First case of parthenogenesis in ladybirds (Coleoptera: Coccinellidae) suggests new mechanisms for the evolution of asexual reproduction
Abstract
Parthenogenesis, the development of unfertilized eggs resulting in the exclusive production of female offspring, is rare in animals relative to sexual reproduction and is mainly reported in invertebrates. It has been hypothesized that polyploidy, hybridization and endosymbiont infections are its major causal events but the mechanisms triggering asexual reproduction remain unclear. Here, we study the proximate causes at the origin of parthenogenesis in the first reported case of asexuality in the Coccinellidae (Coleoptera). The asexual populations were found in the Azores and the Mascarene archipelagos, and were identified as Nephus voeltzkowi Weise, a bisexual species widespread in sub-Saharan Africa. The specimens from both populations are diploid but present different karyotypes and heterozygosities that evoke hybrid origins, commonly associated with parthenogenesis in Coleoptera. However, the close proximity of their genomes (99.8% homology for the complete mitochondrial genome and 99.9% for the complete nuclear ribosomal cluster) together with the congruence between the mtDNA tree and the nuclear tree, and the low heterozygosity levels, suggests that the two populations are not hybrid. We propose that they belong to a single chromosomally polymorphic species undergoing Robertsonian fusions. Furthermore, specimens from both populations are infected with Wolbachia (supergroup B strain), contrary to sympatric bisexual species of the same genus. Although Wolbachia has been shown to induce parthenogenesis in haplodiploid organisms, it has been recently suggested that it could also induce parthenogenesis in hosts with other sex determination systems. Whether chromosome rearrangements and/or Wolbachia infections are post-parthenogenetic events or are at the origin of parthenogenesis still needs to be determined.
1 INTRODUCTION
Asexual reproduction is infrequent in animals, sexual reproduction being the overwhelming trend. The development of unfertilized eggs resulting in the exclusive production of female offspring, usually known as parthenogenesis (Simon, Delmotte, Rispe, & Crease, 2003), is mainly reported in invertebrates (Sinclair, Pramuk, Bezy, Crandall, & Sites, 2010). In insects and other Hexapoda, approximately 1,100 species are thought to reproduce by obligate parthenogenesis (i.e., females lay eggs that all develop into females, without any mating at all) (Normark, 2014).
The prominent hypothesis for the rareness of asexual reproduction is that, although there may be a twofold reproductive advantage in it, by sparing male production, asexual lines cumulate deleterious mutations leading to their long-term extinction. This said, some asexual lines have been present on Earth for millions of years (Heethoff, Norton, Scheu, & Maraun, 2009; Mark Welch, Ricci, & Meselson, 2009; Schön, Rossetti, & Martens, 2009). Therefore, although uni-parental forms of reproduction are generally considered to be disadvantageous, this belief should be qualified: every case of asexuality thrives in a particular ecological context, and is under unique selective processes (Archetti, 2010; Bengtsson, 2009). Several theories discuss the conditions that would allow asexual lines to maintain themselves, to coexist with their sexual counterparts but also to eventually compete and ultimately drive the latter to extinction (Archetti, 2010; Engelstädter, 2017; Rice & Friberg, 2009). The circumstances and mechanisms underlying parthenogenesis but also studies of niche breadth, life histories, competition, predation and parasitism are essential to understanding the evolutionary history of asexual populations. In this study, we are interested in the proximate causes at the origin of parthenogenesis.
Generally, parthenogenesis is considered as developing spontaneously as a result of a mutation within the sexual population or as a consequence of hybridization and/or polyploidy (Bullini, 1994; Kearney, 2005; Lundmark, 2006; Simon et al., 2003). Polyploidy is further subdivided into allopolyploidy and autopolyploidy: in the first case, the chromosomes come from two different species following hybridization, and, in the second, chromosomes result from the same genome (Stenberg & Saura, 2009). Finally, some microorganisms, Wolbachia being the most frequent, can manipulate the reproductive system of arthropods and induce the appearance of parthenogenesis (Werren, Baldo, & Clark, 2008). The precise mechanism at the origin of parthenogenesis and the frequency with which asexuality events occur (the advent in a population of a single clonal line or several lines with different genotypes) lead to different degrees of genetic variability in asexual populations, with important consequences for their evolutionary success. Nevertheless, although there is abundant literature on the subject, the mechanisms triggering asexual reproduction remain unclear (Bullini, 1994; Choleva et al., 2012; Neiman, Sharbel, & Schwander, 2014).
The Coccinellidae is a very diversified family composed of some 6,000 species (Vandenberg, 2002). It has been extensively studied from a systematics, ecological and applied (e.g., biological control, invasion) point of view, both in the field and in the laboratory (see Hodek, van Emden, & Honěk, 2012 for a comprehensive revision). Skewed sex ratios are common in ladybirds and are the result of infections by endosymbionts acting as male killers (Weinert, Tinsley, Temperley, & Jiggins, 2007) but so far, parthenogenesis has never been mentioned for the family. However, during fieldwork in two distantly located biodiversity hot spots, Santa Maria Island—Azores archipelago (Atlantic Ocean, Northern hemisphere) and Reunion Island—Mascarene archipelago (Indian Ocean, Southern hemisphere), we found only females of Nephus voeltzkowi Weise (Weise, 1910), which raises the question of the possible existence of parthenogenesis in the family.
Nephus voeltzkowi belongs to the Scymninae subfamily and is a coccid, particularly mealybug (Homoptera: Pseudococcidae) predator, known in several countries of sub-Saharan Africa and islands in the Indian Ocean (Fürsch, 2007) (Figure 1). Museum collections with N. voeltzkowi from Kenya, the Democratic Republic of Congo, Namibia, South Africa and Madagascar include both male and female specimens (Fürsch, 2007; H. Fürsch pers. obs.). Chazeau, Etienne, and Fürsch (1974) in their fauna of ladybirds from Reunion reported the presence of N. voeltzkowi but did not mention the sex of the collected specimens. Although these authors present an illustration of the genitalia of a N. voeltzkowi male, the first author does not exclude the possibility that the drawing was based on a Madagascar specimen (J. Chazeau pers. com.). In the absence of the original material, we cannot confirm this information. In contrast, it is the first time that N. voeltzkowi is referred for the Azores. Soares, Elias, Resendes, and Figueiredo (2003), in the ladybird fauna of the Azores, had identified it as Nephus hiekei Fürsch but our comparative analyses of external morphology and genitalia (spermatheca) based on Weise (1910), Fürsch (1965, 1967), Chazeau et al. (1974) and Raimundo (1992) show that this was a misidentification.
