Role of Pleistocene climatic oscillations on genetic differentiation and evolutionary history of the Transvolcanic deer mouse Peromyscus hylocetes (Rodentia: Cricetidae) throughout the Mexican central highlands
Contributing authors: Yessica Rico ([email protected]), Jesús A. Fernández ([email protected]), Elizabeth Arellano ([email protected])
Abstract
enHighlands are the most heterogeneous and complex biogeographic regions of Mexico. Species inhabiting these regions have been exposed to geologic events and climatic fluctuations in the past causing limited historical gene flow that resulted in structured genetic variation and high endemism. We examined the genetic variation of the mouse Peromyscus hylocetes throughout its geographic distribution within the Trans-Mexican Volcanic Belt (TVB), to estimate its current environmental suitability, habitat connectivity, and to reconstruct its evolutionary history by inferring the role of past events and abiotic factors. Two main genetic clusters corresponding to the west and east range of the species distribution were detected. Gene flow occurred largely from the west to the east cluster. Peromyscus hylocetes and P. aztecus diverged during the Pliocene–Pleistocene at the central-south region of the TVB. We hypothesized that after this divergence, P. hylocetes colonized the TVB during the Pleistocene and later expanded its distribution to the western TVB. Due to the climatic oscillations in the late Pleistocene, populations were restricted in western TVB during the warmer periods, and displacements occurred during colder periods from the west to the central TVB on several episodes.
Resumen
esLas zonas montañosas son las regiones biogeográficas más heterogéneas y complejas de México. Las especies que habitan estas regiones han sido expuestas a eventos geológicos y fluctuaciones climáticas en el pasado provocando un limitado flujo genético histórico que resultó en la variación genética estructurada y alto endemismo. Examinamos la variación genética del ratón Peromyscus hylocetes a través de su distribución geográfica en la Faja Volcánica Transmexicana (FVT), estimamos su idoneidad ambiental actual, conectividad del hábitat y reconstruimos su historia evolutiva infiriendo el papel de los eventos del pasado y los factores abióticos. Dos principales grupos genéticos fueron detectados correspondientes al oeste y este de su distribución. El mayor flujo genético ocurrió desde el grupo del oste al este. Peromyscus hylocetes y Peromyscus aztecus divergieron durante el Plioceno-Pleistoceno en la región centro-sur de la FVT. Hipotetizamos que después de esta divergencia, P. hylocetes colonizó la FVT en el Pleistoceno y después expandió su distribución al oeste de la FVT. Debido a las oscilaciones climáticas del Pleistoceno tardío, las poblaciones estuvieron restringidas en el oeste de la FVT durante los periodos más cálidos y desplazamientos ocurrieron durante los periodos más fríos desde el oeste al centro de la FVT en varios episodios.
1 INTRODUCTION
Mountainous regions represent approximately 15% of the total land surface area of the world (Körner et al., 2017). However, these areas host a third of the total biodiversity and 40% of the endemism (Myers et al., 2000). These regions have high topographic complexity and climatic stability over a considerable time that are associated with the persistence of the high biodiversity of vertebrates (García-Rodríguez et al., 2021; Quintero & Jetz, 2018). One of these regions is the Trans-Mexican Volcanic Belt (TVB) located in central Mexico. This is the largest Neogene volcanic arc in North America (Bayona-Viveros et al., 2017; Ferrari et al., 2012). The TVB was established as an independent province, in the biogeographic regionalization of Mexico, based on their biotic and abiotic dimensions (Morrone, 2019). This province arose during the early to middle Miocene, as a result of the counterclockwise rotation of the volcanic axis from the NNW-oriented Oligocene Sierra Madre Occidental (Ferrari et al., 2012). Based on tectonic features and orogeny, the TVB is divided into the western, central, eastern, and easternmost sectors (Ferrari et al., 2012; Gómez-Tuena et al., 2005). The west and east districts are recognized based on geology, altitude, climate, vegetation, mammal richness, and endemicity (Gámez et al., 2012).
The TVB is considered one of the most heterogeneous and complex biogeographic regions of Mexico. It is a diversification and biogeographic transition zone for a wide variety of taxa (Gámez et al., 2012; Luna et al., 2007). Mountainous taxa that inhabit this region have been exposed to several geologic events and climatic fluctuations in the past (Mastretta-Yanes et al., 2015). These historical events have limited the gene flow that resulted in structured genetic variation for several species, such as salamanders (Heredia-Bobadilla et al., 2016; Sunny, Duarte-deJesus, et al., 2019), rattlesnakes (Sunny et al., 2015, 2018), and some groups of plants (Pérez-Crespo et al., 2017; Rodríguez-Gómez et al., 2018; Ruiz-Sanchez & Specht, 2013, 2014).
The Transvolcanic deer mouse Peromyscus hylocetes Merriam, 1898 inhabits the central-west TVB in fir, pines, oaks, and cloud forests above 2300 m (Vázquez et al., 2001). This species was described from southern Pátzcuaro, 8000 ft in Michoacán, Mexico. Subsequently, was considered a subspecies of P. aztecus (Saussure, 1860) based on morphological characters (Carleton, 1979). Further molecular studies based on allozymes, chromosomes, and mitochondrial sequences confirmed its taxonomic status as a valid species (Sullivan & Kilpatrick, 1991; Sullivan et al., 1997); however, such evidence came from a few individuals. Currently, it is considered a member of the P. aztecus species complex and it is closely related to P. aztecus (Saussure, 1860), P. cordillerae Dickey, 1928, P. spicilegus Allen, 1897, and P. winkelmanni Carleton 1977 (Kilpatrick et al., 2021; Sullivan et al., 1997).
Sullivan et al. (1997) performed phylogeographic analyses of the P. aztecus species complex using partial mitochondrial sequences from the cytochrome b gene (Cytb) finding reciprocally monophyletic clades between P. hylocetes and P. aztecus. Later, a reexamination of the species complex confirmed these phylogenetic relationships (Kilpatrick et al., 2021). However, in these analyses, less than seven specimens of P. hylocetes from central TVB were included. Monophyly has not yet been tested across the entire geographic distribution of P. hylocetes or whether its populations are geographically isolated and genetically structured. We hypothesize that individuals distributed in the center-west TVB form a monophyletic group. Genetic structure might be the result of gene flow interruption in some regions as a consequence of geologic and climatologic processes at the TVB. Therefore, the aims of this study were (1) to assess the monophyly and genetic variation of P. hylocetes throughout its geographic distribution, (2) to estimate its current environmental suitability and habitat connectivity, and (3) to infer the role of past events and abiotic factors that have influenced the evolutionary history of this species.
2 MATERIALS AND METHODS
2.1 Sampling
A total of 86 individuals were processed for sequencing two mitochondrial and two nuclear markers. From these individuals, 70 were P. hylocetes and the remaining 16 samples belonged to the P. aztecus complex. To complement the taxonomic sampling, 75 sequences for 15 species were downloaded from NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) for the P. aztecus complex and other closely related species, including seven individuals of P. hylocetes (Appendix 1). Additional geographic information for each specimen sequenced in this study is available in Table S1. All available P. hylocetes museum specimens from the TVB were revised in the Colección Nacional de Mamíferos (CNMA) of the Universidad Nacional Autónoma de México (UNAM), Colección de Mamíferos de la Sierra Volcánica Transversal de México (UAM-I) of the Universidad Autónoma Metropolitana, and Colección de Mamíferos del Centro de Investigación en Biodiversidad y Conservación (CMC) of the Universidad Autónoma del Estado de Morelos. Fresh tissue specimens and recent dry skins were requested for DNA extraction. Other tissues samples were granted by the Colección de Mamíferos del Museo de Zoología Alfonzo L. Herrera (MZFC) UNAM and the Mammal Collection of the Texas Tech University (TTU). Sampling was completed with field explorations in central TVB at Michoacán state, and further details on León-Tapia et al. (2020). The 77 total specimens of P. hylocetes encompass 24 different localities throughout the center-west of the TVB (Table S1). To date, this sampling is the best representation of the known geographic distribution of this species.
