Multilocus phylogeny and species delimitation within the genus Glauconycteris (Chiroptera, Vespertilionidae), with the description of a new bat species from the Tshopo Province of the Democratic Republic of the Congo
Abstract
The genus Glauconycteris Dobson, 1875 currently contains 12 species of butterfly bats, all endemic to sub-Saharan Africa. Most species are rarely recorded, with half of the species known from less than six geographic localities. The taxonomic status of several species remains problematic. Here, we studied the systematics of butterfly bats using both morphological and molecular approaches. We examined 45 adult specimens for external anatomy and skull morphology, and investigated the phylogeny of Glauconycteris using DNA sequences from three mitochondrial genes and 116 individuals, which in addition to outgroup taxa, included nine of the twelve butterfly bat species currently recognized. Four additional nuclear genes were sequenced on a reduced sample of 69 individuals, covering the outgroup and Glauconycteris species. Our molecular results show that the genus Glauconycteris is monophyletic, and that it is the sister-group of the Asian genus Hesperoptenus. Molecular dating estimates based on either Cytb or RAG2 data sets suggest that the ancestor of Glauconycteris migrated into Africa from Asia during the Tortonian age of the Late Miocene (11.6–7.2 Mya), while the basal diversification of the crown group occurred in Africa at around 6 ± 2 Mya. The species G. superba is found to be the sister-group of G. variegata, questioning its placement in the recently described genus Niumbaha. The small species living in tropical rainforests constitute a robust clade, which contains three divergent lineages: (i) the “poensis” group, which is composed of G. poensis, G. alboguttata, G. argentata, and G. egeria; (ii) the “beatrix” group, which contains G. beatrix and G. curryae; and (iii) the “humeralis” group, which includes G. humeralis and a new species described herein. In the “poensis” group, G. egeria is found to be monophyletic in the nuclear tree, but polyphyletic in the mitochondrial tree. The reasons for this mito-nuclear discordance are discussed.
1 INTRODUCTION
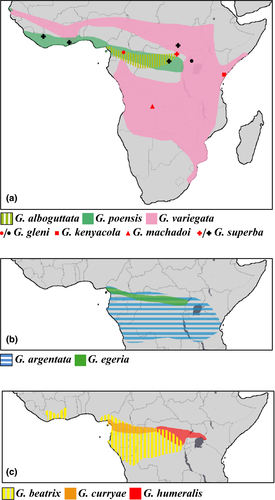
The genus Glauconycteris Dobson, 1875 currently contains 12 species, which are all endemic to sub-Saharan Africa (ACR 2016; IUCN 2016; Figure 1). Their vernacular name, butterfly bats, apparently refers to some resemblance with a large butterfly or moth while in flight, as well as their attractive appearance (Happold & Happold, 2013; Rambaldini, 2010). The pelage of the pied butterfly bat (Glauconycteris superba Hayman, 1939) is probably the most spectacular, with a black body strikingly marked with white spots on the head and shoulders, and white stripes on the throat, along each side of the belly, and on the back. Most species are rarely recorded and poorly known: Glauconycteris curryae Eger & Schlitter, 2001; Glauconycteris egeria Thomas, 1913; Glauconycteris gleni Peterson & Smith, 1973; and G. superba have been collected from only a few localities (respectively, 6, 6, 2, and 6); whereas Glauconycteris machadoi Hayman, 1963 and Glauconycteris kenyacola Peterson, 1982 are known only from the holotype (ACR 2016; Happold & Happold, 2013; Ing et al., 2016).
In early classifications, Glauconycteris was treated as a subgenus of Chalinolobus (e.g., Dobson, 1875; Koopman, 1971, 1994; Ryan, 1966), because the external appearance of butterfly bats was regarded as similar to that of the wattled and pied bats found in Oceania. However, based on skull and baculum differences, many authors have considered Glauconycteris and Chalinolobus as distinct genera (e.g., ACR 2016; Happold & Happold, 2013; Hill & Harrison, 1987; Miller, 1907; Tate, 1942). Molecular studies have shown that Glauconycteris and Chalinolobus are not closely related within the family Vespertilionidae (Hoofer & Van Den Bussche, 2003; Koubínová, Irwin, Hulva, Koubek, & Zima, 2013; Roehrs, Lack, & Van Den Bussche, 2011). On the one hand, Chalinolobus was found to be closely related to Nyctophilus and Vespadelus (two other genera of Oceania) into a larger clade containing four additional genera, that is, Hypsugo, Laephotis, Neoromicia, and Nycticeinops. On the other hand, Glauconycteris was related to the genera Arielulus, Eptesicus (including Histiotus macrotus), Hesperoptenus, Ia, Lasionycteris, Nycticeius, and Scotomanes. However, the monophyly of this group (sometimes referred to as the tribe Nycticeiini) was poorly supported and the position of Glauconycteris remained unresolved. Although only four species of Glauconycteris were represented in previous molecular studies, the genus was found to be monophyletic in the trees of Hoofer and Van Den Bussche (2003) and Roehrs et al. (2011), but Arielulus cuprosus was nested within Glauconycteris in the tree of Koubínová et al. (2013). Because of its unique morphology, the pied butterfly bat (G. superba) has been considered as the sister-group of Glauconycteris sensu stricto and placed into its own genus Niumbaha by Reeder, Helgen, Vodzak, Lunde, and Ejotre (2013). This taxonomic hypothesis, however, has not been tested using molecular data.
