Systematics and evolution of Kibramoa Chamberlin 1924 (Araneae: Plectreuridae) from the California Floristic Province
Abstract
The eight-eyed haplogyne spider family Plectreuridae Simon, 1893 is one of the oldest of spider families, currently comprising two genera (Kibramoa and Plectreurys), 30 extant species and one subspecies. Plectreuridae have not been rigorously examined since 1958, with only three new species added to Plectreurys. This study revisited a subset of taxa from the genus Kibramoa using a combined morphological and multilocus phylogenetic analysis. Species distribution modeling (SDM) was used to investigate the current and paleo distributional patterns of the genus Kibramoa throughout the biodiverse region of the California Floristic Province (CFP). A reexamination of genitalic morphology reflected cryptic species, yet multilocus Bayesian and maximum likelihood analyses of mitochondrial (COI, 16S) and nuclear (ITS1, 28S) markers consistently suggested several divergent lineages. A time-calibrated phylogeny indicated that the most recent common ancestor of Kibramoa appeared in the Mid-Miocene and continued to diversify throughout the Plio-Pleistocene. Xerophilic Kibramoa, as inferred by SDM, had a much wider distribution during the Mid-Holocene, when climate was at its warmest. This body of work uncovered novel findings regarding the evolution of plectreurids, provided the first phylogeny of the family, and demonstrated similar biogeographic patterns displayed in other CFP taxa.
1 INTRODUCTION
The spider family Plectreuridae Simon, 1893 is one of the oldest spider lineages, having diversified as early as the Middle Jurassic (~165 MYA) (Selden & Huang, 2010). These short-sighted, primitive hunting spiders are a small, relictual group that live a semi-sedentary life in silken tubular retreats. This eight-eyed haplogyne family currently comprises two genera (Kibramoa, Chamberlin, 1924 and Plectreurys, Simon, 1893). Plectreurys includes 23 extant species; Kibramoa includes seven species and one subspecies. They are known from western North America, throughout Central America and the Caribbean islands, into northern South America, with a suggested center of origin in the Southwest United States (Gertsch, 1958). Gertsch (1958) published the only comprehensive revision of Plectreuridae, re-describing five species and describing 20 new species and one new subspecies for a total of 25 species.
Since Gertsch's treatment (1958), Plectreuridae has not been rigorously examined, with only a handful of new species descriptions added to Plectreurys (Alayón, 1993; Alayón-García & Víquez-Núñez, 2011; García, 2003). Little is understood regarding factors driving the evolution of the family, which is distributed throughout several regions of high endemism (i.e., California Floristic Province [CFP], Caribbean Islands, Mesoamerica). Because of this, and because they were presumably widespread in the Mesozoic (Selden, 2014; Selden & Huang, 2010), it is likely that the family's true diversity is grossly underestimated. Additionally, no phylogenetic hypotheses exist for plectreurid species.
The genus Kibramoa is distinguished from Plectreurys by the absence of a tibial copulatory spur on leg I and longer, spiny legs. Three of the eight species (including subspecies) of Kibramoa are described from monotypic specimens. According to Gertsch, several species are very similar in morphology, but he indicated that the males may have distinctive leg spine formulas (Gertsch, 1958, p. 33). A key to the male Kibramoa species was provided (Gertsch, 1958), but a study of comparative morphology for females is unavailable.
Kibramoa are relatively large spiders with body size ranging from 5 to 10 mm, and their current distribution is restricted to arid habitats of the southwestern Nearctic. Body coloration can range from chestnut brown to black with an orange-reddish hue. During the day, the preferred habitats of Kibramoa are typically clandestine (i.e., deep in crevices or under several layers of rocks) with surroundings fringed with sporadic lines of silk, but at night they can be completely exposed, presumably for mating or feeding. Females build a protective orb-shaped cell around the egg sac and display partial maternal care by remaining inside the orb until a few days before the hatching of spiderlings. Individuals are commonly solitary. Female plectreurids of both genera are typically domiciliary, and dispersal by “ballooning” has yet to be documented in Kibramoa.
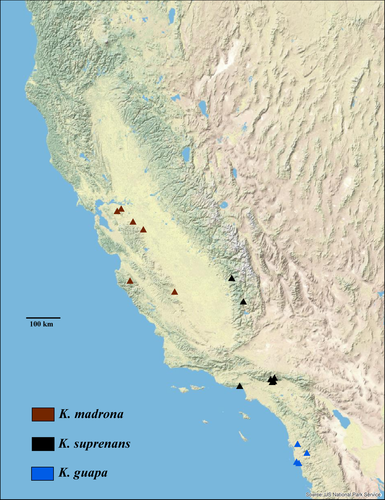
The CFP is recognized for its complex geological history and unique Mediterranean climate—giving rise to species radiations, high biological diversity and endemism (Calsbeek, Thompson, & Richardson, 2003; Myers, Mittermeier, Mittermeier, Fonseca, & Kent, 2000). Moreover, widespread species in the CFP display fine-scaled genetic differences among populations (Calsbeek et al., 2003; Martínez-Solano, Jockusch, & Wake, 2007, Satler, Carstens, & Hedin, 2013). This study focused on populations of Kibramoa from throughout the CFP including three widespread species: K. guapa, K. suprenans and K. madrona (Figure 1). Given the suggested age of Plectreuridae, the distribution of Kibramoa in the CFP, and the significant genetic structure of many exemplar Californian organisms (Feldman & Spicer, 2006; Martínez-Solano et al., 2007), the species diversity of Californian Kibramoa is hypothesized to be underestimated. Moreover, the phylogeography of Kibramoa can be used as a model for understanding the unique geographic history that has influenced population structure of organisms with a similar natural history.