2.2 DNA extraction, amplification, and sequencing
We used a 3 mm2 piece of ventral skin tissue from museum skins. Each sample was washed with sterile distilled water and incubated at 56℃ for 15 minutes three times before DNA extraction following the suggestions to prevent contamination (McCormack et al., 2016; Ruane & Austin, 2017). Genomic DNA from fresh tissue and skin was isolated using the kit ZR Genomic DNA – Tissue MicroPrep (Zymo Research) in a final volume of 30 µl.
We amplified two mitochondrial and two nuclear marker sequences. Mitochondrial markers correspond to 820 pb of the Cytb gene and 657 pb of the cytochrome c oxidase subunit 1 gene (COI); whereas, nuclear markers correspond to 700 pb of intron 7 of the beta fibrinogen polypeptide gene (Fgb-I7) and 567 pb of intron 2 of the alcohol dehydrogenase gene (Adh-I2). The Cytb sequence was amplified using sets of primers in different combinations: MVZ-03, MVZ-06, MVZ-07, MVZ-10, MVZ-11 (Smith & Patton, 1993), L-14115, L-14553, H-14963, and H-14541 (Sullivan et al., 1997). PCR conditions were as follows: an initial denaturing step at 95℃ for 2 min, followed by 30 cycles of denaturing at 95℃ for 1 min, annealing at 45℃ for 1 min, and extension at 72℃ for 1.5 min, with a final extension step at 72℃ for 7 min. The COI sequence was amplified using the primers BatL5310 and R6036R (Ivanova et al., 2012), using the following conditions: initial denaturing step at 94℃ for 2 min, followed by 35 cycles of denaturing at 94℃ for 30 s, annealing at 55℃ for 30 s, and extension at 72℃ for 1 min, with a final extension at 72℃ for 5 min. The Fgb-I7 sequence was amplified with the Fgb-17 U-Rattus and Fgb-17L-Rattus primers (Wickliffe et al., 2003), using an initial denaturing at 93.5℃ for 1 min, followed by 35 cycles of denaturing at 93.5℃ for 40 s, annealing at 53℃ for 45 s, and extension at 72℃ for 1.5 min, with a final extension at 72℃ for 2 min. Finally, the Adh-I2 was amplified with combinations of primers 2340-IF, 2340-II-R, EXON II-F, and EXON II-R (Amman et al., 2006), using an initial denaturing at 95℃ for 2 min, followed by 30 cycles of denaturing at 95℃ for 45 s, annealing at 52℃ for 45 s, and extension at 72℃ for 1.5 min, with a final extension at 72℃ for 4 min. Primers sequences are available in Table S2.
PCR reactions were performed in a final volume of 30 µl with 8 µM dNTPs, 25 µM MgCl2, 3 µl of 5× Green GoTaq buffer (Promega), 8 µM primers, and 0.1 µl of GoTaq® Flexi DNA polymerase (Promega). Successful reactions were purified with a NucleoSpin® Gel and PCR cleanup (Macherey-Nagel GmbH & Co. KG) purification kit and sequenced at the Instituto de Ecología AC (Xalapa, Mexico) and Macrogen Inc. Chromatograms were examined using the software Sequencher v5 (Gene Code Corporation). Sequence data have been submitted to the NCBI GenBank depository under the accession numbers MW264520–MW264605 (Cytb), MW265530–MW235588 (COI), MW264606–MW264684 (Fgb-I7), and MW264685–MW264760 (Adh-I2). Accession numbers for each specimen are available in Appendix 1.
2.3 Phylogenetic inference
All sequences were aligned using the MUSCLE algorithm in AliView v 1.18 (Larsson, 2014). The final matrix was partitioned by marker. PartitionFinder v 1.1 (Lanfear et al., 2012) was used for inferring the best nucleotide substitution and partition scheme based on the Akaike Information Criteria (AIC), unlinked branch lengths, and the greedy algorithm.
Baiomys taylori was selected as outgroup for maximum likelihood (ML) and Bayesian Inference (BI), and this taxon was chosen as member of the sister clade (Baiomyini) of the Peromyscine tribe (Steppan & Schenk, 2017), which was used as node for fossil calibration in the subsequent divergence analyses. For ML, 1000 iterations of rapid bootstrap followed by a thorough ML search were carried out in RAxML v 8.2.12 (Stamatakis, 2014). The BI was performed using two independent Metropolis Markov chains Monte Carlo runs of 10 million generations with a sampling frequency every 1000 generations in MrBayes v 3.2.6 (Ronquist et al., 2012). Likelihood convergence and stability were checked in Tracer v 1.7.1 (Rambaut et al., 2018), and the first 25% of trees were discarded as burn-in and a majority rule consensus tree was built with the remaining trees. Lastly, the mean genetic distances for each marker among the main species clades in the P. aztecus complex and within P. hylocetes were calculated with APE v 5.4.1 package (Paradis & Schliep, 2019) in R software v 3.5.2 (R Foundation for Statistical Computing, Vienna, Austria) using the Kimura 2-parameter (K2p) model for nucleotide substitution to compare our results with other studies.
2.4 Intraspecific genetic differentiation and migration rates
Intraspecific analyses were performed for P. hylocetes samples. The number of haplotypes, haplotype diversity, segregating sites, nucleotide proportion, and nucleotide diversity were estimated for each marker using PEGAS v 0.11 package (Paradis, 2010) and STRATAG v 2.4.905 (Archer et al., 2017) in R software. Based on the concatenated data, we inferred the most likely number of genetic clusters (K) taking into account the geographic information with the non-negative matrix factorization and least-squares optimization method implemented in Tess3r v 1.1.0 (Caye et al., 2016) in R software. This statistical approach was chosen for analyzing the general spatial patterns of genetic variation and to obtain a rapid ancestry coefficient estimation before the Bayesian approach. Tess3r was run from one to ten K with ten repetitions of each K value due to the small number of distant geographic localities and high values of k that can be impractical inferences. The lambda value was set to 1 for the spatial regularization parameter using the method “projected.ls”, the maximum number of iterations of the optimization up to 200 and 1e-05 of tolerance. The optimal value of K was based on the cross-validation criterion, and the individual membership proportion was interpolated on a geographic map of 300 × 300 pixels.
We used the Bayesian Phylogeographic and Ecological Clustering (BPEC) to identify clusters taking into account genetic and environmental information in the BPEC v 1.3.1 package (Manolopoulou et al., 2020) in R software. The results provide estimates and measures of uncertainty on the number of migration events, the geographic distribution of clusters, ancestral locations, and a clustered tree structure (Manolopoulou et al., 2020). For this analysis, only the Cytb dataset was used due to includes information for more individuals and geographic locations. We used nine uncorrelated environmental layers (see ecological niche section), the environmental data were extracted from each individual sampled and the Principal Component Analysis (PCA) was performed in R software. The two first PCs were used as covariates. After several initial short runs, the final analysis was carried out with two maximum numbers of migrations, a strict parsimony level at minimum (zero), and 10 million steps of MCMC with 10,000 posterior samples were saved.
Using the concatenated data, we carried out an AMOVA among the main genetic clusters identified from Tess3r and BPEC using the pairwise distances corrected with K2p and 10,000 permutations in PEGAS v 0.11 package in R software. After removing the ambiguous nucleotide characters and gaps, a haplotype network was built for each marker with the minimum spanning network algorithm implemented in POPART v 1.7.2 (Leigh & Bryant, 2015).
