Just a Vietnamese goldfish or another Carassius? Validity of Carassius argenteaphthalmus Nguyen & Ngo, 2001 (Teleostei: Cyprinidae)
Abstract
Freshwater fishes of the genus Carassius are widespread throughout Eurasia, but notoriously difficult for identification by morphological characters, leaving their systematics in a vague state and the validity of several species is unclear. Consequently, genetic data are used to identify the evolutionary units within this genus. Here, we present an analysis of the genus Carassius in Vietnam and south-eastern China based on phylogenetic reconstruction using the mitochondrial cytochrome b gene and the nuclear S7 gene. We found two lineages in Vietnam with high supports in the nuclear as well as the mitochondrial dataset. One lineage corresponds to the species Carassius auratus, and one lineage represents an independent evolutionary unit. We test whether the lineage may correspond to the species that has once been described as Carassius argenteaphthalmus. The main character for identification, the color of the eye rim, did not distinguish two clades from each other. Considering the absence of genetic data, the imprecision of the original description, and the absence of all type specimens, the name of C. argenteaphthalmus is considered to be a nomen dubium.
1 INTRODUCTION
The freshwater fishes of the genus Carassius are naturally widespread across Eurasia (Nelson, Grande, & Wilson, 2016). The genus includes also one of the best-known fishes in the world, the goldfish. However, the identification of species within the genus Carassius is considered notoriously problematic due to high morphological similarity between species, a general lack of unique characters, and high intraspecific variability (Hensel, 1971; Kalous, Bohlen, Rylková, & Petrtýl, 2012; Lusk & Baruš, 1978; Vasileva, 1990; Vasileva & Vasilev, 2000).
In agreement with Eschmeyer, Fricke, and van Laan (2017), six species are considered as valid within the genus Carassius: Carassius carassius (Linnaeus, 1758) in most of Europe and western Siberia (Kottelat & Freyhof, 2007), Carassius gibelio (Bloch, 1782) in Europe, Siberia, and Northeast Asia (Berg, 1949; Kalous et al., 2012; Kottelat & Freyhof, 2007; Rylková, Kalous, Bohlen, Lamatsch, & Petrtýl, 2013), Carassius cuvieri Temminck & Schlegel, 1842, and Carassius langsdorfii Temminck & Schlegel, 1842 in Japan (Hosoya, 2000; Yamamoto, Takada, Iguchi, & Nishida, 2010) but introduced to Europe (Kalous, Rylková, Bohlen, Šanda, & Petrtýl, 2013; Rylková & Kalous, 2013), Carassius auratus (Linnaeus, 1758) in mainland East Asia but as feral goldfish introduced worldwide (Rylková, Kalous, Šlechtová, & Bohlen, 2010), and recently described Carassius praecipuus Kottelat, 2017, so far known from central part of Laos only.
However, the actual diversity within the genus most likely is significantly larger, as recent genetic studies on material from Europe and Asia suggest. In particular, pertinent are studies of Gao et al. (2012) Takada et al. (2010), Yamamoto et al. (2010), Kalous et al. (2012), Rylková et al. (2013), and Luo et al. (2014) who detected a number of distinct mtDNA lineages with a remarkable phylogenetic and geographic distance. These lineages are presently all referred to as C. auratus or C. gibelio, but the whole group should better be called the C. auratus complex. The observed intraspecific diversity led some authors to use non-taxonomic names for particular lineages, for example, C. auratus var. pengze (Li, Liang, & Zou, 2016). Yet, to corroborate the presence of an independent evolutionary lineage based on the analysis of mtDNA, additional nuclear markers can be helpful. Additional support for the distinctiveness of an evolutionary lineage as a species could stem from the demonstration of diagnostic morphological characters. If such distinctive morphological features could not be found, the lineage could be considered a cryptic species.