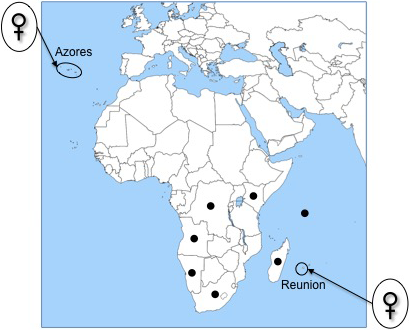
In this work, we carried out a large field survey in the Azores and on Reunion to check the sex ratios of the N. voeltzkowi populations. We also tested for parthenogenetic reproduction in N. voeltzkowi under laboratory conditions. Moreover, we investigated whether individuals from the Azores and Reunion populations are genetically similar, as their similar phenotype would suggest, and tested for their polyploidy and hybrid origin, using both molecular (high-throughput DNA sequencing of the complete mitochondrial genome and of the nuclear ribosomal cluster) and cytogenetic analyses. Finally, we screened the specimens for the presence of bacteria that can manipulate host reproduction, that is Wolbachia, Cardinium, Spiroplasma and Rickettsia, mapping NGS reads from individual ladybirds against the bacterial reference genomes.
2 MATERIAL AND METHODS
2.1 Ladybird field survey
Sampling campaigns in the Azores took place in Santa Maria, São Miguel, Terceira and Pico islands in July 2008 (corresponding to a total of 28 hr of sampling effort) and in São Miguel, São Jorge, Corvo and Flores islands in June 2010 (corresponding to a total of 59 hr of sampling effort). Sampling campaigns in Reunion (Mascarene) were conducted in November 2011 (corresponding to a total of 21 hr of sampling effort) and December 2013 (corresponding to a total of 28 hr of sampling effort). Furthermore, we also had access to all N. voeltzkowi specimens collected from 2006 to 2012 by the Insectarium of Reunion.
Previous field sampling experiences in those regions had shown that ladybird species were rare above 200 m from sea level. Therefore, we concentrated our survey below that altitude. We sampled both natural habitats and organic orchards, insisting on plants infested with mealybugs, Nephus spp main prey. We used a beating tray to inspect trees and bushes, and a sweeping net for lower vegetation.
Individuals were brought to the laboratory, and after dissection, genitalia were observed under a stereomicroscope.
Five specimens of the Santa Maria and five specimens of the Reunion populations were conserved in the scientific collection of the Laboratory Evolution et Diversité biologique—Univ. Toulouse III—Paul Sabatier, under the reference 37Et1B01.
2.2 Experimental test of parthenogenetical reproduction
Around 30 individuals from two populations—Santa Maria Island (Azores) and Reunion island (Mascarene)—were collected in the field in 2008 and 2011, respectively. Separated stock cultures were kept in the laboratory at 23 ± 1°C and a LD 16:8 photoperiod in 175 cm3 plastic boxes containing a piece of corrugated paper. Adults were reared on potato sprouts infested with Planococcus citri Risso (Homoptera: Pseudococcidae). Whenever larvae were present, they were collected and kept in batches of about 20 individuals in new 175 cm3 plastic boxes and reared as described above. When they emerged from pupae, the adults were transferred to new plastic boxes.
In order to determine whether females were able to reproduce without males, 3- to 4th-instar larvae were collected from the stock cultures and were isolated each in a 50-mm Petri dish, and fed ad libitum once a week with P. citri. When adults emerged, they were kept isolated and once a week they were transferred to new 50-mm Petri dishes. The old Petri dishes were then inspected for the presence of offspring. As eggs of N. voeltzkowi are usually laid within the ovisacs of the coccids (lousy structures of wax fibres filled with coccid eggs) and they are very similar in colour, shape and size to coccid eggs, they are difficult to spot. Therefore, we kept the old Petri dishes for 3 weeks and inspected them once a week, under a stereomicroscope, for the presence of ladybird larvae. All Petri dishes were secured with Parafilm to avoid any possible exchange of larvae. We monitored a cohort of 29 Santa Maria adults for 4 months and a cohort of 30 Reunion adults for 2 months, and recorded the rates of parthenogenesis (% of fertile females) as well as the total number of larvae per female. Adults that did not reproduce during the experience were dissected at the end of the experiment for sex determination through genitalia observation.
The all experiment took place at 23 ± 1°C and LD 16:8 photoperiod.
2.3 Santa Maria (Azores) and Reunion (Mascarene) population similarity and tests of hybrid/polyploid origin
2.3.1 Molecular analyses
Field-collected specimens of the two parthenogenetic populations and of Nephus reunioni Fürsch, N. includens (Kirsch) and Nephus sp1 from Portugal, Greece and Reunion, respectively, were preserved in 95% ethanol. Sequences were obtained from two specimens per parthenogenetic population and from one specimen of the remaining species. Total genomic DNA was extracted from the entire individuals using the DNeasy Blood and Tissue Kit (QIAGEN) according to the manufacturer's instructions. The quality and quantity of extracted genomic DNA were evaluated using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and a PicoGreen double-stranded DNA quantitation assay kit (Life Technologies).
The genomic DNA was then sent for library construction and sequencing to the GeTPlaGe core facilities of Genotoul. Libraries were constructed for each specimen using the Illumina TruSeq Nano DNA Sample Prep Kit following the instructions of the supplier (Illumina Inc.). After shearing by ultrasonication with a Covaris M220 (Covaris Ins.), purified fragments were A-tailed and ligated to sequencing indexed adapters. Fragments with an insert size of c. 450 bp were selected with Agencourt Ampure XP beads (Beckman Coulter, In.) and enriched with 8 cycles of PCR before library quantification and validation. The libraries were multiplexed by 24 and hybridized on one lane of Illumina Hiseq 2500 flow cell using the TruSeq PE Cluster Kit v.3, and paired-end reads of 100–150 nucleotides (depending on the specimens; Table 1) were collected using the TruSeq SBS Kit v.3 (200 cycles). Quality filtering was performed by the Consensus Assessment of Sequence and Variation pipeline. The resulting data were stored on the NG6 platform (Mariette et al., 2012), and all computations were performed on the computer cluster of the Genotoul bioinformatics platform.