A coalescent-based method implemented in MIGRATE-N v 4.4.3 (Beerli & Palczewski, 2010) was used for estimating migration patterns among the main genetic clusters with the concatenated data. Several initial short runs were performed with the partitions resulted from PartitionFinder using a Bayesian search strategy to check the convergence of chains and appropriate parameters. We ran three long chains with 1 × 106 steps recording parameters every 100 steps. An adaptive heating scheme with four chains and different temperatures (1, 1.5, 3, and 1 × 106) with a swapping interval of 1 was employed to manipulate the temperatures according to the swapping success and find good values. Four independent runs were carried out to check the consistency and convergence of the estimates that were subsequently averaged. We transformed the bidirectional mutation-scaled immigrants rates (M) into the number of migrants per generation (xNm) multiplying M and the effective population size (Θ).
2.5 Ecological niche modeling and spatial connectivity
The occurrences of P. hylocetes were obtained from fieldwork, museum specimens, and published studies (Amman et al., 2006; León-Tapia et al., 2020; Orduña Trejo et al., 1999; Steppan & Schenk, 2017; Sullivan et al., 1997; Vázquez et al., 2000, 2004). Additionally, a second set of occurrences were downloaded from the Global Biodiversity Information Facility (GBIF; http://www.gbif.org/) and filtered by museum specimens. Occurrences without geographic coordinates and insufficient locality information were discarded. To reduce sampling bias, the occurrences were spatially thinned at 5 km using the SPTHIN v 0.1.0.1 package (Aiello-Lammens et al., 2015) in R software. We used this distance because is sufficient to have altitudinal changes and likely climatic differences. The first dataset was used to build the niche model, whereas the second one was used for data testing.
A total of 19 environmental layers for current conditions (1979–2013) in 30 arc-seconds (Karger et al., 2017) were downloaded from PaleoClim (http://www.paleoclim.org/) and used as variable predictors. The environmental variables were delimited for the TVB and central Sierra Madre del Sur (SMS), merging the TVB biotic province polygon downloaded from the Sistema Nacional de Información sobre Biodiversidad (http://www.conabio.gob.mx/informacion/gis/) and the central SMS proposed by Morrone et al. (2017). We hypothesize these polygons as the likely accessible area for P. hylocetes because all occurrences were restricted to these biogeographic provinces which are delimited by physiographic, morphotectonic, climatic, and vegetation features (Morrone et al., 2017). To minimize redundancy in the bioclimatic layers, values from the 19 layers were extracted and one set of layers was chosen according to the Pearson's correlation threshold of 0.85 in the NTBOX v 0.1.4.5 package (Osorio-Olvera et al., 2020).
Current niche models were built under the maximum entropy algorithm implemented in MAXENT software v 3.4.1 (Phillips et al., 2017) using the KUENM v 1.1.1 package (Cobos et al., 2019) in R software. Levels of model complexity were evaluated, such as over-fitting by varying the regularization multiplier (RM) from 0.5 to 5 every 0.5, and feature classes linear (L), quadratic (Q), product (P), threshold (T), and hinge (H) in five combinations (i.e., L, LQ, LQP, LQPT, LQPTH), and 10,000 background points resulting in 50 candidate niche models. These models were evaluated based on the statistical significance of the partial receiver operating characteristic (pROC; Peterson et al., 2008) with 20% of random point and 500 iterations, 5% omission rates (OR; Anderson et al., 2003) as predictive power. The Akaike information criterion corrected for small sample sizes (AICc; Warren et al., 2010) was used for model complexity. The niche model selected with the best parameters was built and projected in the same polygon with ten bootstrap replicates and logistic output format.
The circuit theory algorithm was used to evaluate spatial connectivity among genetic samples in Circuitscape v 4.0.5 (McRae et al., 2008). This algorithm quantifies movement across multiple possible paths in a landscape, not just a single least-cost path or corridor to predict patterns of gene flow among populations in heterogeneous landscapes (Dickson et al., 2019). The best current ecological niche model was used as a habitat map, wherein each cell value was assigned as conductance. The geographic coordinates for each sampled locality were used as focal node locations. Lastly, the pairwise model was used to calculate connectivity between all pairs of focal nodes. The higher values of the current maps represent ease of movement and major connection schemes that allow gene flow among sampled localities. Lastly, an isolation by resistance (IBR) model was tested by a Mantel test with the resistance pairwise distances matrix output of Circuitscape and the K2p genetic distance matrix using a Pearson correlation and 10,000 permutations in the ADE4 v 1.7-13 package.
2.6 Past inferences: divergence time, historical demography, and paleodistribution
Divergence time estimates were performed in Beast v 2.6.5 (Bouckaert et al., 2019) using the full dataset. Outgroup, partitions, and nucleotide substitution models were as in the phylogenetic analyses. Calibration was performed using Postcopemys repenningi dated at 4.7 ± 0.2 Mya. This taxon is considered as a member of the Neotomine-Peromyscine radiation of the Cricetidae family, with the ancestor-descendent relationship of Copemys-Peromyscus via Postcopemys (Lindsay & Czaplewski, 2011). The relaxed clock lognormal model, Normal distribution for the calibration, and a calibrated Yule model of diversification process were used. Two independent runs were performed, each one with 20 million generations with sampling every 1000 generations. Likelihood scores convergence (ESS >200) and stability were checked in Tracer v 1.7.1 (Rambaut et al., 2018). LogCombiner v 2.6.3 was used to join the two runs and discard the first 25% of trees as burn-in. The final tree with divergence dates was obtained with TreeAnotator v 2.6.3.
To infer the historical demographic change within P. hylocetes, the Tajima's D (Tajima, 1989), Fu’s F (Fu, 1997), and R2 (Ramos-Onsins & Rozas, 2002) were calculated using 10,000 coalescent simulations for the concatenated data and for each marker. Additionally, the concatenated data was used in a mismatch distribution test computed in the PEGAS package. Finally, to infer demographic population fluctuations over time, a Bayesian Skyline Plot was constructed in BEAST v 2.6.1 with the Coalescent Bayesian Skyline. Unlink markers were designated with substitution models as resulting from PartitionFinder. We employed a lognormal distribution and substitution rates with values estimated by BEAST. A relaxed lognormal molecular clock, 20 million generations sampling every 1000 generations, and a final burn-in of 20% were used. The visualizations of final scores and the demographic plot were performed in TRACER v 1.7.1.
Finally, the current niche model was projected to the Last Interglacial (LIG) 120–140 thousand years ago (ka) in 30 arc-seconds (Otto-Bliesner, 2006) downloaded from WorldClim v 1.4 (https://www.worldclim.org/), and the Last Glacial Maximum (LGM) 21 ka in 30 arc-seconds (Karger et al., 2017) downloaded from PaleoClim. Both data are based on the Community Climate System Model simulations. Projections were performed in the KUENM package using the optimal parameters of the best current model with the unconstrained extrapolation method to reduce unrealistic estimations (Guevara & León-Paniagua, 2019). Ten bootstrap replicates were implemented, and the final mean models were projected to the same calibration area with a logistic output format.
3 RESULTS
3.1 Phylogenetic inference
The final matrix comprised 3044 bp. Partition analysis showed that the best scheme was divided into three subsets (−lnL = 18,197.86, AIC = 37,863.72): Cytb with GTR + I + G, COI with GTR + I + G, and Fgb-I7 + Adh-I2 with TVM + I + G as nucleotide substitution model.
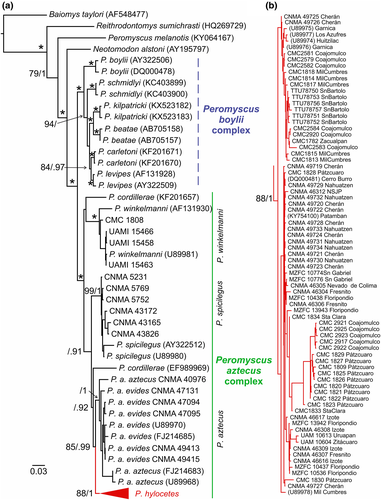
The topology of the ML and BI trees was similar showing high values of nodal support (Figure 1a). The P. boylii complex and P. aztecus complex were highly supported sister clades with 99% bootstrap support (Bt) and a posterior probability (PP) of 1. The phylogenetic interrelationships among the species within the P. aztecus complex received low support values. One disagreement between the analyses was the position of P. cordillerae. In the ML phylogeny (Figure 1a), one of the samples was recovered as the sister lineage of the P. winkelmanni, whereas the other sample was sister to a clade formed by P. aztecus and P. hylocetes. The BI concurred with the non-monophyly of P. cordillerae; however, in this analysis, the position was uncertain recovering each sample in polytomies at different places of the tree. The subspecies P. a. aztecus and P. a. evides, although recovered in a single clade, did not show reciprocal monophyly. Peromyscus hylocetes was recovered as a well-supported monophyletic group (88% Bt, 1 PP), but the phylogenetic structure within this clade was weak; its internal nodes easily collapsed scoring low Bt and PP values (Figure 1b).