To test the hypothesis that there of more than three species in the CFP, the first objective was to re-examine genital morphology to identify possible species differences and use newly acquired molecular data to test Gerstch's hypothesis that only three distinct species exist in the CFP. We used the resulting phylogeny to examine rates and evolutionary processes of diversification; fortuitously, plectreurids have a considerable fossil record, with four putative fossil representatives (Penney, 2009; Selden, 2014; Selden & Huang, 2010; Wunderlich, 2008). Because Gertsch (1958) suggested that morphological data may be inadequate for confidently assigning species limits, a third objective of our study was to use geographic information systems (GIS) and species distribution modeling (SDM) to test the three-species hypothesis, asking the following: (a) What are the distributional limits of the Kibramoa populations? (b) What are the suitable habitats for California Kibramoa? and (c) Do these data suggest a multitude of separately evolving lineages rather than three?
2 MATERIALS AND METHODS
2.1 Taxon sampling
The study included an assemblage of natural history collections from the California Academy of Sciences (CAS), the National Museum of Natural History (Smithsonian), and the American Museum of Natural History. Natural history collections are invaluable because of the historical data that are coupled with the specimen. The data from museum specimens used in this study comprise over 800 exemplars of Plectreuridae, >200 of which pertained to the genus Kibramoa. Specimens stored in ethanol <90% EtOH were used for habitat modeling and morphological study.
Collection dates from museum specimens indicated that plectreurid adults can be found in the CFP throughout the year. Fieldwork locations were selected based on previous collection events, and fieldwork was primarily conducted in the fall and spring months in 2014–2016. All freshly collected specimens were intended for molecular studies and thus preserved in 100% EtOH. As a conservative approach, freshly collected specimens were not assigned to species. Additional specimens for molecular study were supplemented from the San Diego State Terrestrial Arthropod collection.
2.2 Morphology
Gertsch indicated that species-delimiting somatic characters were abstruse, and a dichotomous key to females is lacking. With an augmented sample size, genitalic structures of males and females for all unique populations of adult Kibramoa specimens of both sexes were digitally imaged and examined qualitatively.
Epigyna and palpi were removed from specimens. Epigyna were placed in enzymatic contact lens solution to dissolve proteins and extraneous tissues (Álvarez-Padilla & Hormiga, 2007). Standardized images were obtained using multiple focal plane sources taken with a Leica MZ16, which were then combined with auto-montage Leica Application Suite (LAS) software.
2.3 Molecular phylogenetics
Genomic DNA for species representatives of Kibramoa and Plectreurys (total of 78 specimens; Table S2) were extracted from 2 to 3 legs using the Qiagen DNeasy® extraction protocol for animal tissues. Two mitochondrial (cytochrome c oxidase I (COI) and 16S mitochondrial ribosomal RNA (16S)) and two nuclear (28S ribosomal RNA and internal transcribed spacer I (ITS1)) gene regions were amplified by polymerase chain reaction using primers available in Table S1. DNA sequences can be found on GenBank (MN596215-MN596273, MN603330-MN603376, MN603223-MN603266, MN626641, MN626642, and MN6032367-MN603329) and in Table S2. Gene matrices are available on TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S25130 and http://purl.org/phylo/treebase/phylows/study/TB2:S25131). Amplicons were purified and immediately Sanger-sequenced using an ABI 3130 in the forward and reverse directions at the Center for Comparative Genomics laboratory (CAS, CA). Sequences were assembled into contigs, trimmed, and analyzed in GENEIOUS PRO R7 software (http://www.geneious.com/). Alignments for COI, ITS1, and 16S were performed using the MUSCLE (Edgar, 2004) package in GENEIOUS. For the 28S alignment, the option Q-INS-i in MAFFT v7 (Katoh, Kuma, Toh, & Miyata, 2005) was used because it considers the secondary molecular structure. Final gene alignments were input into ALTER (Glez-Peña, Gómez-Blanco, Reboiro-Jato, Fdez-Riverola, & Posada, 2010) to collapse sequences to haplotypes.
