forno di Zoldo, Italy | 46.3123 | 12.1571 | 20 | S3 | MW248828 | MT969136 | MW248795 | MT912362 | MW292410 |
| Vipera berus marasso | Passo Monte Croce di Comelico, Italy | 46.6605 | 12.4230 | 20 | S4 | MW248829 | MT969137 | MW267747 | MT912363 | MW292411 |
| Vipera berus marasso | Val Mora, Italy | 46.0401 | 9.6207 | 20 | S6 | MW248830 | MT969138 | MW248796 | MT912364 | MW292412 |
| Vipera berus marasso | Val Mora, Italy | 46.0369 | 9.6227 | 20 | S7 | MW248831 | MT969139 | MW248797 | MT912365 | MW292413 |
| Vipera berus marasso | Val Mora, Italy | 46.0388 | 9.6207 | 20 | S10 | MW248827 | MT969135 | MW248794 | MT912361 | MW292409 |
| Vipera berus berus | 1,5 km E of Valtavaara pass, Finland | 66.2052 | 29.2468 | 100 | BEV.10223/ T2992 | MW248822 | MT969129 | MW248788 | MT912355 | MW292404 |
| Vipera berus berus | Topilo, Poland | 52.6345 | 23.6232 | 750 | BEV.7602/ T11978 | - | MT969132 | MW248791 | MT912358 | - |
| Vipera berus berus | Frasne, France | 46.8272 | 6.1740 | 20 | BEV.11592/ T11980 | MW248824 | MT969131 | MW248790 | MT912357 | MW292406 |
| Vipera berus berus | Ågesta, Sweden | 59.2102 | 18.0803 | 10 | T4023 | MW248823 | MT969130 | MW248789 | MT912356 | MW292405 |
| Vipera berus bosniensis | Vitosha, Bulgaria | 42.5940 | 23.2843 | 10 | T4027 | MW248825 | MT969133 | MW248792 | MT912359 | MW292407 |
| Vipera berus bosniensis | Carev Do, Montenegro | 42.8567 | 19.7043 | 1000 | T4031 | MW248826 | MT969134 | MW248793 | MT912360 | MW292408 |
| Vipera seoanei seoanei | 400 m N of Chalet Pedro, France | 43.0404 | -1.0761 | 20 | T10370 | MW248832 | MT969140 | MW248798 | MT912366 | - |
| Vipera seoanei seoanei | Chalets d'Iraty, France | 43.0505 | -1.0647 | 100 | T10371 | MW248833 | MT969141 | MW248799 | MT912367 | - |
| Vipera ursinii rakosiensis | 3 km SSE Asinip, Romania | 46.1617 | 23.5499 | 500 | T5383 (ROU 154) | MW248841 | MT969151 | MW248808 | MT912377 | MW292421 |
| Vipera ursinii ursinii | Pra Mouret, France | 44.0863 | 6.6551 | 10 | T2410 (GCO 14) | MW248842 | MT969152 | MW248809 | MT912378 | MW292422 |
| Vipera ursinii ursinii | Mont Ventoux, France | 44.1812 | 5.2641 | 10 | T2595 (VEN 4) | MW248843 | MT969153 | MW248810 | MT912379 | MW292423 |
| Vipera ursinii ursinii | Montagne de Lure, France | 44.1207 | 5.8038 | 2000 | T2622 (LURE 25) | MW248844 | MT969154 | MW248811 | MT912380 | MW292424 |
| Vipera aspis aspis | Neuvéglise, France | 44.9340 | 2.9982 | 20 | BEV.14511/ T10075 | MW248854 | MT969150 | MW248807 | MT912376 | MW292420 |
| Vipera aspis zinnikeri | Mignoy, France | 44.6943 | -0.6338 | 200 | BEV.14180/ T10692 | MW248838 | MT969147 | MW248805 | MT912373 | MW292417 |
| Vipera aspis zinnikeri | Trachère, France | 42.8090 | 0.3123 | 400 | BEV.8657/ T11981 | MW248839 | MT969148 | MW248806 | MT912374 | MW292418 |
| Vipera aspis zinnikeri | Merdelou, France | 43.7641 | 2.8946 | 20 | BEV.9178/ T754 | MW248840 | MT969149 | MW248817 | MT912375 | MW292419 |
Note
- BEV codes are voucher codes.
- T codes are tissue codes.