Here, we carry out an integrated systematic study to clarify the taxonomic status of butterfly bats collected in 2013 in the Bas-Uele and Tshopo Provinces of the Democratic Republic of the Congo and in southern Central African Republic. A selection of 45 adult specimens is studied for external anatomy and skull morphology. The molecular phylogeny of the genus Glauconycteris is examined based on a data set including three mitochondrial and four nuclear genes (representing 6,179 characters), a diversity of outgroup taxa, and all species of Glauconycteris, except G. gleni and the two species known only from their holotype, G. kenyacola and G. machadoi. Our objectives were to address the following questions: (i) Is the genus Glauconycteris monophyletic? (ii) Which genera are closely related to Glauconycteris? (iii) What are the species relationships within Glauconycteris? (iv) How did Glauconycteris evolve in geological time, specifically during the Neogene and Quaternary, with a particular interest in the biogeographic history and changes in coloration pattern?
2 MATERIALS AND METHODS
2.1 Taxonomic sampling
Most of the specimens analyzed in this study were collected by some of the authors (AH, GCG, PMA, RC, TG, and VTT) using mist-nets (Ecotone, Gdynia, Poland) during field surveys conducted in the Bas-Uele and Tshopo Provinces of the Democratic Republic of the Congo (DRC) (samples identified in Appendix 1 by the code “K13”) and in the Lobaye and Sangha-Mbaéré Provinces of the Central African Republic (CAR) (identified in Appendix 1 by the codes “R08” and “R13,” respectively). After capture, all bats were measured, photographed, and tentatively identified using the keys published in Rambaldini (2010) and Patterson and Webala (2012). For voucher specimens deposited in the MNHN collections (see details in Appendix 1), taxonomic identifications were refined after skull extraction using the data published in Allen (1917), Eger and Schlitter (2001), Happold and Happold (2013), Rosevear (1965), and Thomas (1901, 1913).
A few other specimens or samples were loaned by the following institutions: Ditsong National Museum of Natural History (formerly known as the Transvaal Museum; DNMNH; Pretoria, South Africa), Durban Natural Science Museum (DNSM; Durban, South Africa), Field Museum of Natural History (FMNH; Chicago, USA), Hungarian Natural History Museum (HNHM; Budapest, Hungary), Iziko Museum (also known as the South African Museum [SAM]; Cape Town, South Africa), and Royal Ontario Museum (ROM; Toronto, Canada) (see details in Appendix 1).
2.2 Morphological analysis
A total of 45 adult specimens of Glauconycteris were examined for morphological comparisons (Tables S1 and S2). Besides mass (W, expressed in grams), the following external measurements were taken using digital calipers accurate to 0.01 mm (acronym for each measurement presented in parentheses): forearm length (FA)—from the elbow to the wrist with both joints folded; head and body length (HB)—from the tip of the face to the anus; tail length (Tail)—from the anus to tip of the tail; tibia length (TIB)—from the knee to the ankle; the length of the 1st finger (F1); the lengths of 2nd, 3rd, 4th, and 5th metacarpals (2DM, 3DM, 4DM, and 5DM, respectively)—from the wrist to the end of the respective metacarpals; the lengths of the first and second phalanges of the 3rd digit (3D1P and 3D2P). After skull extraction, nine cranial and six dental measurements were taken using digital calipers accurate to 0.01 mm. Abbreviations and definitions for craniodental measurements include: mandible length (ML)—greatest length of the mandible, measured from the front of incisors to the condylar processes; mandible width (MW)—width across outer points of right and left rami of the mandible; greatest length of skull (GLS)—from anterior-most point of teeth to the most posteriorly projecting point of the occipital region; zygomatic width (ZW)—greatest width of the skull across the zygomatic arches; braincase width (BCW)—greatest width of the braincase; braincase height (BCH)—from the base of the auditory bullae to the highest part of the skull; interorbital width (IOW)—least width of the interorbital constriction; mastoid breadth (MB)—greatest distance across skull at mastoid processes; least width of the palate (LWP); complete upper canine-molar toothrow (C-M3), length from anterior alveolar border of upper canine (C1) to posterior alveolar border of 3rd molar (M3); width across upper canines (C1-C1), taken across the outer alveolar borders of the canines; width across 3rd upper molars (M3-M3), taken across the outer borders of the 3rd molars; complete lower canine-molar toothrow (C-M3), length from anterior alveolar border of lower canine (C1) to posterior alveolar border of 3rd molar (M3); width across lower canines (C1-C1), taken across the outer alveolar borders of the canines; width across 3rd lower molars (M3-M3), taken across the outer borders of the 3rd molars.