Concerning the genus Carassius, the existence of several formally described species with unclear status is worth mentioning. One of the described taxa with an unclear taxonomic state is Carassius argenteaphthalmus Nguyen & Ngo, 2001; originally described as a subspecies of C. auratus from Diên Biên Phu in Diên Biên province in north-western Vietnam and tentatively treated as a synonym of C. auratus (Kottelat, 2013). The morphological description is in general very vague, but the color of the eye rim has been suggested as the main character to distinguish C. argenteaphthalmus from C. auratus: Nguyen and Ngo (2001) state that C. argenteaphthalmus has a silver eye rim while the rim of C. auratus is red. This character has never been tested particularly, but would be of great use in the systematics of Carassius if proven applicable.
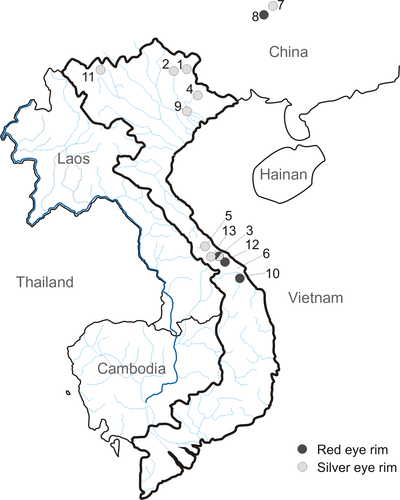
While processing specimens of Carassius from various localities in Vietnam, we found specimens with silver and with red eye rims (Figure 1), often together at the same place. To identify the material, genetic analyses were conducted. The aim of the study was (i) to clarify the taxonomic identity of the Vietnamese individuals, (ii) to evaluate the validity of C. argenteaphthalmus, and (iii) to test whether the color of eye rim could be used as a reliable taxonomic character to differentiate between species or whether it shows morphologic variability within the lineage (Banarescu, 1966).

2 MATERIAL AND METHODS
2.1 Phylogenetic analyses
Fifty-three individuals of Carassius from numerous localities of Eurasia were included in the analysis. Our original dataset consisted of 33 samples coming from 13 localities in Vietnam and adjacent areas in southern China (Figure 2). Investigated material is deposited in fish collection of department of Zoology and Fisheries, Czech University of Life Sciences in Prague, Czech Republic. In addition to these samples, the dataset contains 20 sequences covering described species within the genus Carassius. Common carp, Cyprinus carpio L. was used as an outgroup. Detailed information about the analyzed material is given in Table 1.
| Number of Individuals | Collection No. | GenBank Accession No. (cytochrome b) | GenBank Accession No. (S7) | Ploidy level | Eyerim color | Sample origin | Locality number | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | B1VN | KJ584695 | KP153188 | 2n | Silver | Bang River, CaoBang, Vietnam | 1 | |
| 1 | BG1VN | KJ584696 | KY215825 | – | – | Bang River, Vietnam | 2 | |
| 1 | BG2VN | KJ584697 | KY215826 | – | – | Bang River, Vietnam | 2 | |
| 1 | BG3VN | KJ584698 | – | 2n | – | Bang River, Vietnam | 2 | |
| 4 | H3VN | KJ584699 | KP153194, KP153195a | 2n | Red | Huong River, Hué, Vietnam | 3 | |
| 1 | K1VN | KJ584700 | KP153190 | 3n | Silver | KyCung River, Lang Son, Vietnam | 4 | |
| 2 | TT2VN | KJ584701 | KP153201 | – | Silver | 30 km northwest of Hue, Vietnam | 5 | |
| 1 | V1VN | KJ584702 | KP153191, KP153192a | 2n | Silver | Vihn Dien River, Vietnam | 6 | |
| 1 | VN100 | KJ584703 | – | – | Silver | KyCung River, Lang Son, Vietnam | 4 | |
| 1 | VN168 | KJ584704 | KP153202, KP153203a | 2n | Silver | Bang River, CaoBang, Vietnam | 1 | |