In order to examine potential causes of discordant mtDNA and nuDNA patterns, we tested for possible artifacts caused by the sequencing of nuclear mitochondrial insertions (NUMTs) that are homologous to mtDNA sequences but have diverged after pseudogenization following their insertion into the nuclear genome. To do so, we repeated the analyses of the mitochondrial data from Ghielmi et al. (2016) independently for each amplified mtDNA fragment (five in total). If NUMTs are involved, we do not expect the PCR to amplify selectively the nuclear copies for every primer pair, so concatenated alignments are a mix of nuclear and mitochondrial copies, which can result in flawed phylogenetic inference. This can be detected by examining individual gene trees for every gene fragment amplified (i.e., every primer pair): if they all support the same topology as the concatenated alignment, NUMTs can be excluded as the source of the concatenated topology. Mitochondrial data were the same as those used in Ghielmi et al. (2016).
2.2 Molecular laboratory procedures
Total genomic DNA was extracted using the Dneasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer's recommended procedures. Negative extraction blanks were made by processing tubes in exactly the same way as tissue samples and were used in all PCR reactions to check for the lack of contamination. Five nuclear gene fragments were amplified for all samples: recombination-activating gene [RAG1], neurotrophin 3 [NT3], RNA fingerprint protein 35 [R35], brain-derived neurotrophic factor [BDNF], and ornithine decarboxylase [OD] (Table 2). These fragments were selected after preliminary screening of seven nuclear genes known to be useful in squamates phylogeny and sequenced in one specimen each of Vipera aspis, V. berus, and V. eriwanensis. Two other loci were not retained because they had zero (oocyte maturation factor [CMOS]) or just one (melanocortin 1 receptor [MC1R]) variable site in the three species alignment. PCR were conducted in 20 μl volumes with 2 μl of DNA, 10 μl of Taq Polymerase (Sigma-Aldrich), 0.5 μl of each primer [10 μM], and 7 μl of pure water. Amplifications were done following the same program for all introns: an initiation of 3 min at 94℃; 40 cycles of 30 s at 94℃, 40 seconds at 60℃, 1 min at 72°; and a final elongation of 10 min at 72℃. To test the success of the PCR, 3 μl of the PCR product was migrated on a 1% agarose gel for 30 minutes at 100 V and 80 mA. Successfully amplified DNA fragments were sequenced by Eurofins Genomics (Ebersberg, Germany) using the same primers as for amplification. Sequences were cleaned with Codon Code Aligner v. 9.0.1 (CodonCode Corporation) and aligned visually. Heterozygote positions were identified by eye from the chromatograms and coded with the IUPAC ambiguity codes; private mutations (occurring in one or very few specimens) were also checked by eye on the chromatograms. All new sequences were deposited in GenBank (see Table 1).
| Gene | Primers | Sequence 5’ – 3’ | Source |
|---|---|---|---|
| RAG1 (recombination-activating gene 1) |
R13 R18 |
TCTGAATGGAAATTCAAGCTGTT GATGCTGCCTCGGTCGGCCACCTTT |
Groth and Barrowclough (1999) |
| NT3 (neurotrophin 3) |
NT3-F3 NT3-R4 |
ATATTTCTGGCTTTTCTCTGTGGC GCGTTTCATAAAAATARRGTTTGACC |
modified from Mizsei et al. (2017) |
| R35 (RNA fingerprint protein 35) |
R35-F R35-R |
GACTGTGGAYGAYCTGATCAGTGTGGTGC GCCAAAATGAGSGAGAARCGCTTCTGAGC |
Brandley et al. (2011) |
| BDNF (brain-derived neurotrophic factor) |
BDNF_f BDNF_r |
GACCATCCTTTTCCTKACTATGGTTATTTCATACTT CTATCTTCCCCTTTTAATGGTCAGTGTACAAAC |
Townsend et al. (2008) |
| OD (ornithine decarboxylase) |
OD-F OF-R |
GACTCCAAAGCAGTTTGTCGTCTCAGTGT TCTTCAGAGCCAGGGAAGCCACCACCAAT |
Friesen et al. (1999) |
| CMOS (oocyte maturation factor) |
C08 C09 |
GCTTGGTGTTCAATAGACTGG TTTGGGAGCATCCAAAGTCTC |
Han et al. (2004) |
| MC1R (melanocortin 1 receptor) |
MC1r-F MC1r-R |
GGCNGCCATYGTCAAGAACCGGAACC CTCCGRAAGGCRTAAATGATGGGGTCCAC |
Pinho et al. (2010) |
| BACH1 (BTB and CNC homology 1) |
BACH1_f1 BACH1_r2 |
GATTTGAHCCYTTRCTTCAGTTTGC ACCTCACATTCYTGTTCYCTRGC |
Townsend et al. (2008) |
2.3 Phylogenetic analysis
Sequences from the five nuclear introns were phased with DnaSP v. 6.12.03 (Rozas et al., 2017), using default settings of the PHASE algorithm (Stephens et al., 2001), with 10,000 iterations and 1000 burn-in iterations. The program TCS v. 1.21 (Clement et al., 2006) implemented in PopART v.1.7 (Leigh & Bryant, 2015) was used to infer haplotypes networks using statistical parsimony, for each gene.