| Species | Origin | Number of reads | Read length | Full seq. length (Gb) | Mean coverage mitogenome | Mean coverage ribosomal cluster |
|---|---|---|---|---|---|---|
| Nephus sp1. | Reunion | 18,630,752 | 100 | 1.86 | 309 | 938 |
| Nephus reunioni | Portugal | 19,753,538 | 125 | 2.47 | 763 | 1,931.00 |
| Nephus includens | Greece | 24,432,454 | 125 | 3.05 | 1,031.00 | 611 |
| Nephus voeltzkowi | Reunion | 13,435,826 | 100 | 1.34 | 652 | 2,765.00 |
| Nephus voeltzkowi | Reunion | 74,808,866 | 150 | 11.22 | 3,132.00 | 12,090.00 |
| Nephus voeltzkowi | Santa Maria | 12,650,898 | 100 | 1.27 | 216 | 1,210.00 |
| Nephus voeltzkowi | Santa Maria | 349,364,742 | 150 | 52.4 | 8,600.00 | 43,295.00 |
Sequences were assembled de novo with the ORGanelle ASseMbler software (https://git.metabarcoding.org/org-asm/org-asm/wikis/home). The mitogenome and the nuclear ribosomal cluster of each species were reconstructed separately using an iterative mapping strategy as recently performed for various groups of insects (Cally et al., 2016; Kocher et al., 2016, 2014; Kocher, Guilbert, Lhuillier, & Murienne, 2015). Coverage statistics were computed on the assembled genome with Geneious v.9.1.7, by mapping the reads using the following mapping parameters: a minimum overlap of 100 bp, a minimum overlap identity of 95%, a word length of 50 and a maximum mismatch per read of 5%. The mitochondrial genome was first annotated using MITOS (Berndt et al., 2013), and the annotations were checked using the annotated sequences available in GenBank.
To investigate whether the two parthenogenetic populations belong to the same species, we compared both the genetic similarity of individuals from the two localities and their divergence from congeneric species, using the percent nucleotide identity at mitochondrial and nuclear loci.
To investigate a hybrid origin hypothesis, we tested for (a) incongruence between nuclear and mitochondrial phylogenies; and (b) allelic divergence within bisexual species and parthenogenetic populations using the heterozygosity level (Birky, 1996; Simon et al., 2003). We used the mitochondrial genome (without the control region) of the four Nephus species sequenced in the present study and, as out-group, the available mitogenome sequence of Cryptolaemus montrouzieri, which belongs to the same subfamily as Nephus (i.e., Scymninae), together with those of other Coccinellidae species (Table 2). For the nuclear genes, we used the complete ribosomal rRNA genes 18S and 28S from the ribosomal cluster, as well as seven nuclear single-copy genes (CAD—carbamoyl-phosphate synthetase, HSP90—heat-shock protein 90, Aats-ile—isoleucyl-tRNA synthetase, Ace—acetylcholine esterase, alpha-Spec—alpha spectrin, AP47—adaptor protein complex 1 mu subunit and BOP1—ribosome biogenesis protein BOP1-like protein). We included the sequences of the single Nephus species available in a public database (Nephus sp2) for at least two nuclear loci (ribosomal cluster and CAD genes). According to the sequences available for the single-copy genes, we used different Scymninae species as out-group together with other Coccinellidae species (Table 2). We obtained the nuclear single-copy gene sequences of the Nephus species by mapping the reads onto reference sequences of Scymninae species available in a public database (seven genes; Table 2) using Geneious v.9.1.7 and the default mapping parameters. For each individual, after verification of the quality of the assemblage, the consensus sequence was extracted and we realized a new assemblage by mapping the reads onto the species consensus sequence using the following parameters: a minimum overlap of 50 bp, a word length of 25 and a maximum mismatch per read of 15%. Assemblage statistics, including coverage, were computed, and we used the highest quality consensus calling parameter to obtain the specimen sequences and record the heterozygous sites and coverage statistics.
| Species | Subfamily | Mitogenome | 18S | 28S | CAD | hsp90 | Aats-ile | Ace | Alpha-Spec | Ap47 | BOP1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nephus sp1 | Scymninae | MN164644 | MN168158 | MN168158 | MN185410 | MN185452 | MN185438 | MN185431 | MN185417 | MN185445 | MN185424 |
| Nephus reunioni | Scymninae | MN164643 | MN168157 | MN168157 | MN185411 | MN185453 | MN185439 | MN185432 | MN185418 | MN185446 | MN185425 |
| Nephus includens | Scymninae | MN164642 | MN168156 | MN168156 | MN185412 | MN185454 | MN185440 | MN185433 | MN185419 | MN185447 | MN185426 |
| Nephus voeltzkowi | Scymninae | MN164645–MN164648 | MN168159–MN168162 | MN168159–MN168162 | MN185413–MN185416 | MN185455–MN185458 | MN185441–MN185444 | MN185434–MN185437 | MN185420–MN185423 | MN185448–MN185451 | MN185427–MN185430 |
| Propylea japonica | Coccinellinae | KM244660 | KF792063 | ||||||||
| P. quatuordecimpunctata | Coccinellinae | EF512325 | |||||||||
| Harmonia axyridis | Coccinellinae | KR108208 | KP419116 | FJ687718 | KY695414 | FJ501962 | |||||
| H. octomaculata | Coccinellinae | MG612210 | MG609091 | MG609939 | MG610228 | ||||||
| Rhyzobius sp | Coccidulinae | MG612211 | MG609092 | MG609940 | MG610230 | MG611113 | |||||
| Cryptolaemus montrouzieri | Scymninae | KT874575 | FJ687668 | FJ687709 | KP829398 | HM575418 | |||||
| Nephus sp2 | Scymninae | KP829185 | KP829298 | KP829421 | |||||||
| Sasajiscymnus sp | Scymninae | MG609094 | MG609387 | MG609942 | MG610233 | MG611115 | |||||
| Stethorus sp | Scymninae | EU145617 | EU145676 | KP829423 | |||||||
| Stethorus sp | Scymninae | MG609095 | MG609388 | MG609943 | MG610234 | MG611116 | |||||
| Scymnus sp | Scymninae | EU145603 | EU145665 | KP829405 |
Note
- The sequences of the Nephus species obtained in the present study are indicated at the top of the table. In bold, sequences used as reference to map the Nephus spp reads.