| 3 | H1CN | KJ584705 | KP153215 | 2n | Red | Dongmiaoxiang, China | 7 | |
| 1 | R1CN | KJ584706 | – | – | Silver | RongJiang River, China | 8 | |
| 1 | R3VN | GU942718 | KP153199, KP153200a | – | Silver | Hong River, Hanoi, Vietnam | 9 | |
| 1 | P1VN | GU942716 | KP153196, KP153196a | – | Silver | Huong River, Hué, Vietnam | 3 | |
| 1 | P3VN | GU942717 | KP153198 | – | Silver | Huong River, Hué, Vietnam | 3 | |
| 1 | T1VN | GU942719 | KP153193 | – | Red | Thu Bon River, HoiAn, Vietnam | 10 | |
| 1 | T2VN | GU942720 | – | – | Red | Thu Bon River, HoiAn, Vietnam | 10 | |
| 1 | T3VN | GU942721 | – | – | Red | Thu Bon River, HoiAn, Vietnam | 10 | |
| 1 | VN208 | MF673857 | – | – | Red | Hué, Vietnam | 12 | |
| 1 | VN209 | MF673856 | – | – | Red | Hué, Vietnam | 12 | |
| 1 | VN210 | MF673855 | – | – | Red | Hué, Vietnam | 12 | |
| 1 | VN2011 | MF673858 | – | – | Red | Hué, Vietnam | 12 | |
| 1 | BM1VN | GU942717 | – | – | Silver | Bach Ma, Vietnam | 13 | |
| 1 | V4A | KX601119 | KY215821, KY215822a | – | Silver | Da River, LayChau, Vietnam | 11 | |
| 2 | V5A | KX601120 | KY215819, KY215820a | – | Silver | Da River, LayChau, Vietnam | 11 | |
| 1 | V6A | KX601121 | KY215823, KY215824a | – | Silver | Da River, LayChau, Vietnam | 11 | |
| HQ689875 | – | Tra KhucRiver, Vietnam | Gao et al. (2012) | |||||
| HQ689876 | – | Tra KhucRiver, Vietnam | Gao et al. (2012) | |||||
| HQ689877 | – | Tra KhucRiver, Vietnam | Gao et al. (2012) | |||||
| T3CN | KJ584709 | KP153222 | Tian River, Hunanprovince, China | |||||
| CIGA1 | KX688781 | KX688787 | Ornamental fish market, Czech Republic | |||||
| HGA1 | KX688782 | KX688788 | Ornamental fish market, Czech Republic | |||||
| EU663599 | KP153213, KP153214a | Wuhan, Yangtze, China | Rylková et al. (2010) | |||||
| JN546046 | KP153204, KP153205a | Rokycany, Czech Republic | Kalous et al. (2012) | |||||
| JN546066 | KP153218 | Alma, Topoli, Ukraine | Rylková et al. (2013) | |||||
| RGA2 | KX688786 | KR054639 | Czech Republic | |||||
| PGA1 | KX688785 | KR054650 | Germany | |||||
| DQ399921 | KP153186, KP153187a | AbashiriLake, Hokkaido, Japan | Kalous et al. (2007) | |||||
| GU942710 | KP153182 | RamaLake, Bosnia and Herzegovina | Rylková et al. (2013) | |||||
| JN412529 | KP153183 | Pool at Litvínovice, Czech Republic | Rylková et al. (2013) | |||||
| JN412528 | KP153184, KP153185a | Chornariverbasin, Ukraine | Kalous et al. (2013) | |||||
| JN402304 | KP153216, KP153217a | LakeMikatako, Honsyu, Japan | Kalous et al. (2012) | |||||
| JN412549 | KP153225 | Fishfarm Višňová, Czech Republic | Rylková et al. (2013) | |||||
| GU991400 | KP153224 | Calverton fishfarm, Great Britain | Kalous et al. (2012) | |||||
| LU1CZ | KR131839 | KX688792 | Czech Republic | |||||
| JN412548 | KP153227 | Ångermanälven River, Sweden | Rylková et al. (2013) | |||||
| HM008692 | KP153228, KP153229a | Mekong River, Thailand | Kalous et al. (2012) |
- a Number higher that one states more individuals sampled in the same locality shared the identic haplotype. Multiple S7 protein gene sequences were gained from the same individual. References are only given for sequences not obtained in the course of this study. Numbers of geographic localities correspond to numbers in Figure 2.