3.2 Intraspecific genetic differentiation and migration rates
Comparisons between P. hylocetes and the remaining species of the P. aztecus complex showed that the lowest K2p genetic distance was with P. a. evides (0.36 for Fgb-I7), whereas the highest was with P. winkelmanni (9.44 for Cytb; Table 1). Intraspecific analyses for P. hylocetes encompassed a total of 77 samples. The dataset was cut to the first 811 pb of the Cytb because only two specimens from GenBank had the complete gene. The concatenated matrix consisted of 2697 pb from which 272 were segregating sites (Table 2). The markers with most segregating sites, haplotypes, and haplotype diversity were Cytb, COI, Fgb-I7, and Adh-I2 respectively, whereas for nucleotide diversity were Cytb, COI, Adh-I2, and Fgb-I7 (Table 2).
| Species | Between groups | Within groups | ||||||
|---|---|---|---|---|---|---|---|---|
| Cytb | COI | Fgb-I7 | Adh-I2 | Cytb | COI | Fgb-I7 | Adh-I2 | |
| P. boylii complex | 9.49 | 1.42 | 1.34 | 0.49 | 5.46 | – | 0.38 | 0.22 |
| P. aztecus aztecus | 7.07 | 4.98 | 1.22 | 0.68 | 2.73 | 1.88 | 1.27 | – |
| P. aztecus evides | 6.28 | 2.70 | 0.36 | 1.75 | 2.30 | 0.13 | 0.40 | 2.52 |
| P. cordillerae | 7.92 | – | 1.83 | 1.98 | 7.34 | – | – | – |
| P. hylocetes | – | – | – | – | 4.10 | 2.33 | 0.17 | 0.30 |
| P. spicilegus | 8.19 | 3.11 | 1.10 | 0.63 | 2.61 | 0.13 | 0.82 | 0.53 |
| P. winkelmanni | 9.44 | 1.23 | 1.83 | 0.71 | 1.03 | – | 2.38 | – |
Note
- The gene sequences used correspond to cytochrome b gene (Cytb), cytochrome c oxidase subunit 1 gene (COI), intron 7 of the beta fibrinogen polypeptide gene (Fgb-I7), and intron 2 of the alcohol dehydrogenase gene (Adh-I2).
| Cytb | COI | Fgb-I7 | Adh-I2 | Concatenated | |
|---|---|---|---|---|---|
| Number of sequences | 77 | 60 | 66 | 67 | 77 |
| Pair bases length | 811 | 657 | 663 | 566 | 2697 |
| Segregating sites | 99 | 87 | 26 | 27 | 272 |
| Adenine proportion | 20.6 | 25.8 | 19.6 | 47.3 | 25.5 |
| Cytosine proportion | 43.7 | 29.5 | 43.2 | 16.5 | 34.9 |
| Guanine proportion | 6.7 | 21.7 | 20.1 | 15.2 | 14.2 |
| Thymine proportion | 29 | 23.0 | 17.1 | 21.0 | 25.4 |
| Number of haplotypes | 67 | 39 | 25 | 29 | 77 |
| Haplotype diversity | 0.9948 | 0.9763 | 0.8778 | 0.8575 | 1 |
| Nucleotide diversity | 0.0401 | 0.0366 | 0.0024 | 0.0041 | 0.0249 |
Note
- Molecular markers were cytochrome b gene (Cytb), cytochrome c oxidase subunit 1 gene (COI), intron 7 of the beta fibrinogen polypeptide gene (Fgb-I7), and intron 2 of the alcohol dehydrogenase gene (Adh-I2).
Tess3r analysis suggested the existence of three likely genetic clusters. Ancestry coefficients spatially interpolated onto the map showed that the first genetic cluster encompasses samples from western TVB (west cluster), the second cluster samples from central TVB (east cluster), and the third cluster samples from Pátzcuaro (Figure 2a,b). The inferred percentage of individuals with an ancestry proportion higher than 0.75 for each cluster was: 87.8% for the west cluster, 52% for the east cluster, and 43.75% for the Páztcuaro cluster. Therefore, the Páztcuaro cluster showed the highest admixture.
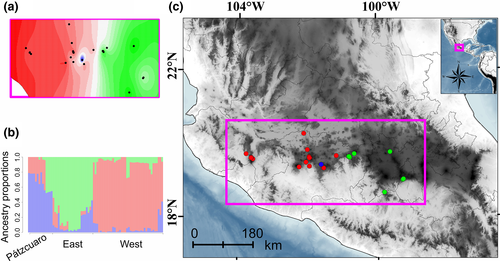
The BPEC for Cytb haplotypes showed that two phylogeographic clusters exist (PP >0.9, low uncertainty; Figure 3a). Localities from the west TVB at the center of the Michoacán state were assigned to the first phylogeographic cluster. The second phylogeographic cluster encompassed samples from the east of Michoacán to the north of Morelos (east geographic distribution). The likely ancestral haplotype was assigned to the locality of Zacualpan at the south-center of the TVB (Figure 3a). The environmental variation showed a slight separation between the two phylogeographic clusters along with the first principal component (Figure 3b). Due to the uncertainty and low ancestry proportions obtained in the Tess3r results, the Pátzcuaro cluster was merged within the west cluster for AMOVA. The results revealed significant structure at all levels with the highest variation among localities (Fst = 0.67, p < 0.001; Table 3). The K2p genetic distances between and within the genetic clusters (west, east) for each marker were Cytb 5.5% (2.13%, 3.96%), COI 3.12% (0.76%, 3.16%), Fgb-I7 0.18% (0.17%, 0.18%), and Adh-I2 0.31% (0.3%, 0.3%).

| Source of variation | df | Sum of squares | Variance components | Percentage of covariation | Fixation index |
|---|---|---|---|---|---|
| Among genetic clusters | 1 | 0.2258273 | 0.0053026 | 32.98967 | Fct = 0.3299 |
| Among localities | 24 | 0.4846616 | 0.0055354 | 34.43806 | Fst = 0.6743 |
| Localities within genetic clusters | 51 | 0.2670128 | 0.0052355 | 32.57227 | Fsc = 0.5139 |
| Total | 76 | 0.9775017 |
Note
- Localities were divided into west and east TVB genetic clusters. All values were significant at a p < 0.001.
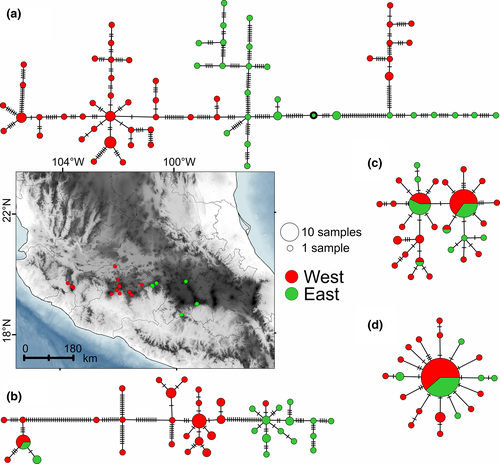
The haplotype networks for Cytb (Figure 4a) and COI (Figure 4b) showed few shared haplotypes and several mutational changes. In the Cytb, COI, and Fgb-I7 networks (Figure 4c), multiple haplogroups were visible, whereas the Adh-I2 network displayed a star pattern (Figure 4d). In general, the two previously inferred genetic clusters could be identified in the networks, but with several shared haplotypes. Migration analyses reached stationarity in each parameter and every chain (average correlation >0.93 and ESS >2000). Results showed asymmetric gene flow between clusters: from west to east was 43.28 (0–83.83; 95% confidence interval) and from east to west 20.85 (0–57.83) individuals per generation. The effective population size with a 95% confidence interval was 0.015 (0.008–0.018) for the east cluster, whereas for the west cluster was 0.045 (0.038–0.051).