For Bayesian phylogenetic inference methods, best-fit models of molecular evolution were selected using bModeltest (Bouckaert et al., 2014) which implements reversible rate jump Bayesian estimation of substitution models, simultaneously estimating rate heterogeneity and invariant sites (Table 1). Individual mitochondrial and nuclear matrices, as well as a concatenated matrix were assembled in SequenceMatrix (Vaidya, Lohman, & Meier, 2011), and gene trees were reconstructed using BEAST v1.7 and MrBayes 3.2.6 (Drummond, Suchard, Xie, & Rambaut, 2012; Ronquist et al., 2012) on the Cyber Infrastructure for Phylogenetic Research (CIPRES; Miller, Pfeiffer, & Schwartz, 2010). MrBayes analyses were run for 50 million generations and sampled every 5,000 generations. The standard deviation of split frequencies was evaluated after each analysis to assess convergence.
| Locus | #Seq | BP length | #Informative | jModelTest/PartitionFinder (BIC) | bModeltest |
|---|---|---|---|---|---|
| CO1 | 59 | 697 | 259 | TrN+I+G, F81+I+G, TrN+G | TN93+I+G, K81+I+G,TIM+G |
| 16S | 44 | 439 | 144 | TIM2 + G | TN93 + G |
| 28S | 63 | 810 | 78 | TrN+G | TN93+G |
| ITS1 | 49 | 714 | 187 | K80+G | TN93+G |
The programs jModeltest 2 (Darriba, Taboada, Doallo, & Posada, 2012) and PartitionFinder (Lanfear, Calcott, Ho, & Guindon, 2012) were used and the Bayesian information criterion (BIC) was implemented to select nucleotide substitution models for subsequent maximum likelihood phylogenetic analyses (Table 1). Maximum likelihood analyses were performed using RAxML-HPC BlackBox 8.0.24 (Stamatakis, Hoover, & Rougemont, 2008), also run on CIPRES, and the GTRGAMMA model was used for each of six partitions: 1) ITS1, 2) 28S, 3) 16S, 4–6) CO1 (1st codon position + 2nd codon position + 3rd codon position).
Coalescent-based analyses and time calibrations (Yang & Rannala, 2006) were implemented in BEAST 2 (Bouckaert et al., 2014). Each gene was initially analyzed using an uncorrelated lognormal (UCLN) relaxed clock model (Drummond, Ho, Phillips, & Rambaut, 2006) using the bModeltest template. Analyses were run for 20 million generations sampled every 2,000 generations. Parameter estimates associated with the clock rate and substitution models for each gene were then examined using Tracer v1.6.0 (Rambaut, Suchard, Xie, & Drummond, 2014) to assess convergence, appropriate burn-in, and to verify that all effective sample size (ESS) values were over 200. If the 95% highest posterior density of the coefficient of variation for any individual gene was close to zero, a strict molecular clock was specified for that locus (Drummond et al., 2006) and ESS values were reexamined after subsequent analyses. Initial BEAST analyses using recovered nucleotide substitution models from bModeltest for all six partitions resulted in low ESS values, so all partitions were subsequently analyzed using a simple HKY+G model (Grummer & Bryson, 2014; Leavitt, Starrett, Westphal, & Hedin, 2015) and a lognormal relaxed clock.
2.4 Divergence estimation
Divergence times were estimated using three alternative scenarios. First, a fossil calibration date considering the youngest plectreurid fossil from Miocene Dominican amber (16 Ma; Penney, 2009) was specified as a tMRCA prior under a Birth–Death model with plectreurid taxa recovered for all four loci treated as a crown group. A gamma-distributed prior was assigned with an alpha of 2 and a beta of 30. Second, the same approach was applied as the first scenario, but estimated with a lognormal distribution with a mean value of 20, a standard deviation of 0.75 and offset of 16 Ma. Third, a well-calibrated araneomorph COI clock rate (ucld.mean = 0.0199 or 3.98% per million years; Bidegaray-Batista & Arnedo, 2011) was specified implementing a normal distribution prior and standard deviation of 0.0045. LOGCOMBINER (Rambaut & Drummond, 2014) was used to combine tree files and also to apply a resample frequency to those tree files to obtain a product of 10,000 trees. TREEANNOTATOR (Rambaut & Drummond, 2014) was used to reconstruct a maximum clade credibility (mcc) tree.
2.5 Habitat modeling
Kibramoa specimens were identified to genus and isolated for study. Nearly all older collection data maintain a daunting legacy of a lack of geographic coordinates, often specifying the locality only by the name of the nearest town. This specifies a large, ill-defined area that may cover a variety of habitat types (Crawford, 1983). Therefore, specimens with limited locality information were recorded but not considered for further study.
To couple locality descriptions with geographic coordinates, the point-radius method for georeferencing was used because it encompasses the full locality description and its associated uncertainties (Wieczorek, Guo, & Hijmans, 2004). GEOlocate (Rios & Bart, 2010), a platform for georeferencing biodiversity collections data, was used to reduce the ambiguity of collection descriptions (e.g., CA: Sierra Nevada Mountains, 2.5 mi S, 2.5 mi W of Big Pine). GEOlocate uses a series of lookups and displacement calculations to estimate geographic coordinates in decimal degrees using the World Geodetic Survey 1984 (WGS84) datum and associated uncertainty. Localities with ambiguous or dubious locality strings [e.g., table 1 of Wieczorek et al. (2004) (i.e., “Isla Boca Brava?”, “presumably central Chile,” or “San Jose, Mexico”)] were not considered for georeferencing.