Unphased nuclear sequences were concatenated for subsequent phylogenetic analyses. The best-fitting substitution model in terms of both BIC and AICc, T92+Γ, was found using Mega-X (Kumar et al., 2018) with default settings and was used to build maximum-likelihood (ML) and neighbor-joining (NJ) trees with Mega-X. The support of the clades was estimated by 1000 bootstrap repetitions. Bayesian inference (BI) was conducted with MrBayes v. 3.2 (Ronquist & Huelsenbeck, 2001) using the GTR +Γ evolution model (the closest model from T92+Γ among the models implemented in Mr Bayes, as suggested in Mr. Bayes v. 3.2 manual). We ran the program for 3,000,000 generations, a value selected after checking that the standard deviation of split frequencies was below 0.01, that the potential scale reduction factor (PSRF) was reasonably close to 1.0 for all parameters and that the effective sample sizes (ESSs) were above 100 for all parameters, as suggested in Mr. Bayes v. 3.2 manual. Burn-in parameters were kept as default, that is, discarding 25% of samples as burn-in.
To root the nuclear tree, we searched GenBank for sequences of the nuclear fragments used here obtained in species from the sister genera to Vipera (Daboia, Montivipera, and Macrovipera, see Šmíd & Tolley, 2019). None of the possible outgroups is represented by more than three of our five nuclear fragments, so we selected the following species (GenBank accession numbers in parentheses): Montivipera raddei (NT3: KX695031, RAG1: KX169144, and BDNF: KX694739) and Daboia russellii (NT3: EU390916, R35: HQ876367, and BDNF: EU402636). In both ML and BI analyses, the evolutionary models used were the same as without outgroups. The length of the run (number of generations) for the BI analyses was determined as explained above; in the ML analyses, the support of the clades was estimated by 100 bootstrap repetitions. In BI trees, we tried to run the models with and without outgroup constraints and with and without a relaxed-clock model. Enforcing or not an outgroup constraint did not affect the result (root position). Enforcing a relaxed-clock model (as opposed to the default model without clock) in MrBayes affected the position of the root; the non-clock model was significantly better than the relaxed-clock model (as assessed by comparing the means of the marginal likelihoods, see MrBayes manual), so we selected the non-clock model as the BI result for the root position. In all analyses (ML or BI), the addition of outgroups lowered the resolution of the trees and the clades supports, probably because the amount of information in outgroups was always lower than for the in-group taxa. We thus retained the results of the phylogenetic analyses with the in-group taxa only (the Vipera species) and simply indicated on the BI tree in Figure 1 the various positions of the root inferred by different approaches.
Last, we built a NJ population tree using a pairwise between-groups net average distance matrix computed with Mega-X (T92 + G model) to represent the patterns of nuclear divergence between populations based on nuclear data. Groups were defined as species or subspecies for polytypic ones. The clade supports were not evaluated as there is no commonly available way to evaluate support for distance-based population trees.
We also retrieved the five mtDNA gene fragments used in Ghielmi et al. (2016) to produce five single-gene trees with the ML method and 1,000 bootstraps repetitions in Mega-X and the BI method with MrBayes using 200,000 generations. The best substitution models were selected for each gene using default settings in Mega-X and used for ML and BI analyses (for BI the closest model available in MrBayes was selected, as explained in the MrBayes manual). The T92+Γ model was selected for 16S ribosomal RNA [16S], the T92+I model was selected for Control Region 1 [CR1], the HKY+Γ model was selected for Control Region 2 [CR2], and NADH dehydrogenase subunit 4 [ND4] and the TN+Γ model were selected for Cytochrome B [CYTB].