Genomic DNA was isolated from ethanol preserved or fresh tissue using DNeasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany) according to manufacturer's instructions. The mitochondrial cytochrome b gene was amplified using primers Glu L. Ca14337-14359: GAA GAACCA CCG TTG TTA TTC AA and Thr H. Ca15568-15548: ACC TCC RAT CTY CGG ATT ACA (Šlechtová, Bohlen, Freyhof, & Ráb, 2006). PCR amplification was performed as described in Rylková et al. (2013). The ribosomal protein S7 (RPS7) gene was amplified using the primers S7RPEX1F (TGG CCT CTT CCT TGG CCG TC) and S7RPEX2R (AAC TCG TCT GGC TTT TGC CC) (Chow & Hazama, 1998). PCR reaction contained 3 μl of template DNA; 0.5 mM of each primer; 25.5 μl of Combi PPP Master Mix (Top-bio) and ddH2O up to a total volume 50 μl. The PCR profile (carried out on MJ Mini™ thermal cycler) started with 5 min period of initial denaturation at 95°C, followed by two cycles each consisting of: 94°C for 1 min, 60°C for 1 min 30 s, and 72°C for 2 min; two cycles: 95°C for 1 min, 58°C for 1 min 30 s, and 72°C for 2 min; two cycles: 94°C for 1 min, 56°C for 1 min 30 s, and 72°C for 2 min; 30 cycles: 94°C for 1 min, 54°C for 1 min 30 s, and 72°C for 2 min. PCR was terminated by a final elongation period of 72°C for 7 min. All PCR products were purified and sequenced by Macrogen Inc., Seoul, Korea. The raw chromatograms were manually assembled and checked by eye for potential mistakes using the computer software BioEdit 5.0.9 (Hall, 1999); the same program was used to align sequences using the ClustalW algorithm. The phylogenetic relationships were estimated from aligned sequences using the method of maximum parsimony (MP) performed in PAUP* version 4.0b10 (Swofford, 2000) and Bayesian analysis (BAY) using the program MrBayes ver. 3.0 (Huelsenbeck & Ronquist, 2001) as described in Šlechtová et al. (2006).
2.2 Ploidy level determination
The ploidy level of seven individuals was determined as relative DNA content of ethanol-preserved tissue cells by means of flow cytometry (Partec CCA I; Partec GmbH, Münster, Germany) according to Vindeløv and Christensen (1994) using 4′,6-diamidino-2-phenylindol (DAPI) for nuclear DNA staining (Otto, 1994). Ethanol preserved fin tissue samples of a diploid (2n = 100) ornamental Goldfish C. auratus (EU663582) was used as a reference standard. Samples were analyzed separately at the speed 0.4–0.3 μl/s. The geographic origin and collection numbers of the analyzed specimens are given in Table 1.
3 RESULTS
3.1 Phylogenetic analyses
The final matrix of 53 cytochrome b sequences consisted of 1,087 basepairs containing 269 variable characters with 174 parsimony informative sites. Both employed methods have recovered trees of very similar topologies consist in minor differences within marginal branches. Trees had high statistical supports and sorted sequences into six well-supported lineages (Figures 3 and 4). The final matrix of 48 nuclear RPS7 gene sequences consisted of 734 basepairs containing 213 variable characters with 155 parsimony informative sites.