3.3 Ecological niche modeling and spatial connectivity
A total of 49 different records with complete geographic information were obtained, and after thinning, 38 were used as training data to construct the niche model. Sequencing and revised specimens confirmed 80% of the localities, whereas the remaining 20% were by published records. After thinning from GBIF, 27 records were obtained and used as testing data (Table S3). Nine variables were selected after the correlation analysis of the environmental variables: bio1 (annual mean temperature), bio3 (isothermality), bio4 (temperature seasonality), bio7 (temperature annual range), bio12 (annual precipitation), bio14 (precipitation of driest month), bio15 (precipitation seasonality), bio18 (precipitation of warmest quarter), and bio19 (precipitation of coldest quarter).
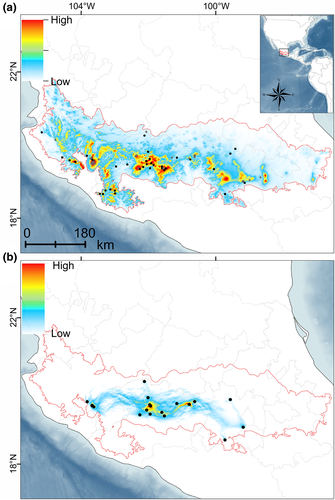
Based on the levels of model complexity, the best-fit model has a RM = 1, linear feature class with a pROC p < 0.001, OR = 0.037, and AICc = 1425.37. The main environmental variables that contributed to the current niche model were bio 1 (42.7%), bio15 (16.1%), bio4 (13%), and bio19 (11.4%). The geographic projection of the model showed that the highest suitability values were distributed in several fragmented areas along the center-west TVB (Figure 5a). The current climatic spatial resistance surfaces model revealed high spatial connectivity among localities at the center TVB (Figure 5b). Finally, the isolation by resistance test showed a positive and significant relationship (r = 0.299, p = 0.01).
3.4 Past inferences: divergence time, historical demography, and paleodistribution
The chronogram (Figure 6) was consistent with the ML and BI phylogenies. The main difference was that P. aztecus was not recovered as monophyletic. The sample P. cordillerae (KF201657) was the sister taxon of P. aztecus (FJ214683–U89968) within the P. aztecus clade. The chronogram showed that the split between the complexes P. aztecus and P. boylii occurred 4.13 Mya with a 95% HPD that ranged from 3.14 to 5.16 Mya. The divergence between P. winkelmanni and its sister clade (P. spicilegus + P. codillerae + P. aztecus + P. hylocetes) was dated 3.58 Mya (2.67, 4.5). The split between P. spicilegus and P. codillerae + P. aztecus + P. hylocetes occurred 3.42 Mya (2.41, 4.44). Finally, the divergence time between P. hylocetes and P. aztecus occurred 3.18 Mya (2, 4.44).
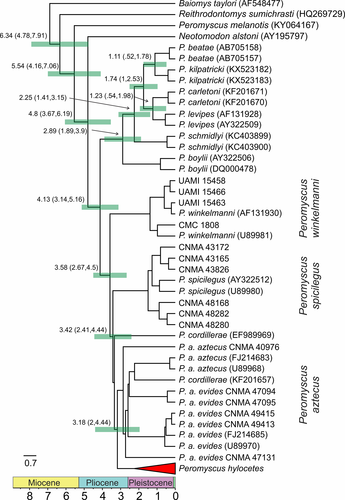
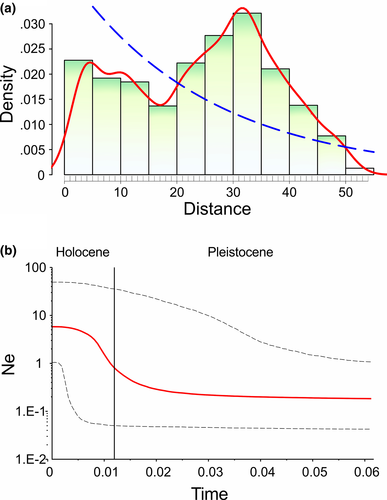
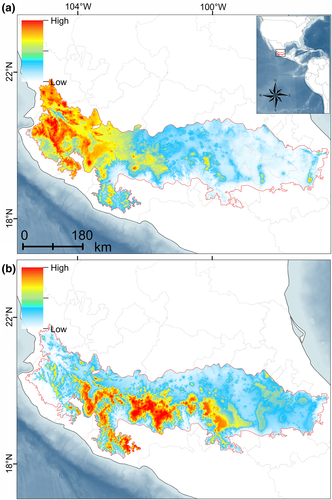
Historical demography analyses revealed negative values for the concatenated dataset and for each marker (Table 4). Fu's F was ranged between −1.99 (COI) and −38.23 (Fgb-I7). This result was consistent with the other descriptors used (i.e., Tajima´s D and R2). The mismatch distribution analysis displayed a multimodal distribution (Figure 7a) and the Bayesian Skyline Plot showed a constant equilibrium of effective population size with a demographic increment around the transition between the late Pleistocene and early Holocene (Figure 7b). The past projections of the ecological niche model revealed that the high suitability values during the LIG were distributed at the west TVB with scarce isolated areas at the southwest of the TVB (Figure 8a). Finally, during the LGM the high suitability values were extended at the center-west of the TVB (Figure 8b).
| Test | Cytb | p | COI | p | Fgb-I7 | p | Adh-I2 | p | Concatenated | p |
|---|---|---|---|---|---|---|---|---|---|---|
| Fu's F | −34.63 | −1.99 | −38.23 | −19.27 | −17.71 | |||||
| Tajima's D | 0.47 | NS | 0.94 | NS | −3.29 | *** | −1.96 | 0.05* | −3.29 | *** |
| R 2 | 0.121 | NS | 0.1324 | NS | 0.0737 | NS | 0.0388 | 0.039* | 0.0054 | *** |
- Abbreviation: NS, no significative.
4 DISCUSSION
4.1 Phylogenetic relationships and interspecific genetic differentiation
Our phylogenetic reconstruction was congruent with the evolutionary interrelationships proposed in previous studies (Platt et al., 2015; Sullivan et al., 2017). Therefore, the paraphyly of the genus Peromyscus in our study was not a surprise. Neotomodon alstoni was recovered more closely related to the Peromyscus core clade than the sample of P. melanotis. Other phylogenetic and taxonomic problems also exist at inter and intraspecific ranks within the Peromyscus core clade. For instance, rearrangements for the Peromyscus aztecus species complex have been suggested (Kilpatrick et al., 2021; Sullivan et al., 1997). As stated by Kilpatrick et al. (2021), further studies are needed to clarify its phylogenetic relationships and taxonomic status of the P. aztecus subspecies (i.e., P. a. aztecus, P. a. evides, and P. a. oaxacensis). Individuals of P. hylocetes were recovered in a clade that was closely related to P. aztecus. These two groups were separated by a long genetic gap (6.2%–7.9% genetic distance) supporting their species-level rank (Baker & Bradley, 2006), common ancestry, and their distinctiveness as separate evolutionary units (Kilpatrick et al., 2021; Sullivan et al., 1997).