3 RESULTS
The final dataset (provided as Supporting information) included sequences of five nuclear DNA introns for each individual: RAG1 (amplicon length approx. 1150 bp, trimmed length 1036 bp, 39 variable sites), R35 (amplicon length approx. 700 bp, trimmed length 635 bp, 35 variable sites), NT3 (amplicon length approx. 750 bp, trimmed length 697 bp, 20 variable sites), BDNF (amplicon length approx. 750 bp, trimmed length 670 bp, 5 variable sites), and OD (amplicon length approx. 600 bp, trimmed length 542 bp, 42 variable sites). Despite several attempts, we failed to amplify the OD intron for the two V. seoanei samples (see Table 1).
3.1 Phylogenetic reconstruction: nuclear sequences
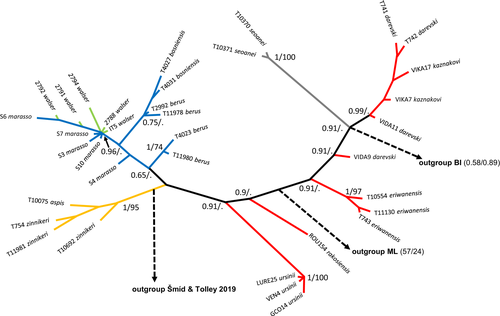
Phylogenetic trees reconstructed using BI, ML, and NJ methods from the concatenation of the five nuclear introns without outgroups resulted in concordant topologies (Figure 1; Figures S1 and S2). In all nuclear trees, low bootstrap values or posterior probabilities for internal nodes indicate a lack of support for the relationships between species based on our five nuclear introns. As expected for the reasons explained above, the position of the root was always poorly supported (Figures S3 and S4). Even though the species relationships of the tree are poorly resolved, the clade made of V. ursinii and the Caucasian taxa (V. eriwanensis, V. darevskii, V. kaznakovi) to the exclusion of V. aspis and V. berus receives a high support in BI with a posterior probability (pp) of 0.91. Surprisingly, V. seoanei is included in the clade made of V. ursinii and the Caucasian taxa in all three possible rooting options rather than excluded from it and does not group with V. berus, although these two taxa have been recovered as sister species by Alencar et al. (2016), Šmíd and Tolley (2019), and Freitas et al. (2020).
On the contrary, valid species are usually recovered as monophyletic clades with high support: V. aspis (V. a. aspis + V. a. zinnikeri, pp = 1), V. seoanei (pp = 1), V. eriwanensis (pp = 1), and V. ursinii (pp = 1). The only exceptions to this pattern are the two Caucasian species, V. kaznakovi and V. darevskii, which are not recovered as reciprocally monophyletic in any of the BI, ML, or NJ (Figure 1; Figures S1 and S2) trees, and our single sample of V. ursinii rakosiensis, which either does not group with V. u. ursinii (BI) or groups with low support (ML and NJ). All these results are valid under any of the three possible positions of the root of our nuclear tree (Figure 1).
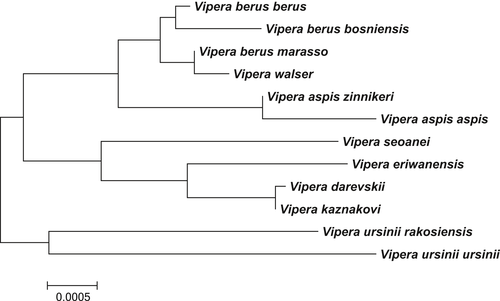
However, whatever the phylogenetic inference method used or the root position, V. walser is not recovered as a distinct cluster but all its samples are mixed with samples of V. berus marasso in a highly supported clade (BI: pp = 0.96) that includes all V. b. marraso samples except S4. All samples of V. berus (inc. V. b. berus and V. b. bosniensis), together with the samples of V. walser and V. b. marasso, form a monophyletic clade in all three methods even though this clade receives a low support (pp = 0.65, ML bootstrap = 48, NJ bootstrap = 43). The genetic proximity of V. walser with the Italian populations of V. berus is also supported by the distance-based population tree that groups V. walser and V. berus marasso within a monophyletic V. berus (Figure 2).