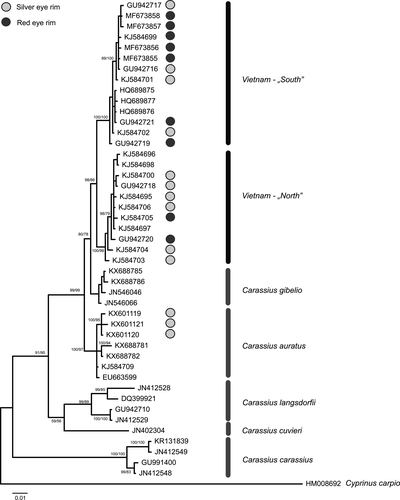
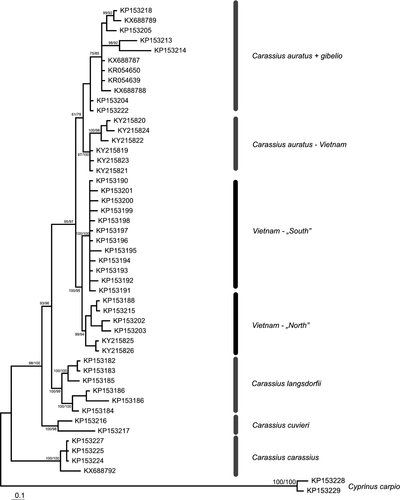
The phylogenetic reconstructions obtained from both markers were highly congruent and identified six lineages within the analyzed material. Five of them refer to C. carassius, C. cuvieri, C. auratus, C. gibelio, and C. langsdorfii lineages as observed in former studies (e.g., Gao et al., 2012). Moreover, some of the samples from Diên Biên Phu (Vietnam), we sequenced, clustered within C. auratus. The sixth lineage is composed of Vietnamese samples exclusively, but is clearly isolated from other lineages. Within the newly detected Vietnamese lineage, two sublineages are visible. One lineage gather all samples from northern Vietnam (except those that cluster with C. auratus) and another gathering samples from central Vietnam.
3.2 Ploidy level determination
Results showed that samples sharing “Vietnamese” haplotype are diploids (2n) with one exception of sample KJ584700 which was determined as triploid (3n). Particular values measured via flow cytometry are listed in Table 1.
4 DISCUSSION
Both our phylogenetic reconstructions indicate the existence of six major lineages within the analyzed material. The relationships observed here are very similar to those reported in former studies (Gao et al., 2012; Kalous et al., 2012; Luo et al., 2014; Rylková et al., 2013; Takada et al., 2010 and Yamamoto et al., 2010). Five of recovered lineages correspond to five of six valid species in the genus Carassius (C. carassius, C. cuvieri, C. langsdorfi, C. gibelio and C. auratus, respectively), while the sixth lineage is independent from all species presently considered as valid. This new lineage contains mainly samples from Vietnam; therefore, we will refer to it as the “Vietnamese lineage.” The detailed congruence between the phylogenies based on the nuclear as well as on the mitochondrial marker indicate a genetic isolation of the Vietnamese lineage from the other lineages over a prolonged period of time with no recent gene flow. This allows us to conclude that the Vietnamese lineage is an own evolutionary unit within the genus Carassius. The clear genetic isolation between the Vietnamese lineage and the recognized species of Carassius and the fact that the Vietnamese lineage and C. auratus co-occur in northern Vietnam strongly suggest that it represents a distinct species of Carassius. The species could be the C. argenteaphthalmus already mentioned in Gao et al. (2012) as lineage C1, including also four individuals from central Vietnam. The identification of this species is presently hampered by uninformative descriptions and lack of type material. The individuals that have been collected at the type locality of C. argenteaphthalmus all appeared in the C. auratus lineage.