The high genetic diversity (Table 2) and the genetic structure observed in P. hylocetes showed a similar pattern to that reported for other vertebrates within the TVB. For example, in rattlesnakes of the Crotalus triseriatus species complex, several species were recognized for the west, center, and east regions of the TVB (Blair et al., 2019). The salamander Pseudoeurycea robertsi also showed two divergent lineages at the center of the TVB (Sunny, Duarte-deJesus, et al., 2019). The woodrat Nelsonia goldmani has sympatric localities and similar geographic distribution at the center-west TVB (León-Tapia, 2021; León-Tapia & Cervantes, 2019). The genetic study of N. goldmani revealed that the diversification of four lineages distributed at the western-central TVB occurred mainly by climatic changes during the late Miocene–Pliocene (León-Tapia & Cervantes, 2021). Although the lineages of N. goldmani and the two clusters of P. hylocetes have the same geographic limit at the east of the Michoacán state, the genetic differentiation in P. hylocetes is apparently more recent (Pleistocene or Holocene).
In P. hylocetes, the geographic limit between the two genetic clusters was set at the east of the Michoacán state. This limit is congruent with the geologic sector between the central and eastern TVB (Ferrari et al., 2012). Nonetheless, the geological origin of these sectors is older than our estimated split time of the genetic clusters observed in P. hylocetes. The estimated age for the western TVB is ~10 Mya, for the central TVB is 11–17.6 Mya, and for the eastern TVB is 14.5–17.5 Mya (Ferrari et al., 2012). Therefore, other factors such as climatic changes could have fragmented the habitat of P. hylocetes populations in a relevant time to lead to their differentiation. The low genetic differentiation between the two clusters of P. hylocetes suggests that its ongoing differentiation is a recent process. It is likely that the climatic fluctuations during the Quaternary, in which over 20 extensive glaciations have occurred (Augustin et al., 2004; Ehlers & Gibbard, 2011), have had an important role in such process. This hypothesis is supported by the abrupt climatic changes that took place in the past in central Mexico. Dry conditions at the late Pleistocene and early Holocene favored the appearance of grassland vegetation, followed by moist conditions during the middle Holocene with the recovery of the forests (Ortega et al., 2010). Also, evidence of soil composition changes suggested high volcanic activity during these periods and likely transformed the landscapes in central Mexico (Ibarra-Arzave et al., 2019). Therefore, climatic and landscape changes likely modified the habitat of P. hylocetes restricting genetic flow during dry periods with the fragmentation of forests by the dominance of grasslands and reconnecting it during the moist periods after the recovery of the forests.
High altitude specialists tend to be more vulnerable to the reduction of their habitat due to anthropogenic effects. At the TVB, there is a constant environmental pressure by overexploitation of natural resources, pollution, land-use changes, introduction of exotic species, fires, pests, and diseases (Arriola-Padilla et al., 2014). These can be particularly harsh for small mammals like the mice endemic to the TVB, such as the Nelsonia goldmani (León-Tapia & Cervantes, 2019), Reithrodontomys chrysopsis (Barrera-Moreno et al., 2011), Neotomodon alstoni (Williams et al., 1985), Peromyscus kilpatricki (Bradley et al., 2016), P. purepechus (León-Tapia et al., 2020), and P. hylocetes (Sullivan et al., 1997).
Areas along the TVB with high environmental suitability for P. hylocetes were scarce and isolated. Unfortunately, these suitability areas are reduced by the current anthropogenic activity (Arriola-Padilla et al., 2014). Other vertebrate studies have revealed similar patterns of isolation throughout the TVB. The rattlesnake C. triseriatus (Sunny et al., 2019), the alligator lizard Barisia imbricata (Sunny et al., 2017), and the garter snake Thamnophis scalaris (González-Fernández et al., 2018) are well-documented examples. Our analyses suggest low connectivity maintained mainly at the center of the TVB in small areas that could represent scarce routes for gene flow. Previous reports stated that P. hylocetes inhabits at altitudes above 2300 m; however, our sampling demonstrates that P. hylocetes can be found from medium elevations (>1288 m) at western TVB to high mountains (3000 m) at central TVB. Certainly, along such altitudinal gradient, there are habitats in which this species fulfills its biological requirements. Vázquez et al. (2004) recorded a high population density of P. hylocetes in a mature cloud forest at western TVB. These rodents mainly fed on fruits, leaves, stems, seeds, and a few insects. It is likely that populations of P. hylocetes had more interconnection in medium mountainous areas through cloud forest than at coniferous forest along the TVB. Definitely, populations at high altitudes are susceptible to the effect of past events that could have produced a barrier to gene flow between the west and the east clusters.
4.2 Biogeographic and demographic history
The phylogenetic relationship between P. hylocetes and P. aztecus, as well as the ancestral Cytb haplotypes located at southern TVB inferred by BPEC, suggest the possibility that these two species diverged from a common ancestor during the Pliocene-Pleistocene at the north-central part of the Sierra Madre del Sur and central-south TVB. During this period several climate changes are reported (Lourens et al., 2004) resulting in the modification from forest to open vegetation in North America (Prescott et al., 2018). Independent studies for other taxa have hypothesized similar biogeographic histories that occurred during the Pliocene-Pleistocene; for example, the harvest mouse Reithrodontomys sumichrasti (Hardy et al., 2013), rattlesnakes of the C. triseriatus species complex (Blair et al., 2019), tree frogs of the Sarcohyla bistincta species complex (Caviedes-Solis & Leaché, 2018), the bark beetle Dendroctonus mexicanus (Anducho-Reyes et al., 2008), the mistletoe Psittacanthus calyculatus (Pérez-Crespo et al., 2017), and the weevil Trichobaris soror (De-la-Mora et al., 2015).
Once P. hylocetes diverged from P. aztecus, their populations settled at the TVB going through a period of demographic stability until its effective population size increased during the late Pleistocene and early Holocene. The paleodistribution showed that during the warmer periods at the LIG (120–140 ka), the environmental suitability for P. hylocetes was restricted to areas at the west of the TVB suggesting that at that zone its populations encountered more stable environmental conditions. More recently, during the LGM (21 ka) the environmental suitability extended to the central TVB. This hypothetic biogeographic scenario would imply that at warm periods during the Pleistocene, populations of P. hylocetes were isolated due to forest fragmentation after its altitudinal rise. Later, during cold periods bridges between forest patches were formed favoring the migration between the west and central areas of the TVB. This hypothesis is supported by the large gene flow from western to eastern TVB, and the expansion contractions of forests documented in Mexico during this period (Domínguez-Vázquez et al., 2019; Mastretta-Yanes et al., 2018; Ramírez-Barahona & Eguiarte, 2013).
Nonetheless, alternative hypotheses have been proposed. Sullivan et al. (1997) concluded that P. hylocetes and P. aztecus probably have had contact in the TVB due to a geographic expansion of their populations during the Pleistocene cold periods. However, the conclusions drawn from the suitability and the ecological niche models projected to the past suggest that that is unlikely. More samples of P. a. aztecus and P. a. evides are needed to contrast those hypotheses. At least, a good representation from the west distribution of P. a. aztecus in the Sierra Nevada volcanic range is required. Apparently, this mountainous system restricted the contact between P. a. aztecus and P. hylocetes due to the volcanic activity registered from 1.8 Mya that continues until today (Macías et al., 2012). Additional samples from central-west SMS will provide further support to the hypotheses proposed in this study. The environmental suitability connection between western TVB and central-west SMS regions is supported by the paleodistribution of P. hylocetes during the LIG and LGM. The complex geologic and climatic histories, together with the environmental heterogeneity of the TVB and SMS make it difficult for reaching precise inferences of the evolutionary history for endemic taxa. This highlights the need for more studies within those complex biogeographic regions to improve our understanding of the processes and mechanisms that occurred in the past that might explain the high endemism currently observed in the central highlands of Mexico.