3.2 Nuclear haplotype networks and patterns of alleles sharing
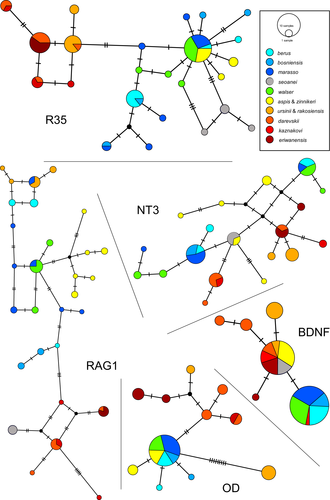
The nuclear gene alignments revealed various levels of diversity: six alleles for BDNF, 11 for OD, 19 for NT3, 24 for R35, and 29 for RAG1. Despite extensive lineage sharing, most valid species are entirely separated in the network (no allele sharing) in at least one of the five markers, such as RAG1 and BDNF for V. aspis and V. berus or R35 and OD for the western European species versus V. ursinii + Caucasian taxa (see Figure 3 for all haplotype networks). However, none of the five amplified markers was, considered individually, able to separate all the different valid species. Nevertheless, V. walser shares alleles with V. berus marasso for all five loci (sometimes also with other clades of V. berus, V. aspis, or V. kaznakovi), and those which are not shared (private alleles) are without exceptions linked to some V. berus marasso alleles in the network. The five allele networks reconstructed from the phased introns are therefore concordant with the relationships outlined by the species tree reconstructions and group V. walser with specimens of V. berus marasso.
3.3 Mitochondrial trees
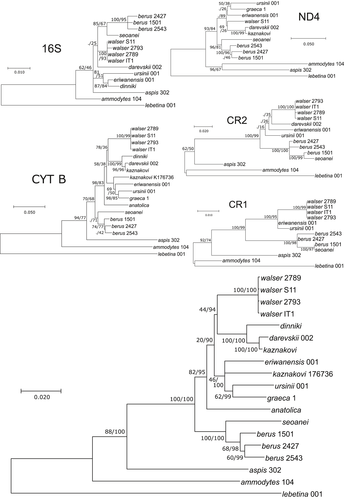
All of the five single gene trees reconstructed from each independently amplified mtDNA gene fragments are congruent with the concatenated mitochondrial tree of Ghielmi et al. (2016) even if they generally lack support. Most importantly, none of the gene fragment groups V. walser with V. berus as a monophyletic clade and most gene fragments support a position for V. walser closer to the Caucasian species than to V. berus (Figure 4).
4 DISCUSSION
4.1 Nuclear and mitochondrial data suggest incongruent relationships for Vipera walser
The nuclear data we generated unambiguously place Vipera walser inside the genetic diversity of Vipera berus. Both concatenated trees and nuclear haplotype networks mix V. walser individuals with the specimens of V. berus marasso and fail to recover two distinct clusters corresponding to V. walser and V. b. marasso. The pattern of population-level divergence confirms that these two taxa are closely related and are grouped with the other V. berus subspecies that we analyzed. This contrast with the other species included in our dataset, which are all recovered as well-supported monophyletic clusters and exhibit higher levers of genetic divergence from the other species (except for V. darevskii and V. kaznakovi, discussed below). This confirms that there is enough information in our nuclear data to separate the taxa that correspond to well-established and fully valid species, even if our nuclear data are clearly not powerful enough to resolve the evolutionary history of the genus Vipera (see below). We were not able to amplify the nuclear gene BTB and CNC Homology 1 [BACH1] that exhibited distinct alleles separated by 12 substitutions in one sample of V. walser and one sample of V. berus from France in Ghielmi et al. (2016). However, in our data set, the markers RAG1 or NT3 showed levels of divergence similar to BACH1 between the most different alleles of V. b. berus and V. walser (compare our Figure 2 with Ghielmi et al., 2016). It is thus unlikely that adding BACH1 to our data set would have affected our results, as this marker does not seem to behave differently from the markers we analyzed. In conclusion, we can be confident that V. walser and V. berus marasso are not recovered as distinct lineages based on the nuclear data, and these two taxa have a much lower genetic divergence than most other pairs of well-supported European vipers in our nuclear markers, as shown by the population tree (Figure 2).