Sequences were aligned using MAFFT (Katoh & Standley, 2013) default parameters. We inferred maximum-likelihood trees and bootstrapping with RAxML 8.2.10 (Stamatakis, 2014) under the GTR + G model. Congruence between the mitochondrial and nuclear datasets as well as between nuclear loci was assessed using the incongruence length difference (ILD) test (Farris, Källersjö, Kluge, & Bult, 1994) implemented in PAUP*v4.0a165 (Swofford, 2003) as partition homogeneity tests, using a heuristic search with 1,000 replicates. To compare the allele divergence between species, we recorded the proportion of heterozygous sites over a homologous sequence corresponding to the minimum sequence length available for all species.
2.3.2 Cytogenetic analyses
Adults from each of the two parthenogenetic populations were collected from the stock cultures and kept in 50-mm Petri dishes with a P. citri infested potato sprout. Having noticed that females accepted to lay a part of their eggs in cotton fibres, we added a piece of dental roll made of pure absorbent cotton wool and cellulose wadding rolled in thin layers (Celluron®, Hartmann) to the Petri dishes. This made the collection of eggs much easier. Every day, adults were transferred to new Petri dishes with new food and rolls and the old rolls were inspected under a stereomicroscope for the presence of the ladybird eggs. Adults and eggs were kept in the same conditions as described above. Two to three days after being laid, 12–30 eggs were carefully collected from the thin layers of the rolls and dropped into an Eppendorf tube (1.5 ml) filled with 0.7 ml of Earle's balanced salt solution (Gibco®) added with 15 μl of a solution of colchicine (4 mg/L). The eggs were squashed with a piston pellet (Eppendorf®) adapted to the inside shape of the tube and left for 45 min. After a 6-min. centrifugation (700 g), the supernatant was removed and the cell pellet was suspended for 10 min in diluted foetal calf serum (serum::distilled water, 1 vol.::3 vol.). One drop of Carnoy fixative (ethanol::acetic acid, 3 vol.::1 vol.) was added, and the cell suspension was centrifuged for 6 min. The supernatant was replaced by Carnoy fixative, and the cells in suspension were left for at least 30 min. After a further centrifugation, only two drops of fixative were left, in which cells were suspended, before deposition on microscope superfrost slides (Thermo Scientific, Braunschweig). Giemsa staining, silver staining (to locate the nucleolus organizer region—NOR) and C-banding were performed as described in Dutrillaux, Pluot-Sigwalt, and Dutrillaux (2010).
2.4 Reproductive endosymbiont screening
To determine the potential presence of bacteria, the paired-end reads from the different populations and species were mapped onto reference genomes of Wolbachia (AM999887, NZ_AP013028, NC_018267, NC_012416), Cardinium (NC_018605), Spiroplasma (NZ_CP010899) and Rickettsia (CP002428, CP004889).
Wolbachia supergroup affiliation was determined by a phylogenetic analysis of the five housekeeping genes (gatB, coxA, hcpA, ftsZ and fbpA) used in the multilocus sequence typing (MLST) scheme that was recently developed for Wolbachia (Baldo et al., 2006). According to the MLST protocol, we assigned a unique number to each unique allele of each of the five loci; Wolbachia strain was thus characterized by the combination of the five alleles (i.e., allelic profile). Wolbachia supergroup affiliation was determined by a phylogenetic analysis of the five MLST loci sequences from the Nephus samples, together with 22 Wolbachia strains belonging to the main known supergroups (A, B, C, D, E and F) retrieved from the Wolbachia MLST and NCBI databases (Table 3). For the supergroups A and B, we included strains found in Coccinellidae, other groups (including some Coleoptera families) as well as parthenogenesis inducing strains. We inferred a maximum-likelihood tree and bootstrapping with RAxML 8.2.10 (Stamatakis, 2014) under the GTR + G model.
| Host | Supergroup | Accession numbers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Phylum | Order | Family | coxA | fbpA | ftsZ | gatB | hcpA | |
| Drosophila melanogaster | Arthropoda | Diptera | Drosophilidae | A | DQ842304 (1) | DQ842340 (1) | DQ842378 (1) | DQ842415 (1) | DQ842452 (1) |
| Muscidifurax uniraptor | Arthropoda | Hymenoptera | Pteromalidae | A | DQ842293 (7) | DQ842367 (1) | DQ842329 (6) | DQ842441 (8) | DQ842404 (8) |
| Nasonia giraulti | Arthropoda | Hymenoptera | Pteromalidae | A | DQ842294 (17) | DQ842368 (17) | DQ842330 (3) | DQ842442 (11) | DQ842405 (18) |
| Acromis sparsa | Arthropoda | Coleoptera | Chrysomelidae | A | DQ842271 (2) | DQ842308 (26) | DQ842345 (21) | DQ842382 (23) | DQ842419 (26) |
| Ceutorhynchus obstricus | Arthropoda | Coleoptera | Curculionidae | A | HQ602875 (33) | HQ602879 (122) | HQ602877 (32) | HQ602878 (32) | HQ602876 (42) |
| Xylosandrus germanus | Arthropoda | Coleoptera | Scolytidae | A | (47) | (154) | (76) | (99) | (97) |
| Nephus voeltzkowi | Arthropoda | Coleoptera | Coccinellidae | B | MN168163 (4) | MN168164 (230) | MN168165 | MN168166 | MN168167 |
| Naupactus ambiguus | Arthropoda | Coleoptera | Curculionidae | B | (14) | (181) | (96) | (9) | (13) |
| Coccinella septempunctata | Arthropoda | Coleoptera | Coccinellidae | B | (20) | ||||
| Tribolium confusum | Arthropoda | Coleoptera | Tenebrionidae | B | (5) | (7) | (18) | (6) | (6) |
| Culex pipiens | Arthropoda | Diptera | Culicidae | B | DQ842277 (3) | DQ842313 (4) | DQ842351 (22) | DQ842388 (4) | DQ842425 (3) |
| Chelymorpha alternans | Arthropoda | Coleoptera | Chrysomelidae | B | DQ842274 (14) | DQ842348 (14) | DQ842310 (12) | DQ842422 (9) | DQ842385 (15) |
| Trichogramma deion | Arthropoda | Hymenoptera | Trichogrammatidae | B | DQ842302 (22) | DQ842376 (21) | DQ842338 (19) | DQ842450 (21) | DQ842413 (25) |
| Dirofilaria immitis | Nematoda | Spirurida | Onchocercidae | C | KU255267 | KU255327 | KU255351 | KU255376 | |
| Onchocerca cervipedis | Nematoda | Spirurida | Onchocercidae | C | JX075222 (179) | JX075224 (247) | JX075226 (157) | JX075228 (192) | |
| Brugia malayi | Nematoda | Spirurida | Onchocercidae | D | DQ842273 (29) | DQ842341 (30) | DQ842347 (26) | DQ842384 (28) | DQ842421 (33) |
| Litomosoides brasiliensis | Nematoda | Spirurida | Onchocercidae | D | KU255273 | KU255333 | KU255354 | KU255381 | |
| Folsomia candida | Arthropoda | Collembola | Isotomidae | E | KT799590 (148) | KT799595 (227) | KT799605 (135) | KT799610 (171) | KT799615 (179) |
| Mesaphorura italica | Arthropoda | Collembola | Tullbergiidae | E | AJ575103 | ||||
| Adalia sp. | Arthropoda | Coleoptera | Coccinellidae | F | (409) | ||||
| Cimex lectularius | Arthropoda | Heteroptera | Cimicidae | F | DQ842275 (27) | DQ842349 (28) | DQ842311 (24) | DQ842423 (26) | DQ842386 (31) |
| Apoica pallens | Arthropoda | Hymenoptera | Vespidae | F | (63) | (78) | (61) | (65) | (73) |
| Supella longipalpa | Arthropoda | Dictyoptera | Ectobiidae | F | (147) | (414) | (87) | (113) | (273) |
Note
- Sequences are referenced with their GenBank accession numbers and/or, in parentheses, with their allele numbers in the Wolbachia PubMLST database.