Most collection events maintain considerable sampling bias toward easily accessible areas. To reduce bias, spatial filtering was applied. Previous studies suggest that filtered data sets lead to better quality data for calibration (Anderson, 2012; Hijmans, 2012; Veloz, 2009) and unfiltered data can overfit the modeling resulting in falsely inflated estimates in performance (Radosavljevic & Anderson, 2014). Only unique localities were maintained, and the dataset was filtered to a 1 km2 radius.
Species distribution modeling for Kibramoa was conducted with MaxEnt version 3.3.2k (Phillips, Anderson, & Schapire, 2006; Phillips & Dudík, 2008). MaxEnt represents an approximation of a species' realized niche (Hutchinson, 1957) by estimating the probability of a potential distribution of maximum entropy, based on presence data and environmental data (Phillips et al., 2006). It is commonly employed, has proven to outperform other methods, is least sensitive to small sample sizes, and recently has been playing an increasingly important role in evolutionary research (Dekarabetian, Burns, Starrett, & Hedin, 2016; DiDomenico & Hedin, 2016; Elith, 2006; Hernandez, Graham, Master, & Albert, 2006; Leache et al., 2009; Raxworthy, Ingram, Rabibisoa, & Pearson, 2007; Raxworthy et al., 2003; Stockman & Bond, 2007; Townsend Peterson, Papeş, & Eaton, 2007; Wiens & Graham, 2005).
In total (including fresh samples), 132 presence records (25% of which were used as testing points; Table S3) were utilized to estimate current (1960–1990), Mid-Holocene (~6,000 years ago), and Last Glacial Maxima (LGM; ~22,000 years ago) distributions, respectively (see Table S3). Bioclimatic variables (19 in total) and elevation for current environmental data were downloaded at 30-arc seconds (WorldClim v1.4; Hijimans, Cameron, Parra, Jones, & Jarvis, 2005), while the Community Climate System Model (CCSM4) datasets for Mid-Holocene and LGM were used at a downscaled resolution of 2.5 arc-min (WorldClim v1.4; Hijimans et al., 2005).
In addition to sampling bias, correlations between environmental variables can also lead to false predictions and overparameterization (Boria, Olson, Goodman, & Anderson, 2014; Rissler, Hijmans, Graham, Moritz, & Wake, 2006). To identify and remove redundant variables, bioclimatic information for all environmental variables for each collection locality was collected. The 19 bioclimatic variables were added as layers to ArcMap in ArcGIS Desktop v 10.3 (ESRI), and values were extracted using the “Extract Values to Points” function. A Pearson's correlation coefficient test was implemented in Rstudio (RStudio Team, 2014). Two different correlation thresholds were considered to remove correlated variables. First, bioclimatic variables >0.75 were removed following the methods of Rissler et al. (2006), resulting in a model that includes six bioclimatic variables (Table 2) and elevation. Second, bioclimatic variables >0.90 were recognized as highly correlated and consequently extracted (Jezkova, Olah-Hemmings, & Riddle, 2011), resulting in a second model which includes 13 bioclimatic variables and elevation (Table 2).
| ID | Variable | >0.75 Pearson's Correlation |
|---|---|---|
| BIO2 | Annual Mean Temperature | ✓ |
| BIO3 | Mean Diurnal Range | ✓ |
| BIO4 | Temperature Seasonality | |
| BIO5 | Max Temperature of Warmest Month | |
| BIO6 | Minimum Temperature of Coldest Month | ✓ |
| BIO8 | Mean Temperature of Wettest Quarter | |
| BIO9 | Mean Temperature of Driest Quarter | |
| BIO10 | Mean Temperature of Warmest Quarter | |
| BIO12 | Annual Precipitation | ✓ |
| BIO14 | Precipitation of Driest Month | |
| BIO15 | Precipitation Seasonality | ✓ |
| BIO17 | Precipitation of Driest Quarter | |
| BIO18 | Precipitation of Warmest Quarter |
Due to the interest in general distributional limits, MaxEnt models were run under a liberal fixed threshold (tenth percentile training presence) versus a conservative threshold (minimum training presence threshold) (Papeş & Gaubert, 2007). The tenth percentile training presence also yields better performance than the minimum training presence when identifying areas of endemism (Escalante, Rodríguez-Tapia, Linaje, Illoldi-Rangel, & González-López, 2013).
ASCII Maxent result files were transferred to ArcMap and were converted to Boolean presence/absence maps according to the tenth percentile minimum training logistic threshold using the Raster Calculator function in the Spatial Analyst toolbox. To identify regions of overlap between the two paleoclimate results, binary maps of Mid-Holocene and LGM were added together also using the Raster Calculator.