On the contrary, our results support unambiguously that the mtDNA of V. walser is not closely related to the mtDNA of V. berus (Figure 4). Vipera walser shares no mtDNA haplotype with V. berus in any of the five gene fragments sequenced by Ghielmi et al. (2016), and it never groups inside the V. berus +V. seoanei clade that is recovered in all five single-fragment trees (Figure 4). None of the single-fragment trees recovers well-supported relationships, which is not surprising since they are all very short. However, they are all in agreement with the concatenated mitochondrial tree of Ghielmi et al. (2016; repeated here, Figure 4) in placing V. walser haplotypes closer to members of the Caucasian clade than to Vipera berus. The fact that all five mitochondrial fragments, amplified independently with five different primer pairs, support the same phylogenetic placement for V. walser excludes the possibility of artifacts resulting from the sequencing of NUMTs as it would be extremely unlikely to amplify preferentially the nuclear copy in V. walser for every primer pair. We can thus safely exclude NUMTs as explanation for the unexpected mtDNA relationships of V. walser. The different relationships of V. walser in mtDNA and nuDNA are thus both genuine and constitute a striking instance of cyto-nuclear discordance.
4.2 Nuclear relationships within Vipera
The nuclear data that we have generated are clearly not adequate to address the phylogeny of the genus Vipera, as evidenced by the low support for several nodes and the unexpected positions of some species, including outgroups. Indeed, the position of V. seoanei in the concatenated nDNA tree seems counterintuitive, as this species has always been considered as the sister species of V. berus (based on morphology and mtDNA analyses, Alencar et al., 2016; Freitas et al., 2020) and not related to Caucasian species. Similarly, the position of outgroups (hence the rooting of the tree) is always poorly supported, sometimes in a highly unexpected position (Figure S3), and always differs from the root position established by the most recent phylogeny of the Viperidae (Figure 1; Šmíd & Tolley, 2019): the BI analyses placed the root on the branch between V. eriwanensis and V. darevskii +V. kaznakovi (Figure S3), while the ML tree placed it between a clade made of these species +V. seoanei on the one hand and the rest of the vipers on the other hand (Figure S4), while Šmíd and Tolley (2019) would place it between V. aspis and all the other species. The difficulties in placing the root probably stem partly from missing data (at most three loci out of five were available for outgroups), but it is nevertheless clear that, at least for Eurasian vipers, obtaining robust phylogenies from nuclear sequence data would require a larger number of loci than what we used here.
Despite their lack of resolution, our nuclear data generate some valuable results that would justify further investigation. While this is outside the scope of this study, the high divergence between our single sample of V. u. rakosiensis and V. u. ursinii in nuclear DNA, mirrored in the mtDNA of these two taxa (Mizsei et al., 2017; Zinenko et al., 2016), reinforces the need for more detailed study in this species. On the contrary, the lack of reciprocal monophyly between V. darevskii and V. kaznakovi, together with the high divergence between some of our V. darevskii samples, could be partly due to a lack of resolution of our markers but also suggests the effects of admixture. These species appear as prime candidates for further genomic studies, such as done by Zinenko et al. (2016) or Mizsei et al. (2017) on other Caucasian taxa. Last, the position of V. seoanei in the nuclear dataset is poorly supported, but it never groups with V. berus under any possible position of the root of the Vipera tree. This calls for a re-examination of its relationships, which entirely rested on mtDNA and morphological data before our study.
All the nuDNA loci that we analyzed here in Eurasian vipers have a weak phylogenetic signal individually. In addition, as shown here and recently by Freitas et al. (2020) on a much larger dataset, allele sharing in nuclear loci is widespread between Eurasian viper species, even between species that are highly divergent in morphology, ecology, or mtDNA (as between V. aspis and V. berus in our dataset for example). Such allele sharing and/or weak phylogenetic signal of single-locus sequences are not restricted to vipers but are instead typical of nuclear gene sequences among closely related reptile species (e.g., Miralles et al., 2020; Vasconcelos et al., 2020) and can result from a combination of interspecific gene flow and/or incomplete lineage sorting (e.g., Pinho et al., 2008). In European vipers, it would be interesting to test whether lineage sharing is more extensive between sympatric or parapatric species than between fully allopatric species, once accounting for divergence time. This would allow assessing the respective role of incomplete lineage sorting and hybridization (see for instance Tarroso et al., 2014 or Guiller et al., 2017) in generating this large amount of allele sharing.