5 CONCLUSIONS
Phylogenetic relationships among P. aztecus group were low supported, and the subspecies of P. aztecus were not recovered as monophyletic. Samples of P. hylocetes were monophyletic and two main genetic clusters from west and east of its distribution in the TVB were detected, likely correlated with the fragmented environmental suitability and low connectivity found along the TVB. Gene flow occurred largely from the west to the east cluster, probably throughout small mountainous areas with cloud forests at the center of the TVB. The diversification of P. hylocetes occurred during the Pliocene-Pleistocene after diverging from P. aztecus at the northern-central part of Sierra Madre del Sur and central-southern TVB. After their divergence, populations of P. hylocetes settled at the TVB with demographic stability until the effective population size increased during the late Pleistocene and early Holocene. We hypothesized that after colonizing the TVB, P. hylocetes expanded its distribution to the western TVB. With the climatic oscillations during the Pleistocene, populations were restricted at western TVB during the warmer periods, and displacements occurred during colder periods from the west to central TVB on several episodes. The climatic changes and volcanic activity across the central TVB during the late Pleistocene and early Holocene likely modified the habitat of P. hylocetes restricting gene flow that resulted in the genetic differentiation observed in this study.
ACKNOWLEDGMENTS
We thank Consejo Nacional de Ciencia y Tecnología for the graduate student grant (346631 to MALT). To curators and associated people for their help for accessing the museum specimens, data, and tissue samples: Fernando A. Cervantes, Julieta Vargas Cuenca and Yolanda Hortelano Moncada at the Colección Nacional de Mamíferos (CNMA); Livia León Paniagua and Giovani Hernández Canchola at the Museo de Zoología "Alfonso L. Herrera" (MZFC); Francisco Xavier González Cózatl at the Colección de Mamíferos of the Universidad Autónoma de Morelos (CMC); José Ramírez Pulido and Noé González Ruiz at the Colección de Mamíferos of the Universidad Autónoma Metropolitana Unidad Iztapalapa (UAMI); Caleb Phillips and Robert Bradley at the Mammal Collection of the Texas Tech University (TTU). We also thank to Janet Nolasco-Soto for providing logistic support for laboratory work and Cristina Bárcenas Pazos for sequencing some PCR products at the Instituto de Ecología AC. Celia López-González, Francisco Ornelas, Andrés Lira, and Francisco González-Cózatl provided comments that improved the manuscript. This manuscript was enriched with the final comments and observations of Ulyses Pardiñas and one anonymous reviewer.
APPENDIX 1
GenBank accession numbers for the sequences used in this study
| Species | Cytb | COI | Fgb-I7 | Adh-I2 | Museum | Catalog |
|---|---|---|---|---|---|---|
| Baiomys taylori | AF548477 | JQ600060 | AY274213 | KT361508 | ||
| Reithrodontomys sumichrasti | HQ269729 | JQ600068 | HQ269793 | JX910117 | ||
| Peromyscus melanotis | KY064167 | JF446185 | FJ214711 | FJ214673 | ||
| Neotomodon alstoni | AY195797 | JF446056 | AY274202 | AY994210 | ||
| Peromyscus boylii | DQ000478 | – | AY274208 | AY994227 | ||
| Peromyscus boylii | AY322506 | – | – | AY994226 | ||
| Peromyscus beatae | AB705158 | ROM 98290 | FJ214696 | AY994223 | ||
| Peromyscus beatae | AB705157 | ROM 98291 | – | AY994222 | ||
| Peromyscus kilpatricki | KX523183 | – | – | – | ||
| Peromyscus kilpatricki | KX523182 | – | – | – | ||
| Peromyscus carletoni | KF201671 | – | – | – | ||
| Peromyscus carletoni | KF201670 | – | – | – | ||
| Peromyscus levipes | AY322509 | – | FJ214707 | KT361507 | ||
| Peromyscus levipes | AF131928 | – | – | AY994224 | ||
| Peromyscus schmidlyi | KC403900 | – | FJ214718 | KT318182 | ||
| Peromyscus schmidlyi | KC403899 | – | – | AY994229 | ||
| Peromyscus winkelmanni | U89981 | – | – | – | ||
| Peromyscus winkelmanni | AF131930 | – | FJ214721 | FJ214678 | ||
| Peromyscus winkelmanni † | MW264520 | MW265530 | MW264606 | – | CMC | 1808 |
| Peromyscus winkelmanni † | MW264521 | – | MW264607 | – | UAMI | 15458 |
| Peromyscus winkelmanni † | MW264522 | – | MW264608 | – | UAMI | 15463 |
| Peromyscus winkelmanni † | MW264523 | – | MW264609 | – | UAMI | 15466 |
| Peromyscus spicilegus | AY322512 | ROM ASK1888 | FJ214719 | AY994234 | ||
| Peromyscus spicilegus | U89980 | ROM ASK1889 | – | AY994233 | ||
| Peromyscus spicilegus † | MW264524 | MW265531 | MW264610 | MW264685 | CNMA | 48282 |
| Peromyscus spicilegus † | MW264525 | MW265532 | MW264611 | MW264686 | CNMA | 48280 |
| Peromyscus spicilegus † | MW264526 | MW265533 | MW264612 | MW264687 | CNMA | 48168 |
| Peromyscus spicilegus † | MW264527 | – | – | MW264688 | CNMA | 43165 |
| Peromyscus spicilegus † | MW264528 | – | MW264613 | – | CNMA | 43826 |
| Peromyscus spicilegus † | MW264529 | – | – | – | CNMA | 43172 |
| Peromyscus cordillerae | KF201657 | – | FJ214714 | FJ214675 | ||
| Peromyscus cordillerae | EF989969 | – | – | – | ||
| Peromyscus aztecus aztecus | FJ214683 | ROM YHM230 | FJ214695 | – | ||
| Peromyscus aztecus aztecus | U89968 | ROM 100795 | – | – | ||
| Peromyscus aztecus aztecus † | MW264530 | – | MW264614 | MW264689 | CNMA | 40976 |
| Peromyscus aztecus evides | FJ214685 | – | FJ214700 | FJ214670 | ||
| Peromyscus aztecus evides | U89970 | – | – | – | ||
| Peromyscus aztecus evides † | MW264531 | MW265534 | MW264615 | MW264690 | CNMA | 47131 |
| Peromyscus aztecus evides † | MW264532 | MW265535 | MW264616 | MW264691 | CNMA | 47094 |
| Peromyscus aztecus evides † | MW264533 | MW265536 | MW264617 | MW264692 | CNMA | 47095 |
| Peromyscus aztecus evides † | MW264534 | MW265537 | MW264618 | MW264693 | CNMA | 49415 |
| Peromyscus aztecus evides † | MW264535 | MW265538 | MW264619 | MW264694 | CNMA | 49413 |
| Peromyscus hylocetes | DQ000481 | – | FJ214705 | AY994235 | TTU | TK45309 |
| Peromyscus hylocetes | U89974 | – | – | – | ZTNHC | CWK 2040 |
| Peromyscus hylocetes | U89975 | – | – | – | ZTNHC | CWK 2781 |
| Peromyscus hylocetes | U89976 | – | – | – | ZTNHC | CWK 2035 |
| Peromyscus hylocetes | U89977 | – | – | – | ZTNHC | CWK 4229 |
| Peromyscus hylocetes | U89978 | – | – | – | ZTNHC | CWK 2853 |