3 RESULTS
3.1 Ladybird field survey
All field-collected ladybirds in the Azores (40 individuals) and Reunion (37 individuals) were females, and we never found males despite a huge sampling effort. Specimens collected during 7 years by the Insectarium of Reunion were also females.
All the collected individuals were present near the coast and were associated with lower vegetation, mainly with plants infested with Pseudococcidae, which can be found on a variety of plants. The populations were rare and generally composed of a small number of individuals. Nevertheless, there were exceptions: one of the Santa Maria and one of the Reunion populations represented, respectively, 87.5% and 94.5% of the total individuals collected in the Azores and Reunion.
3.2 Experimental test of parthenogenesis reproduction
The laboratory study to test for parthenogenesis shows that, during the first 2 months of monitoring, 27.6% and 36.7% of the individuals from Santa Maria (Azores) and Reunion (Mascarene) cohorts, respectively, were fertile. This ratio increased with time: there were 62.1% fertile individuals in the Santa Maria cohort at the end of the 4 months monitoring. The mean number of larvae per female during the first 2 months of the experiment was of 3.1 (SD: 3.0) and 1.9 (SD: 1.4), respectively, for the Santa Maria and Reunion cohorts; it attained 7.7 (SD: 5.2) larvae per female for the Santa Maria cohort after 4 months monitoring. The observation of the genitalia of the individuals that did not reproduce during the experience indicated that they were also all females.
3.3 Santa Maria (Azores) and Reunion (Mascarene) population similarity and possible hybrid/ polyploid origin
To investigate whether the two parthenogenetic populations are genetically similar, we sequenced individuals from the two localities together with three conspecific species. We obtained, in a first Illumina run, between 12,650,898 and 24,432,454 reads per specimen and, in another run, 74,808,866 and 349,364,742 reads for a second specimen of the Santa Maria and Reunion populations, respectively (Table 1). Reads were assembled into circular mitochondrial genomes ranging from 16,430 to 17,060 bp in size (GenBank accession numbers MN164642–MN164648) and into nuclear ribosomal cluster ranging from 6,172 bp to 6,779 bp in size (GenBank accession numbers MN168156–MN168162) with high coverage (min ~600X, see Table 1). The mitogenome organization consisted of the typical set of genes (13 protein-coding genes, 22 transfer RNA genes and two ribosomal RNA genes) and one control region. The nucleotide composition was A + T biased (79.5%).
The comparative analyses of the complete mitochondrial genome (17,060–17,072 bp) and ribosomal cluster sequences (6,172 bp) in individuals from the two parthenogenetic populations show a very high similarity in both their mitochondrial (99.77% ± 0.14%, including the control region) and nuclear (99.96% ± 0.01%) genomes. We found very little genetic variation between specimens from focal localities as only three and two substitutions were recorded, respectively, in the mitochondrial control region and in the nuclear ribosomal cluster. The Santa Maria and Reunion parthenogenetic populations appear to be clearly divergent from the three congeneric bisexual related species—Nephus includes, N. reunioni and Nephus sp1—with a genetic similarity of 78.94% ± 0.65% for the complete mitochondrial genome and a similarity of 77.89% ± 0.78% for the nuclear ribosomal cluster. These divergences are comparable to or even higher than the divergence observed between these three species (similarity of 83.59% ± 0.33% and 85.63% ± 3.6%, for mitochondrial genome and ribosomal cluster, respectively). This confirms the close genetic proximity of the two parthenogenetic populations, suggesting they belong to a single species. Based on the mitochondrial nucleotide substitution rate calculated by Pons, Ribera, Bertranpetit, and Balke (2010) for the Cucujiformia (Coleoptera) and the generation time of Nephus under laboratory conditions (10–12 generations per year), we estimated that the two parthenogenetic populations diverged very recently (i.e., about 8,000 years ago). However, due to the high heterogeneity in the substitution rates found within this lineage (Timmermans et al., 2015; Yuan et al., 2016), including Coccinellidae, the sparse taxonomical sample used here, and the possible higher generation time in natural conditions, these dating estimates should be treated cautiously.
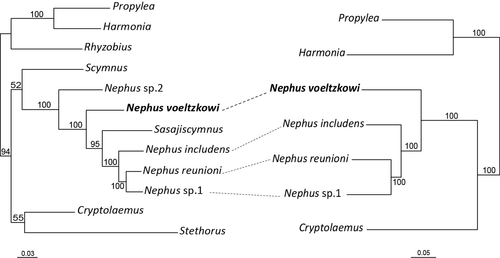
To investigate whether there is a hybrid origin of the parthenotes, we first reconstructed phylogenetic relationships using mitochondrial and nuclear loci. The mitochondrial tree, based on an alignment of 14,845 bp, recovered with high support (100% BP) the parthenogenetic N. voeltzkowi as the sister group of the three bisexual Nephus species—N. reunioni, N. includens and Nephus sp1 (Figure 2, Data S1). The nuclear tree, based on the 9 nuclear genes concatenated (10,812 bp aligned, Data S2), recovered the same relationships with high support (100% BP), with the species Nephus sp2 from the public database situated at the base of the clade (Figure 2). Similarly, the nuclear ribosomal genes (4,190 bp aligned) and the single-copy genes, either independently (from 609 to 1,653 bp aligned) or concatenated (6,622 bp aligned), yielded a congruent topology. For the five gene trees where a Sasajiscymnus sp. was included, we found this species within the Nephus clade, as the sister group of the three bisexual species, despite a low support in individual single gene phylogenies. When Sasajiscymnus sp. was excluded from the dataset, the topology recovered was congruent to the remaining nuclear genes or mitogenome trees. The ILD test revealed no significant differences between phylogenies either based on mitogenome or nuclear genes (ribosomal and single-copy nuclear genes; p-value = 1.0), or based on the ribosomal and single-copy nuclear genes (p-value = 1.0). The topological congruence between mitochondrial and nuclear loci as well as between nuclear loci is not consistent with phylogenetic predictions under a hybrid origin of the parthenogenetic populations.