4.3 Possible causes of cyto-nuclear discordance
Incomplete lineage sorting (ILS) can generate gene trees that do not agree with the species tree and is thus a well-known cause of discordance between mitochondrial gene trees and species trees. However, ILS would seem like an extraordinary explanation in this case. ILS happens when internodes in a tree are short enough or effective population sizes are large enough for species divergence to be initiated before ancestral polymorphism is lost; as a result, ILS usually affects species that are closely related (Funk & Omland, 2003). It thus seems unlikely that ILS would affect patterns of divergence to the extent seen here. If ILS had caused cyto-nuclear discordance, V. walser would probably be identified as a divergent cluster in both mtDNA and nuDNA datasets and would be closer to V. berus in both datasets, even if its relationships were different when inferred from mtDNA and nuDNA. However, an unequivocal rebuttal of the ILS hypothesis would require formal testing, such as simulation of mtDNA data under a species history based on the Vipera phylogeny, with a wide range of biologically possible population sizes, to see whether such level of discordance and such amount of genetic divergence of mtDNA lineages within V. berus can be generated. This would first require a robust assessment of species relationships in Vipera independently of mtDNA data, which is still not available.
Consequently, the most credible hypothesis to explain the observed pattern of nuclear and mitochondrial divergence of V. walser relative to V. berus is introgression. Mitochondrial introgression (here defined as the occurrence of mtDNA lineages originating from another species in the genetic background of a given species) is a common phenomenon in animals, resulting in most extreme cases in mitochondrial capture (the fixation of the foreign mtDNA in the receiving population). Introgression is the main cause of cyto-nuclear discordance (reviewed in Funk & Omland, 2003 and Toews & Brelsford, 2012), and mitochondrial introgression has been detected in several European reptiles and amphibians (see Dufresnes et al., 2018; Renoult et al., 2009; Wielstra & Arntzen, 2020; Zieliński et al., 2013).
Evolutionary mechanisms leading to mitochondrial introgression have been reviewed by Petit & Excoffier (2009) and Excoffier et al. (2009). Spatial expansion in particular can lead to massive introgression of local genes into the genome of an invading species, while male-biased dispersal seems to favor higher introgression rate of mtDNA compared with nuDNA. These situations can result in populations where the original mtDNA remains in place but occurs in a nuclear genetic background that has been massively swamped by the invading species (as recently proposed for Iberian hares by Seixas et al., 2018). Studies of sex-biased dispersal in Viperidae are rare but Clark et al. (2008, Crotalus horridus), Zwahlen et al. (2021, Vipera aspis) and François et al. (2021, V. berus) reported male-biased dispersal. Male-biased dispersal has also been reported in the Colubrid species Coronella austriaca (Pernetta et al., 2011) and Thermophis baileyi (Hofmann et al., 2012) and seems to be the general pattern in Squamates (reviewed in Ferchaud et al., 2015). It thus seems likely that male-biased expansion of V. berus marasso into the area inhabited by V. walser generated the current situation where populations have retained the original V. walser mtDNA in a nuclear background heavily admixed with V. berus alleles.
Under this hypothesis, Vipera walser's mitochondrial lineage would really be the footprint of the existence of a highly divergent lineage of Vipera in the western Alps with no close relative in Western Europe and sharing common ancestors with the Meadow–Caucasian group of taxa (as suggested in Ghielmi et al., 2016). More recently, nuclear introgression from expanding V. berus marasso resulted in extensive allele sharing and loss of divergence for the five nuclear introns we sequenced, and thus probably for a substantial part of the nuclear genome. As V. walser and V. berus marasso are currently not in contact, the genetic exchange should have taken place during the last glaciation (or before), when both species were probably at a lower altitude and possibly in contact. We checked a recently published, extensive review of Viperidae fossil material (table S3 in Šmíd and Tolley, 2019) and none of the material from Italy can be attributed to the Meadow–Anatolian–Caucasian viper group, which would support this scenario (but note that no fossil material of V. berus from Italy could be found either).
A better understanding of the precise mechanisms that generated this pattern will require studying more nuclear loci, as a precise estimation of nuclear admixture is a prerequisite to distinguish between various possible scenarios. Bonnet et al. (2017) have shown that massively discordant mitochondrial introgression, where all or nearly all individuals of a species harbor the mtDNA of the other species with very little nuclear introgression, is extremely unlikely without positive selection of the foreign mtDNA or multiple barriers to introgression in the nuclear genome. The threshold to define a massive discordance, in case where the mtDNA is not admixed, is that nuclear admixture is less than 20% (in our case, less than 20% of nuclear alleles would need to be of V. walser origin). This seems to be the case for most of the five markers we have sequenced (only one allele of the locus BDNF is shared between walser and kaznakovi) but is not necessarily true for the whole nuclear genome.