3 RESULTS
3.1 Molecular phylogenies of 16S, COI, ITS1, and 28S
Four gene MrBayes and RaxML analyses recovered similar topologies resulting in five well-supported lineages (Figure 2). Exemplars of K. madrona and K. guapa used in this study were named as such due to proximity to the type locality. Three other clades were informally named based on geographic region and used herein (Figure S1). First, a well-supported (posterior probability (PP) = 1/100 bootstrap value) Northern Sierran clade including members distributed from Shasta County near Crystal Lake to Tuolumne and Calaveras Counties, crossing the Central Valley to Stanislaus County at Del Puerto Canyon located at the foothills of the South Coast Ranges. Next, sister to the Northern Sierran Clade, a strongly supported (PP = 0.96/46 bootstrap value) South Coast Ranges clade with a distribution west of the San Andreas Fault from San Benito County in Pinnacles National Park. The sister relationship of the Northern Sierran clade and the South Coast Range clade, together forms a higher-level Trans-Valley clade. Following, the Trans-Valley clade, is a well-supported (PP = 1/79 bootstrap value) K. madrona clade, sister to the Northern Sierran + South Coast Ranges clades, that comprises an exemplar from the madrona type locality (Monterey County: Hastings Natural History Reservation), a specimen from Santa Clara County near Alum Rock Canyon Park, and a specimen from Butte County near Lake Oroville. Conversely, the RaxML analysis produced adequate support (79%) for the Butte County taxon (G1501) to be nested within K. madrona. Finally, a Kingston Range lineage is recovered with a PP of 0.92 and a bootstrap value of 63; this clade is distributed in Inyo County near the Kingston Range, located east of the Sierra Nevada Mountains. Lastly, a basal K. guapa lineage is recovered from San Diego County near Otay Mesa (PP of 0.98/ bootstrap value of 49).
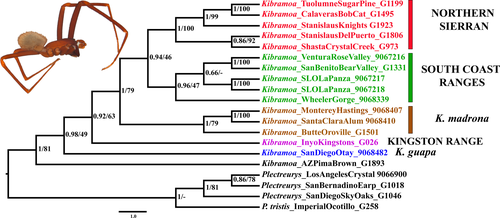
The combined COI, 16S, and 28S Bayesian analysis allowed a complete taxon sampling of 30 taxa (seven outgroup and 23 ingroup taxa) and offered an alternative phylogenetic hypothesis with six lineages (Figure S2). A consistent Northern Sierran clade is supported, but the placement of the Butte County specimen (G1501) shifted from the K. madrona clade to the Northern Sierran clade. The South Coast Ranges clade was split into two putative clades (Figure S2): South Coast Ranges and South Coast Ranges 2. A Trans-Valley clade was again recovered. In this case, results were consistent with the topology recovered in Figure 2 in which the Northern Sierran clade is sister to the South Coast Ranges clade (comprising the Trans-Valley clade). The South Coast Ranges 2 clade, sister to the Northern Sierran and South Coast Range clades, consisted of San Benito County near Pinnacles National Monument, Monterey County near White Oaks Campground, extending south to Ventura County near Rose Valley Falls. Next, there is a divergent Kingston Range lineage that branches from the Northern Sierran and both South Coast clades, with strong support. A K. madrona clade was maintained in this analysis with an increased taxon set ranging from Santa Clara County to the foothills of the Southern Sierras in Kern County near Caliente. Finally, a K. guapa clade with two representatives from San Diego County was also well supported.
3.2 Kibramoa morphology
Although a considerable amount of work has contributed to advancing the knowledge of haplogyne genitalia (Pérez-González, Rubio, & Ramírez, 2016), plectreurids have received little to no attention. Exemplars of both sexes representing 24 populations were dissected and imaged. The genitalia of both sexes are morphologically conserved, and specimens could not confidently be assigned to species based on morphology alone.
Palpal variation is tenuous (Figure S4), displaying a spherical bulb and a long, thin coiled embolus—with the exception of a specimen from the Fort Irwin area in San Bernardino, California (Figure S4o). In contrary to the Kibramoa examined here, the representative from the Fort Irwin population displayed an elongated spherical bulb with a shortened spiraled embolus. Among the other geographically unique populations, the spherical bulb and length/curvature of the embolus was shown to vary subtly in shape, yet significant morphological patterns that corroborate main clades recovered in any phylogeny (Figures 2 and 3; Figures S2 and S3) are absent.
Female genitalic morphology offered some insight for species designations (Figure S5). Exemplars representing the K. madrona clade reveal a similar shape of the spermathecae being distally elongated in exemplars representing the K. madrona clade (Figure S5m–7p). The comparative morphology of representative populations within this clade suggests a synapomorphy for this clade. Morphological characters of the female genitalia are puzzling due to the high morphological variability in the exemplars examined and the lack of currently existing homologs for comparison. Understanding Kibramoa female genitalia will benefit from more detailed studies of plectreurids, and well as of other Synspermiata (Michalik & Ramírez, 2014), particularly other families of the “lost tracheae clade,” that is, Diguetidae, Pacullidae, and Tetrablemmidae (Griswold & Ramírez, 2017; Ramírez, 2000; Wheeler et al., 2017).