The screening of allele divergence in N. voeltzkowi detected heterozygosity in all of the eight nuclear gene regions investigated (Table 4). The average heterozygosity level was low, ranging from 0.057% in the ribosomal genes to 0.329% in the alpha-Spec, reflecting reduced allelic divergence within parthenote genomes. In bisexual species, we also detected heterozygosity at six over the eight nuclear genes, for at least one species per gene (up to 0.563% in the CAD; Table 4). When we consider all Nephus species, the heterozygosity level is low for all species, despite a slightly higher level in asexual individuals (mean asexual = 0.17% ± 0.15% vs. bisexual 0.10% ± 0.25%, Kruskal–Wallis test p-value = .012).
| Specimens | Ribosomal cluster | CAD (length = 1,007 pb) | HSP90 (length = 1,710 pb) | Aast-ile (length = 792 pb) | Ace (length = 1,035 pb) | alpha-Spec (length = 761 pb) | AP47 (length = 1,125 pb) | BOP1 (length = 1,363 pb) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % He | Coverage | % He | Coverage | % He | Coverage | % He | Coverage | % He | Coverage | % He | Coverage | % He | Coverage | % He | Coverage | |
| N1 | 0.065 | 2,692.7 | 0.298 | 4.36 | 0.058 | 8.1 | 0.126 | 6.6 | 0.097 | 7.1 | 0.394 | 5.2 | 0.089 | 6.2 | 0.220 | 8.1 |
| N2 | 0.032 | 12,350.7 | 0.099 | 53 | 0.175 | 44 | 0.126 | 59.8 | 0.580 | 42.5 | 0.394 | 50.8 | 0.089 | 46.9 | 0.367 | 58.4 |
| N3 | 0.065 | 1,495.1 | 0.397 | 7.2 | 0.058 | 7.3 | 0.253 | 4.4 | 0.097 | 5.2 | 0.263 | 9.2 | 0.089 | 6.9 | 0.147 | 6.8 |
| N4 | 0.065 | 44,271.9 | 0.298 | 238.3 | 0.000 | 264.6 | 0.000 | 288.6 | 0.000 | 200.6 | 0.263 | 277.5 | 0.000 | 252.5 | 0.147 | 271.3 |
| Mean for N. voeltzkowi | 0.057 | 0.273 | 0.073 | 0.126 | 0.193 | 0.329 | 0.067 | 0.220 | ||||||||
| N5 | 0.030 | 957.5 | 1.192 | 8.6 | 0.175 | 6.9 | 0.000 | 4.5 | 0.000 | 6.2 | 0.263 | 5.4 | 0.000 | 4.6 | 0.000 | 3.3 |
| N6 | 0.000 | 2,207.4 | 0.199 | 7.9 | 0.000 | 9.1 | 0.126 | 2 | 0.097 | 8.3 | 0.000 | 5.5 | 0.000 | 3.4 | 0.000 | 5.7 |
| N7 | 0.015 | 1,209.9 | 0.298 | 7.8 | 0.000 | 9.8 | 0.000 | 8.3 | 0.000 | 7.8 | 0.000 | 8.2 | 0.000 | 6.4 | 0.000 | 11 |
Note
- N1 and N2 = N. voeltzkowi (Reunion); N3 and N4 = N. voeltzkowi (Santa Maria); N5 = Nephus sp1 (Reunion); N6 = N. reunioni (Portugal); N7 = N. includens (Greece).
- In bold, mean heterozygosity levels for N. voeltzkowi based on N1-N4 specimens values.
Concerning the cytogenetic analyses and given the very small size of the biological material, we had to pool at least 12 eggs for the hypotonic shock method. Consequently, the population of cells analysed represented an unknown number of specimens.
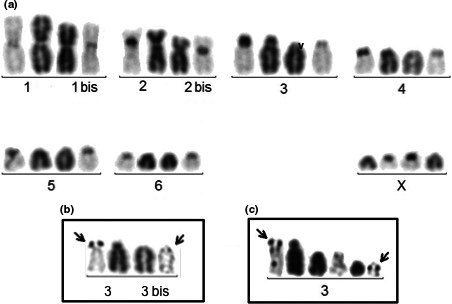
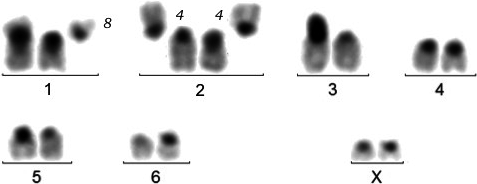
In the Santa Maria specimens (Figure 3), two different karyotypes were observed. Karyotype I (25/32 cells) is composed of fourteen chromosomes of which four sub-metacentrics (chromosomes 1, 1bis, 2 and 2bis) could not be paired (Figure 3a). All the other chromosomes are acrocentric and were paired without certainty of the homology. In “pair” 3 (Figure 3b), chromosome 3 carries a large amount of C-banded heterochromatin on its short arm, which harbours an amplified NOR, as shown by silver staining; chromosome 3bis harbours a small NOR proximally on its long arm. Karyotype II (7/32 cells) is composed of fifteen chromosomes. Compared to karyotype I, chromosome 3bis is replaced by two small ones, of which one carries the NOR at the same position as chromosome 3bis (Figure 3c).