4.4 Systematic implications
Our results suggest that the status of Vipera walser needs further investigation. Its ranking as a valid species or not will depend on the extent of nuclear admixture from V. berus. Our five nuclear sequences indicate that a substantial part of its genome is now shared with Vipera berus marasso, but the number of loci we used is limited and represents a tiny proportion of the nuclear genome. If most of the nuclear genome of V. walser is indeed shared with V. berus marasso, as is likely if our five nuclear markers are representative of the rest of the genome, V. walser cannot be recognized as a valid species. Even if its mitochondrial DNA constitutes the past footprint of an ancient evolutionary history, few species concept would recommend treating these populations as a distinct species based on their mitochondrial and weak morphological differences only. On the contrary, if substantial parts of the nuclear genome have resisted introgression from V. berus and large tracks of V. walser original genome remain, it could be argued that V. walser constitutes a valid species. In other cases, new species have been recognized despite massive nuclear introgression as long as a substantial fraction of the genome resists introgression (e.g., Nater et al., 2017; Toews et al., 2016). We thus recommend screening of a larger fraction of the genome of V. walser, for example, with genomic approaches (RAD sequencing or whole genome sequencing) to understand its history and decide on its validity as a species. Whatever the outcome of these future studies, these populations are the remnants and last testimony of a fascinating evolutionary history that requires to be preserved. Future investigations to understand this issue better will only be possible if adequate strategies are implemented to ensure the persistence of V. walser populations, which should at least be regarded as constituting an evolutionary significant unit (Moritz, 1994). Unfortunately, only a small fraction of the populations is currently included in Italian protected areas, while most of them fall in areas without specific protection and are increasingly exposed not only to the effects of climate change, but also to direct anthropic pressures that jeopardize their medium/long-term persistence.
On a more general note, our results illustrate the pitfalls of mitochondrial-based species delimitations. The risks of single-locus studies have long been known (e.g., Balloux, 2010; Moore, 1995; Renoult et al., 2009), and the need to use multiple markers for molecular taxonomy, phylogeography, and phylogenetic studies is now consistently advocated (e.g., Dupuis et al., 2012; Jacobs et al., 2018). However, because of the smaller number of informative sites per nuclear sequence compared with mtDNA, combining mtDNA and one or two nuclear introns in a single tree (as is still often done) usually results in the same topology as the mtDNA tree alone. A proper multilocus approach should employ enough nuclear markers to provide a truly independent topology and should report it (Dool et al., 2016). Despite these well-known issues with mtDNA-alone approaches, many recent studies still rely mostly on mtDNA for phylogenetic or taxonomic analyses because of its easy access and low cost, and this includes several proposed new species (for recent examples in European reptiles, see Bassitta et al., 2020; Senczuk et al., 2019). We concur with Speybroeck et al. (2020) that the status of such candidate species whose recognition rests primarily on mtDNA divergence should be evaluated with other lines of evidence before they are widely accepted.
ACKNOWLEDGEMENTS
We thank Philippe Geniez and Anne-Laure Ferchaud for their help to access the BEV samples and Dragan Arsovski, François Bole, Marc Cheylan, Michel Geniez, Philippe Geniez, Ioan Ghira, Bayram Göcmen, Nasit Igci, Arnaud Lyet, Anil Mehmet Oguz, Jean-Louis Plazanet, Jean-Luc Poitevin, Gilles Pottier, Bernard Ricau, Adrien Sprumont, Alexandre Teynié, Frédéric Veyrunes, Alexander Westerström and Stephan Zamfirescu for their help in collecting some of the samples used here. KM and MK were supported by the Scientific and Technical Research Council of Turkey (TÜBİTA K) under Grant 111T338 and 114Z946, the Mohamed bin Zayed Species Conservation Fund, project no. 13057971 and 150510677, and the Wilhelm Peters Fund 2013 of the German Herpetological Society DGHT (Deutsche Gesellschaft für Herpetologie und Terrarienkunde).