3.3 Divergence time estimation
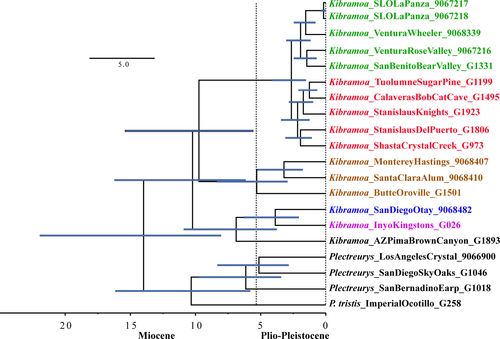
Results from the four gene BEAST analyses are summarized in Table 3. First, the gamma-distributed Miocene amber fossil calibration estimates suggest that the split between North American Plectreurys and Kibramoa is relatively old with median time to most recent common ancestor (tMCRA) being ~27 Ma during the Oligocene (33.9–22.9 Ma). A major genetic lineage radiation appears to have occurred in the Miocene (23.0–5.2 Ma), with the diversification of K. guapa, the Kingston Range lineage, and K. madrona. The Trans-Valley clade appears to have emerged in the very late Miocene (5.7 Ma), but very close to the demarcated Plio-Pleistocene geological age of 5.3 Ma. The Trans-Valley clades in these analyses favored a west to eastward colonization of the Sierras, since the South Coast clades are dated to be older than Northern Sierran clade (Table 3).
| Divergences | DR Fossil calibration (16 Ma) gamma distribution | DR Fossil calibration (16 Ma) lognormal distribution | Bidegaray-Batista and Arnedo (2011) rate |
|---|---|---|---|
| tMRCA Plectreurys + Kibramoa | 25.68 (17.09–41.07) | 20.19 (16.08–30.31) | 13.99 (8.04–21.95) |
| tMRCA Kibramoa guapa + Kingston Range | 10.26 (6.3–17.09) | 6.87 (4.66–10.77) | 3.88 (2.08–6.29) |
| tMRCA Kibramoa madrona + Trans-Valley | 15.95 (10.29–25.71) | Monophyletic relationship not supported | 9.74 (5.28–15.42) |
| tMRCA Kibramoa madrona | 10.72 (6.86–17.53) | w/o G1501 3.58 (2.34–5.65) | 5.3 (2.94–8.37) |
| tMRCA Trans-Valley | 5.46 (3.42–9.00) | 3.26 (2.34–5.09) | 2.65 (1.53–4.15) |
| tMRCA South Coast Ranges | 4.52 (2.84–7.50) | 2.78 (1.83–4.29) | 1.94 (1.18–3.04) |
| tMRCA Northern Sierran | 3.97 (2.47–6.61) | 2.57 (1.74–3.99) | 2.17 (1.25–3.43) |
Second, the lognormal distributed Miocene amber fossil calibration suggested later lineage divergence than that of the gamma distributed, with most lineages diversifying in the Plio-Pleistocene. The cladogenic event between North American Plectreurys and Kibramoa occurred ~20.8 Ma during the Miocene. Nodal estimates for a K. madrona + Trans-Valley clade were not recovered in the chronogram (Table 3; Figure S6), where K. madrona is recovered as the most basal lineage. Another inconsistency was the position of G1501 (Butte Co.), which was placed as sister to the Trans-Valley clade with PP of 1. The Trans-Valley clades in this analysis favored an east to west directionality because Northern Sierran clades are an older clade than the Coast Ranges clade.
Finally, the third divergence estimate scenario using the calibrated clock results also favored a later Plectreurys and Kibramoa diversification at ~13.99 Ma (summarized in Table 3). The chronogram produced an identical topology to the first divergence scenario, and most cladogenesis of major lineages occurred during the Miocene (Figure 3), with a recent Trans-Valley clade at ~2.65 Ma.
3.4 Habitat modeling
According to the 132 presence coordinates, Kibramoa favors climatic conditions that include an average mean annual temperature ranging from 7.9°C in Riverside County near Andreas Canyon to the warmest at 22.2°C in Mariposa County in Yosemite National Park; average mean annual precipitation ranged from 10.2 cm in San Bernardino County near Fort Irwin to 150.3 cm in Shasta county near Crystal Lake; elevation ranged between 2 m above sea level in Contra Costa County, 2 mi SW Marsh Creek Springs, to 1,650 m in Mariposa County in Yosemite National Park.