In the Reunion specimens, three different karyotypes were found. Karyotype I (18/35 cells) is composed of seventeen chromosomes (Figure 4a), with “pair” 1 formed by a sub-metacentric (chromosome 1) and an acrocentric (chromosome 1bis), assumed to be the homolog of the sub-metacentric long arm. “Pair” 3 (Figure 4b) has the same characteristics as “pair” 3 of the Santa Maria specimens. In these karyotypes with seventeen chromosomes, there is a single chromosome placed in position 8 (Figure 4a). Karyotype II (16/35 cells) is composed of eighteen chromosomes. It differs from karyotype I by the addition of a minute chromosome arbitrarily placed in “pair” 8 (Figure 4c). It is strongly stained after C-banding, which indicates that it is largely composed of heterochromatin, and it may be considered as a B chromosome. Karyotype III (1/35 cells) is composed of nineteen chromosomes. It differs from Karyotype II by duplication of this minute chromosome.
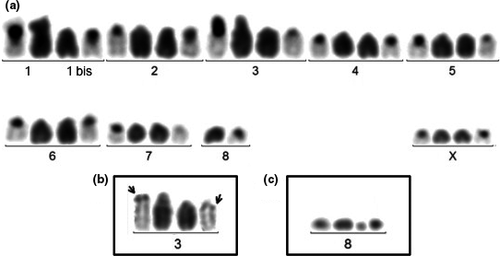
The karyotypes with fourteen (Santa Maria specimens) and seventeen (Reunion specimens) chromosomes look quite different at first glance, but they have the same fundamental number (i.e., number of chromosome arms) and it is possible to reconstruct the karyotype of the Santa Maria specimens with the chromosomes of those of the Reunion (Figure 5). Therefore, both karyotypes may simply derive from a common ancestral karyotype, composed of eighteen acrocentrics, by one and four Robertsonian translocations (fusions), respectively. Possibly, the largest sub-metacentric in “pair” 2 of the Santa Maria specimens (Figure 3a) and the sub-metacentric in “pair” 1 of the Reunion specimens are homologous (Figure 4a).
3.4 Presence of endosymbionts
Between 1% and 2.8% of the paired-end reads from individuals of the two parthenogenetic populations were successfully mapped onto the genomes of Wolbachia (coverage between 6.6 X and 362 X), but not of Cardinium, Spiroplasma and Rickettsia, suggesting infection only by a Wolbachia strain. The resulting consensus sequences of the Wolbachia MLST genes gatB (369 bp), coxA (402 bp), hcpA (444 bp), ftsZ (435 bp) and fbpA (430 bp) were all identical for the two parthenogenetic populations, suggesting an infection by the same strain (strain sequences were deposited in the NCBI database; Table 3). All Wolbachia supergroups were recovered as monophyletic with high statistical support in the phylogenetic analyses for concatenated MLST loci (2,079 bp in total; Data S3; Figure 6). The Wolbachia strain infecting parthenogenetic populations clustered within supergroup B, with high bootstrap support. Strains infecting Coccinella septempunctata and Adalia sp. (Coccinellidae), included in the phylogenetic reconstruction, were, respectively, assigned to supergroups B and F. For the three sexual species of the Nephus genus (N. includens, N. reunioni and Nephus sp1), using the same settings, no read was mapped onto any of the bacterial genomes.

4 DISCUSSION
We report here, for the first time, the existence of all-female populations in the Azores and the Mascarene, genetically similar and identified as N. voeltzkowi. Ladybirds are commonly affected by male-killing endosymbionts, leading to a scarcity of males in some populations (Weinert et al., 2007). However, in our field surveys in the Azores archipelago and on Reunion Island (Mascarene archipelago) we did not find a single N. voeltzkowi male, suggesting that in natural conditions these ladybirds are able to reproduce asexually. Thelytokous parthenogenesis, the development of female offspring from unfertilized eggs (Normark, 2014), was confirmed by our laboratory experiments. This finding is significant because despite the large amount of basic and applied research work on the Coccinellidae, asexuality had never been detected in the family before. Furthermore, although parthenogenesis appears across the tree of life (Parker et al., 2019), it is proportionally rare in the most species-rich animal order, the Coleoptera (Hunt et al., 2007).
Why transition rates from sexuality to asexuality vary among taxa is largely unknown (Neiman et al., 2014). New cases of parthenogenesis can help address the evolutionary reasons and mechanisms at the origin of asexuality, for which there is still no consensus (Meirmans, 2009; Neiman et al., 2014; Schön, Martens, & Dijk, 2009). Hereafter, we discuss our results on the proximate causes of parthenogenesis in N. voeltzkowi.
Transitions from sexual reproduction to asexuality are often of genetic origin (Bullini, 1994; Neiman et al., 2014). Polyploidy, for example, is well documented in parthenogenetic animals (Choleva & Janko, 2013), and 95% of asexual Coleoptera are polyploid (Dutrillaux, Pluot-Sigwalt, & Dutrillaux, 2014). However, our cytological results show that the chromosome numbers of individuals from the two populations examined are in the range of those of diploid Coccinellidae (Lyapunova, Vorontsov, Yadav, Korablev, & Yanina, 1984; Smith, 1960). That is to say, these karyotypes are among the rare non-polyploid karyotypes in parthenogenetic Coleoptera.
A hybrid origin is also often associated with asexuality (Bullini, 1994), including in Coleoptera (Gómez-Zurita & Cardoso, 2019). In our case, we might be in the presence of (a) a single hybridization, followed by a population split (Azores vs. Mascarene); (b) two different hybridization events sharing one of the parental species; or (c) two different hybridization events involving two different pairs of parental species. The comparison of the mitogenome alone only allows the exclusion of hybridogenesis (maternal inheritance) if two different pairs of parental species are involved. Our results show that the mitogenome and ribosomal cluster are highly similar; therefore, we can already exclude hypothesis c, two different hybridization events involving two different pairs of parental species .
In the two remaining hypotheses (a and b), it would be expected that at least half of the chromosomes were shared between the two populations. Moreover, hybrids are predicted to possess high heterozygosity and alleles typical of the parental species (high allelic divergence) as both the maternal and paternal genomes are transmitted clonally. Both the odd number of chromosomes and the presence of different unpaired chromosomes in the karyotypes from the two populations indicate heterozygosities that could be the result of independent hybrid origins (hypothesis b) but we cannot exclude a single hybrid origin with the original karyotype suffering subsequent independent chromosome rearrangements (hypothesis a). The congruence between the mtDNA tree and the nuclear tree suggests that the two parthenogenetic populations belong to a single species and are not of a hybrid origin (i.e., refuting hypotheses a and b), although we cannot totally discard the possibility that these results may be an artefact of limited taxonomic sampling. In addition, we found low heterozygosity levels within asexual individuals, in the range of those observed for three sexual Drosophila species (i.e., 0.4%–2%; Moriyama & Powell, 1996), which is congruent with a non-hybrid origin of the parthenotes. The heterozygosity levels may be biased due to the small number of individuals analysed and the difference in gene coverage between species but evidence of high allelic divergence in asexual species has even been demonstrated using a single individual (Mark Welch & Meselson, 2000).