Up to here, our results are not contradicting with the assumption of Nguyen and Ngo (2001) that two species of Carassius occur in Vietnam. Nguyen & Ngo further postulated that the color of the eye rim is the diagnostic character to differentiate between them. However, mapping the color of the eye rim on our phylogenetic tree (Figure 3) reveals that red as well as silver eye rims are found in the Vietnamese lineage, while the individuals of C. auratus are usually said to possess silver eye rims (e.g., Kottelat & Freyhof, 2007) contrary to Nguyen and Ngo (2001). Consequently, we consider the color of the eye rim as unsuited to diagnose species of Carassius.
The known distribution of the Vietnamese lineage reaches from the Pearl River basin and Fujian province in southeast China southwards to Quang Nam province in Central Vietnam. However, the samples for the present analysis originated from two separated geographic regions, first the very north of Vietnam and adjacent regions of southeast China and second from Central Vietnam. In accordance with this geographic sampling pattern, the samples from northern and Central Vietnam form separate sublineages in both phylogenies. Since there is a considerable geographic distance between these areas from where no samples have been included (about 600 km), it stays speculative if these sublineages represent separate genetic units in nature or if they are an artifact of the sampling pattern. Moreover, clustering of samples from South China and North Vietnam is not surprising since the area of North Vietnam and mainland China forms a zoogeographic region where North Vietnam represents the southern limit for mainland China species (Mai, 1985; Orsi, 1974). The fish fauna of North Vietnam is more closely related to that of southern China and Hainan than to southern parts of Vietnam (Mai, 1985). To avoid over-interpretation at this point, we will restrict the discussion to the Vietnamese lineage in general and not discuss its sublineages.
It is well-known that representatives of the genus Carassius may bear various ploidy levels, most often diploids and triploids (Apalikova, Eliseikina, Kovalev, & Brykov, 2008; Iguchi, Yamamoto, Matsubara, & Nishida, 2003; Lusková, Halačka, Vetešník, & Lusk, 2004; Rylková et al., 2013; Takada et al., 2010). Tetraploids were also recorded (Abramenko, Nadtoka, Makhotkin, Kravchenko, & Poltavtseva, 2004; Xiao et al., 2011) but these individuals could be the result of hybridization (Knytl, Kalous, Symonová, Rylková, & Ráb, 2013; Mezhzherin, Kokodii, Kulish, Verlatii, & Fedorenko, 2012). In the case of the Vietnamese lineage, almost all analyzed individuals were diploids with only one exception. This information showed that also in this lineage ploidy level shift occurs as they do in other polyploid members of the genus.
Our findings led us to the following conclusions: (i) color of eye rim does not match the genetic background of Carassius fishes, thus this character cannot be considered as a distinctive marker at any taxonomic level; (ii) a newly identified genetic lineage within the C. auratus complex does not match C. auratus sensu stricto; (iii) analysis of mitochondrial DNA suggests that the newly identified clade belongs to the C. auratus complex, but integrity of the clade within the nuclear analysis shows its outstanding position. It is most probably preserved due to geographical conditions unlike in the case of C. auratus and C. gibelio, in which nuclear monophyly is lost by secondary hybridization of both previously separated taxa. Further (iv) the name C. (a.) argenteaphthalmus is considered nomen dubium, and (v) the genetic structure within the “Vietnamese” samples shows clear geographical pattern, which was reported in a broader scale by Gao et al. (2012) and Takada et al. (2010) in Asia.
ACKNOWLEDGEMENT
We owe thanks to Joerg Bohlen for several discussions and various suggestions during the preparation of the manuscript, Pavlína Kuříková for help with processing of genetic samples. We are also grateful to the editors Thomas Stach and Elisabeth Haring for their careful work. Their comments, and suggestions significantly improved our manuscript. This work was supported by project of Official Development Cooperation between the Czech Republic and the Socialist Republic of Vietnam financed by the Ministry of Agriculture of the Czech Republic (No. 29/MZe/B/08-10), the Czech Development Agency (No: 22/2015/12) and by the grant CIGA, No. 20182013.