Current SDM utilizing 14 bioclimatic variables and elevation results communicated an area under the curve (AUC) score of 0.809 for the Test samples gain and 0.899 for Training and a Logistic Threshold Value of 0.310 under the tenth percentile threshold. LGM results produced AUC scores of 0.994 for Test and 0.995 for Training with a 0.341 Logistic Threshold Value. Ultimately, the Mid-Holocene SDM presented a 0.994 Test and Training, accompanied with a 0.341 threshold value.
Combined SDMs identified a large climatically stable area and possible refugia after the last major glaciation event (Figure 4a). During the LGM, habitats were latitudinally favorable toward the southern part of North America. Consecutively, the warming climate during the Mid-Holocene suggested a northward shift into present Oregon territory. During the Mid-Holocene, SDMs recovered most of San Joaquin Valley as unsuitable habitat with suitable habitat latitudinally near the San Francisco Bay Area. Habitats east of the Sierras (>1,000 m during this time, see Discussion) are mostly unsuitable for Kibramoa populations.
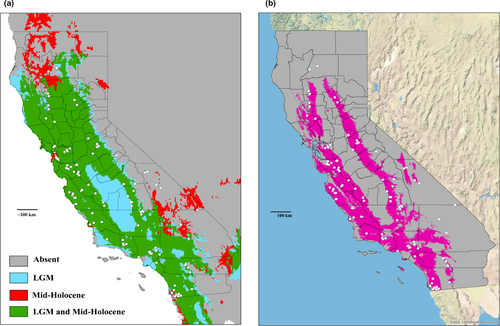
Current SDM results display a smaller distribution than historically predicted (Figure 4b), with several outlier localities in Shasta and San Bernardino Counties. Visible geographic barriers include the Peninsular Ranges in Southern California, the High Sierra separating the western populations from populations in the White Mountains of Inyo County, the Salinian Block, the San Francisco Bay, and on a larger scale—the Central Valley.
4 DISCUSSION
4.1 Kibramoa systematics
As Gertsch (1958) originally revealed, many Kibramoa species were difficult to demarcate based on morphological characters. A qualitative re-assessment of the genitalia of representatives of K. guapa, K. suprenans, K. madrona, and unique geographically distributed populations were insufficient to confidently diagnose species using morphology. Some exceptions include a male specimen from San Bernardino Co. near Fort Irwin that includes a dramatically shorter embolus (Figure S4o) and female genitalia that seemed to have morphological consistency in the spermathecae that delimited K. madrona (Figure S4m–p), but further quantitative methods are warranted to assess certainty of these patterns.
Molecular phylogenetics reveal that there are indeed more than three independently evolving lineages, where the type distribution of K. madrona allude to a morphological species complex since molecular analyses suggest many independent lineages within this region (Figures 2 and 3; Figure S3). Both four gene and three gene topologies suggest the Central Valley, historically, was suitable habitat which allowed for panmixia between populations east and west of the Central Valley. With a changing climate, the Central Valley currently is a vicariant barrier for Kibramoa populations and is majorly unsuitable for Kibramoa populations.
Gertsch hypothesized that the center of origin for this family was in the southwestern United States (Gertsch, 1958). All molecular analyses can provide additional evidence for this hypothesis as populations from the southern United States are being recovered as basal and northern populations of plectreurids are more recently diverged (Figures 2 and 3; Figures S2 and S3).
Current SDMs suggest that Kibramoa favors a geographic distribution that circumscribes the Central Valley, does not extend further North into Oregon, or East into the Mojave Desert. Extrinsic barriers include the Peninsular Ranges in Southern California, the High Sierra separating the western populations from populations in the White Mountains of Inyo County, the Salinian Block, and the San Francisco Bay. The separation of populations by these geographic barriers has shaped the evolution of Kibramoa resulting in the genetic divergences displayed in all phylogenetic analyses.
4.2 California historical biogeography and evolution
Much of California was inundated for most of the Cenozoic and the Great Valley of California (Central Valley) was a large sea (Bartow, 1991; Schierenbeck, 2014). The Central Valley experienced periods of submerging fluctuations, especially in the San Joaquin Valley or San Joaquin Embayment (Bartow, 1991). A San Joaquin marine breach occurred in the southern part of the San Joaquin valley near the end of the Oligocene (Bandy & Arnal, 1969) until a final retreat at the end of the Pliocene—synchronous with tectonic events surrounding the valley and diminution of sea level (Bartow, 1991).