| Peromyscus hylocetes | KY754100 | – | – | – | LSUMZ | 25106 |
| Peromyscus hylocetes † | MW264536 | MW265539 | MW264620 | MW264695 | CNMA | 46305 |
| Peromyscus hylocetes † | MW264537 | MW265540 | MW264621 | MW264696 | CNMA | 46304 |
| Peromyscus hylocetes † | MW264538 | MW265541 | MW264622 | MW264697 | CNMA | 46306 |
| Peromyscus hylocetes † | MW264539 | MW265542 | MW264623 | MW264698 | CNMA | 46307 |
| Peromyscus hylocetes † | MW264540 | MW265543 | MW264624 | MW264699 | MZFC | 10437 |
| Peromyscus hylocetes † | MW264541 | MW265544 | MW264625 | – | MZFC | 10438 |
| Peromyscus hylocetes † | MW264542 | MW265545 | MW264626 | MW264700 | MZFC | 10536 |
| Peromyscus hylocetes † | MW264543 | MW265546 | MW264627 | MW264701 | MZFC | 13942 |
| Peromyscus hylocetes † | MW264544 | MW265547 | MW264628 | MW264702 | MZFC | 13943 |
| Peromyscus hylocetes † | MW264545 | – | MW264629 | MW264703 | CNMA | 46308 |
| Peromyscus hylocetes † | MW264546 | MW265548 | MW264630 | MW264704 | CNMA | 46309 |
| Peromyscus hylocetes † | MW264547 | MW265549 | MW264631 | MW264705 | CNMA | 46616 |
| Peromyscus hylocetes † | MW264548 | MW265550 | MW264632 | MW264706 | CNMA | 46617 |
| Peromyscus hylocetes † | MW264549 | MW265551 | MW264633 | MW264707 | MZFC | 10774 |
| Peromyscus hylocetes † | MW264550 | MW265552 | MW264634 | MW264708 | MZFC | 10776 |
| Peromyscus hylocetes † | MW264551 | MW265553 | MW264635 | MW264709 | CNMA | 46312 |
| Peromyscus hylocetes † | MW264552 | – | MW264636 | – | UAMI | 10604 |
| Peromyscus hylocetes † | MW264553 | – | MW264637 | – | UAMI | 10613 |
| Peromyscus hylocetes † | MW264554 | MW265554 | MW264638 | MW264710 | CNMA | 49734 |
| Peromyscus hylocetes † | MW264555 | MW265555 | MW264639 | MW264711 | CNMA | 49733 |
| Peromyscus hylocetes † | MW264556 | MW265556 | MW264640 | MW264712 | CNMA | 49732 |
| Peromyscus hylocetes † | MW264557 | MW265557 | MW264641 | MW264713 | CNMA | 49731 |
| Peromyscus hylocetes † | MW264558 | MW265558 | – | MW264714 | CNMA | 49729 |
| Peromyscus hylocetes † | MW264559 | MW265559 | MW264642 | MW264715 | CNMA | 49730 |
| Peromyscus hylocetes † | MW264560 | MW265560 | MW264643 | MW264716 | CNMA | 49720 |
| Peromyscus hylocetes † | MW264561 | MW265561 | MW264644 | MW264717 | CNMA | 49721 |
| Peromyscus hylocetes † | MW264562 | – | MW264645 | MW264718 | CNMA | 49722 |
| Peromyscus hylocetes † | MW264563 | MW265562 | – | – | CNMA | 49723 |
| Peromyscus hylocetes † | MW264564 | – | MW264646 | MW264719 | CNMA | 49724 |
| Peromyscus hylocetes † | MW264565 | – | MW264647 | MW264720 | CNMA | 49725 |
| Peromyscus hylocetes † | MW264566 | MW265563 | – | MW264721 | CNMA | 49726 |
| Peromyscus hylocetes † | MW264567 | MW265564 | MW264648 | MW264722 | CNMA | 49727 |
| Peromyscus hylocetes † | MW264568 | MW265565 | – | MW264723 | CNMA | 49719 |
| Peromyscus hylocetes † | MW264569 | MW265566 | – | MW264724 | CNMA | 49728 |
| Peromyscus hylocetes † | MW264570 | MW265567 | MW264649 | MW264725 | CMC | 1833 |
| Peromyscus hylocetes † | MW264571 | MW265568 | MW264650 | MW264726 | CMC | 1834 |
| Peromyscus hylocetes † | MW264572 | MW265569 | MW264651 | MW264727 | CMC | 1809 |
| Peromyscus hylocetes † | MW264573 | MW265570 | MW264652 | MW264728 | CMC | 1820 |
| Peromyscus hylocetes † | MW264574 | MW265571 | MW264653 | MW264729 | CMC | 1821 |
| Peromyscus hylocetes † | MW264575 | MW265572 | MW264654 | MW264730 | CMC | 1822 |
| Peromyscus hylocetes † | MW264576 | MW265573 | MW264655 | MW264731 | CMC | 1823 |
| Peromyscus hylocetes † | MW264577 | MW265574 | MW264656 | MW264732 | CMC | 1825 |
| Peromyscus hylocetes † | MW264578 | – | MW264657 | MW264733 | CMC | 1826 |
| Peromyscus hylocetes † | MW264579 | MW265575 | MW264658 | MW264734 | CMC | 1827 |
| Peromyscus hylocetes † | MW264580 | MW265576 | MW264659 | MW264735 | CMC | 1828 |
| Peromyscus hylocetes † | MW264581 | – | MW264660 | MW264736 | CMC | 1829 |
| Peromyscus hylocetes † | MW264582 | MW265577 | MW264661 | MW264737 | CMC | 1830 |
| Peromyscus hylocetes † | MW264583 | MW265578 | MW264662 | MW264738 | CMC | 1813 |
| Peromyscus hylocetes † | MW264584 | MW265579 | MW264663 | MW264739 | CMC | 1814 |
| Peromyscus hylocetes † | MW264585 | MW265580 | MW264664 | MW264740 | CMC | 1815 |
| Peromyscus hylocetes † | MW264586 | MW265581 | MW264665 | MW264741 | CMC | 1817 |
| Peromyscus hylocetes † | MW264587 | MW265582 | MW264666 | MW264742 | CMC | 1818 |
| Peromyscus hylocetes † | MW264588 | MW265583 | MW264667 | MW264743 | TTU | 78750 |
| Peromyscus hylocetes † | MW264589 | MW265584 | MW264668 | MW264744 | TTU | 78751 |
| Peromyscus hylocetes † | MW264590 | MW265585 | MW264669 | MW264745 | TTU | 78752 |
| Peromyscus hylocetes † | MW264591 | MW265586 | MW264670 | MW264746 | TTU | 78753 |
| Peromyscus hylocetes † | MW264592 | MW265587 | MW264671 | MW264747 | TTU | 78756 |
| Peromyscus hylocetes † | MW264593 | MW265588 | MW264672 | MW264748 | TTU | 78757 |
| Peromyscus hylocetes † | MW264594 | MW265589 | MW264673 | MW264749 | CMC | 1782 |
| Peromyscus hylocetes † | MW264595 | MW265590 | MW264674 | MW264750 | CMC | 2579 |
| Peromyscus hylocetes † | MW264596 | MW265591 | MW264675 | MW264751 | CMC | 2581 |
| Peromyscus hylocetes † | MW264597 | MW265592 | MW264676 | MW264752 | CMC | 2582 |
| Peromyscus hylocetes † | MW264598 | MW265593 | MW264677 | MW264753 | CMC | 2583 |
| Peromyscus hylocetes † | MW264599 | MW265594 | MW264678 | MW264754 | CMC | 2584 |
| Peromyscus hylocetes † | MW264600 | MW265595 | MW264679 | MW264755 | CMC | 2917 |
| Peromyscus hylocetes † | MW264601 | – | MW264680 | MW264756 | CMC | 2920 |
| Peromyscus hylocetes † | MW264602 | – | MW264681 | MW264757 | CMC | 2921 |
| Peromyscus hylocetes † | MW264603 | MW265596 | MW264682 | MW264758 | CMC | 2922 |
| Peromyscus hylocetes † | MW264604 | MW265597 | MW264683 | MW264759 | CMC | 2923 |
| Peromyscus hylocetes † | MW264605 | MW265598 | MW264684 | MW264760 | CMC | 2925 |
- Molecular markers are cytochrome b gene (Cytb), cytochrome c oxidase subunit 1 gene (COI), intron 7 of the beta fibrinogen polypeptide gene (Fgb-I7), and intron 2 of the alcohol dehydrogenase gene (Adh-I2). Symbol † indicate samples sequenced for this study, otherwise, sequences were downloaded from GenBank. Additional information is available in Table S1.