In this context, how can the Azores and Mascarene karyotype heterozygosities be explained? A close look at these karyotypes shows that although the number and the shape of their chromosomes differ, their fundamental number is the same. We suggest they belong to a single chromosomally polymorphic species undergoing, respectively, 4 and 1 Robertsonian (Rb) fusions. Chromosomal rearrangements and changes in chromosome numbers have been reported in other parthenogenetic insects and are considered as post-parthenogenesis events (Blackman, Spence, & Normark, 2000; Dutrillaux et al., 2009; Manicardi, Nardelli, & Mandrioli, 2015). It is therefore possible that following an event leading to parthenogenesis descendants of N. voeltzkowi disseminated overseas. Indeed, a short-term advantage of asexual organisms is their superior colonizing ability (Hörandl, 2009); a single individual can potentially engender a new population. On each colonized island, parthenotes independently accumulated a few DNA alterations, including those leading to chromosome rearrangements, which is possible because insularity may favour Rb evolution, as shown for the house mouse in Madeira (Britton-Davidian et al., 2000). In addition to insularity, the lack of meiotic selection against chromosomal changes, due to parthenogenesis, might have allowed an unusually fast accumulation of chromosome rearrangements (Birky, 1996). The presence of specimens with different karyotypes within the same parthenogenetic population supports this hypothesis.
Besides polyploidy and hybridization, we can consider the possibility of a mutational origin. Facultative parthenogenesis, where females can reproduce either sexually or asexually, is rare in animals (van der Kooi & Schwander, 2015) and has never been reported in Coleoptera (e.g., recent revision papers by Normark, 2014 and Gokhman & Kuznetsova, 2018). Nevertheless, although we cannot discard this hypothesis, testing it for N. voeltzkowi requires additional studies, including breeding experiments impossible to perform at this stage due to the absence of collected males. A more widespread phenomenon is tychoparthenogenesis, an accidental spontaneous hatch of unfertilized eggs that develop into females (Bullini, 1994—“spontaneous theory”). In this accidental parthenogenesis, the proportion of unfertilized eggs and their hatching rates are very low, but could be magnified by mate limitation (the “mate scarcity hypothesis,” Rhainds, 2010; Schwander, Vuilleumier, Dubman, & Crespi, 2010) in small effective populations or after bottleneck events (Little, Chapuis, Blondin, Chapuis, & Jourdan-Pineau, 2017). This could explain the well-known relationship between parthenogenesis and certain habitats such as islands (“geographical parthenogenesis” Vandel, 1928). We found parthenogenetic lineages of N. voeltzkowi on islands in the Azores and Mascarene archipelagos, and the number of fertile females in our experiment shows that parthenogenesis capability in N. voeltzkowi is not an accidental phenomenon. Therefore, N. voeltzkowi could be an example of transition between sexual and obligate parthenogenetical reproduction through tychoparthenogenesis.
Additionally, chromosome rearrangements are mutations and we cannot exclude the possibility that they started before parthenogenesis and continued afterwards. Although the relatively high stability of sexual Polyphaga beetle karyotypes (Smith & Virkki, 1978) does not support this hypothesis, cytological polymorphisms (both in number and shape) have been reported for some ladybird species (Lyapunova et al., 1984; Smith, 1960; Yadav & Pillai, 1979). It would be interesting to evaluate to what extent Rb translocations could be at the very origin of parthenogenesis, as a dysgenic process affecting the regular progress of meiosis (Vasco et al., 2012).
Finally, we cannot exclude the possibility of an infectious origin of parthenogenesis because we detected the presence of Wolbachia in all the parthenotes, but not in the sexual specimens tested. This Wolbachia strain belongs to the B supergroup, one of the two most common supergroups present in arthropods (Duron et al., 2008). There is no established link between a particular Wolbachia supergroup and parthenogenesis in the hosts (Lindsey, Werren, Richards, & Stouthamer, 2016), and the B supergroup is present in taxa both with and without asexuality. Furthermore, infectious induction of obligate parthenogenesis has only been confirmed in organisms with haplodiploid sex determination and not in diplodiploid organisms like ladybirds (Ma & Schwander, 2017; Normark, 2003; Werren et al., 2008). Nevertheless, Rodriguero, Confalonieri, Guedes, and Lanteri (2010) show a significant correlation between Wolbachia infection and parthenogenesis in weevils, and Pike and Kingcombe (2009) demonstrated that the parthenogenetic springtail Folsomia candida cured of Wolbachia by antibiotics lays eggs that fail to hatch. These studies question the exclusive infectious origin of parthenogenesis in haplodiploid organisms (Ma & Schwander, 2017). That is to say, N. voeltzkowi may constitute an objective example of Wolbachia-induced parthenogenesis in diplodiploid organisms.
We have reported here the first case of parthenogenesis in the Coccinellidae and combined different approaches to investigate the proximate mechanisms at its origin. Our results reject polyploidization and strongly support a non-hybrid origin, two of the most common phenomena responsible for parthenogenesis. In contrast, we identified Rb translocations and a Wolbachia infection in individuals from the two parthenogenetic populations. These may be post-parthenogenesis events but we cannot exclude the possibility that they are proximal causes of parthenogenesis, which would be a novelty but still needs to be investigated by further studies. N. voeltzkowi may constitute a good model to tackle the conditions underlying the transition to asexual reproduction.
ACKNOWLEDGEMENTS
We are extremely grateful to our late friend, Serge Quilici (CIRAD, St Pierre), for his logistic support during AM visits to Reunion and more than everything for his kindness. We also would like to thank J. Rochat for access to samples from the Insectarium de La Réunion and N. Dardenne for technical support. Last but not least, we are grateful to Drs E. Haring, A. Blanke and four anonymous referees for their comments and suggestions to the first version of this manuscript that helped us to improve it deeply. AM, EL, JM and JLH were supported by the “Laboratoires d'Excellence” LabEx TULIP (ANR-10-LABX-41) and CEBA (ANR-10-LABX-25-01).