The Miocene was arguably the zeitgeist of California in which this region endured significant geologic and climatic transitions coinciding with major lineage diversification of Californian Kibramoa. The elevation of the Sierra Nevada was greater than 1,000 m, and the well-recognized Mediterranean climate of the CFP stabilized (Schierenbeck, 2014). The breadth of Monterey County existed as islands (Yanev, 1980), and a very apparent strike-slip of the San Andreas Fault suggested an increased rate of fault displacements during this time (Harding, 1976). Thick accumulation of siliceous sediment near the Monterey Formation, submerged channels in Monterey Bay, and the absence of equivalent channels near the San Francisco Bay indicate that the area around Monterey Bay (especially Salinas and Pajaro Valleys) was the San Joaquin Valley's predominant drainage outlet (Bartow, 1991; Fairbanks, 1901). The formation of the southern Coast Ranges in Monterey ~5–3 Ma (Hall, 2002; Sims, 1993) closed off the San Joaquin drainage outlet. Preceding this orogeny, the San Joaquin Valley was supplemented by alpine glaciers to ultimately form the ephemeral freshwater lake, Lake Corcoran, near the mid-Pleistocene (Bartow, 1991). The exodus of Lake Corcoran approximately coincided with the formation of the modern Carquinez Strait and the San Francisco Bay drainages approximately 0.6 Ma (Sarna-Wojcicki et al., 1985). The SDMs estimated at the LGM suggest that the San Joaquin Valley might have been suitable for Kibramoa around ~22,000 years ago but became inhospitable at the time of the Mid-Holocene (Figure 4a).
The San Joaquin Embayment did not extend far north (Figure 4a; Davis & Coplen, 1989), and there is no direct evidence that pertains to paleochannels crossing the Sierra Nevada south of the San Joaquin River (Wakabayashi, 2013). Genetic similarity of freshwater ichthyofauna and geological evidence from the Sacramento drainage, San Francisco Bay, and Monterey Bay suggests an ancient river assemblage connection (Aguilar & Jones, 2009; Dupre, 2000; Fairbanks, 1901). This paleochannel network may be a reasonable explanation for the recovery of the Butte County (G1501) specimen nested within taxa from the southern Coast Ranges (Figure 2; Figure S3) and for Shasta County G973 and western Stanislaus County G1806 to show close relationships (Figure 2). Ancestral populations of Kibramoa could have passively rafted from the northern latitudes to newly available habitat in the Coast Ranges, as hypothesized by Hedin, Starrett, and Hayashi (2013) for turret spiders. Prior to 5 Ma, much of the Coast Range terrestrial habitat was unavailable. Divergence estimates for Kibramoa are harmonious with geological uplift estimates of the Coast Ranges (5–3 Ma) with the K. madrona clade (that includes Butte County G1501), which diverged from the coast range taxa ~5.3 Ma, and a most recent clade Trans-Valley clade (that includes South Coast clade and Northern Sierran clade) with an estimated cladogenesis event at ~2.65 Ma (Table 3; Figure 3). The Fork and Yuba Rivers that drain into the Sacramento River have recently been calibrated to have developed around 3 Ma (Wakabayashi, 2013), but these were originally proposed to have originated 5 Ma (Wakabayashi & Sawyer, 2001). Following this range calibration of the Fork and Yuba Rivers, and assuming that the Sacramento River was a main drainage source during the Pliocene, then this additional geological evidence corroborates the passive rafting hypothesis.
Many exemplar dispersal limited salamanders, such as Batrachoseps attenuatus (Martinez-Solano et al., 2007), Ensatina eschscholtzii xanthoptica (Kutcha, Parks, Mueller, & Wake, 2009), and Aneides lugubris (Lapointe & Rissler, 2005) and mygalomorph spiders like Calisoga (Leavitt et al., 2015), Antrodiaetus (Hedin et al., 2013; Starrett & Hedin, 2007), and Aliatypus (Satler et al., 2013) exhibit a disjunct distribution at latitudes near the San Francisco Bay dated to around the Mid-Pleistocene. Additionally, present-day populations of Batrachoseps persist on the valley floor north of the San Francisco Bay (Jockusch & Wake, 2002). Although it is apparent that the San Francisco area was dynamic for many taxa and an obvious genetic leak was present around this time, the directionality shown by vertebrates and invertebrates is discordant. While the salamanders suggest a west to east colonization of habitats, spiders suggest an east to west mode of dispersal. Kibramoa, as suggested by the SDMs, had a much wider distribution during the Mid-Holocene when climate was at its warmest. This distribution is expected because Kibramoa has an affinity for xeric habitats.
This study is the first to examine the evolution and phylogeny of the family Plectreuridae and study provides further evidence that many taxa in the CFP reflect fine-scaled genetic differences and a dynamic evolutionary history, and that, conservation for these habitats should be considered.
ACKNOWLEDGEMENTS
Funding for this project was provided by the American Arachnological Society's Vince Roth Fund to E.G. Thanks to Daniel Palmer, Marshal Hedin, Susan Kennedy, Darrell Ubick and Jeff Hodson for supplementing this study with plectreurid specimens. Thanks to Teta Charernsuk for helping with databasing and imaging specimens. We are grateful to Anna Sellas and Brian Simison for the tools and resources in the CCG lab at CAS. For providing helpful discussions and tips in phylogenetic analyses and approaches to this project, thanks to Shahan Derkarabetian, Kristen Emata, and Sean Harrington. Thanks to Paula E. Cushing for advice and edits to the manuscript. We also recognize CAS Arachnology: Darrell Ubick and Sarah Crews for their undying foundational support, curiosity, and fostering nature.