Based on our observations, we also described the following 11 categorical variables: wing colour (WC), with four character-states (A: brown; B: pale; C: black; D: pale yellowish-orange with dark brown pigment); dorsal general color (DGC), with seven character-states (A: blackish brown; B: gray/golden-fawn; C: reddish-brown; D: curry/reddish-fawn; E: brown; F: piebald pattern; G: yellow); dorsal hairs (DH), with three character-states (A: unicolor; B: bicolor; C: tricolor); ventral general color (VGC), with three character-states (A: lighter than dorsal color; B: similar to dorsal color; C: piebald pattern); shoulder spot (SS), with three character-states (A: conspicuous white spot; B: no white spot; C: faint white spot); white flank stripes (FS), with three character-states (A: flank stripe close to the wing membrane; B: flank stripe more dorsally located; C: no flank stripe); inner margin of tragus (IMT), with two character-states (A: straight; B: strongly concave); outer margin of tragus (OMT), with four character-states (A: convex; B: slightly convex; C: circular; D: straight); tragus color (TC), with four character-states (A: brown; B: pale; C: dark brown; D: black); ears color (EC), with two character-states (A: identical to that of the tragus; B: different); and skull profile (SP), with four character-states (A: strongly concave; B: concave; C: weakly concave; D: straight).
Continuous and categorical variables were analyzed together using the Factor Analysis of Mixed Data (FAMD) method in the FactoMineR package (Lê, Josse, & Husson, 2008) in R version 3.2.1. (R Core Team 2015).
2.3 DNA extraction, amplification, sequencing
Total genomic DNA was extracted from muscle or patagium samples (preserved in 95% ethanol) using QIAGEN DNeasy Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol with a final elution of 50 μl.
Three mitochondrial genes were sequenced for this study: the 5′ fragment of the CO1 gene, the complete cytochrome b (Cytb) gene, and the 5′ fragment of the 12S rRNA gene (12S). The primers used for PCR amplification of mitochondrial genes were UTyrLA and C1-L705 for CO1 (Hassanin et al., 2012), CB-GLU-CH2 and CB-LTHR-CH for Cytb (Hassanin, 2014) and 12S-U1230M2-CH (5′-GCA-CTG-AAA-ATG-CYT-AGA-TG-3′) and 12S-L2226M1 (Hassanin et al., 2012) for 12S. Four nuclear genes were sequenced for a subset of samples: intron 10 of HDAC2 (histon deacetylase 2 gene), intron 6 of RIOK3 (RIO kinase 3 gene), intron 6 of ZFYVE27 (zinc finger, FYVE domain containing 27), and the recombination activating gene 2 (RAG2). The primers used for amplifying the nuclear introns are detailed in Hassanin, An, Ropiquet, Nguyen, and Couloux (2013), those used for RAG2 were specifically designed for this study: RAG2-CHU (5′-CTT-CGC-TAC-CCA-GCC-ACT-TGC-A-3′) and RAG2-CHL (5′-GGC-AGG-CTT-GTT-TAG-CTC-AGT-TG-3′). Amplifications were performed in 20 μl using 3 μl of Buffer 10X with MgCl2, 2 μl of dNTP (6.6 mM), 0.12 μl of Taq DNA polymerase (2.5 U, Qiagen, Hilden, Germany) and 0.5–1 μl of the two primers at 10 μM. The PCRs were run using the C1000 Touch thermal cycler (BIO-RAD, Hercules, California, USA) with the following standard conditions: 4 min at 94°C; five cycles of denaturation/annealing/extension with 45 s at 94°C, 1 min at 60°C, and 1 min at 72°C, followed by 30 cycles of 30 s at 94°C, 45 s at 55°C, and 1 min at 72°C, followed by 10 min at 72°C. PCR products were resolved by electrophoresis on a 1.5% agarose gel stained with ethidium bromide and visualized under UV light. Both strands of PCR products were sequenced by Eurofins MWG Operon (Ebersberg, Germany) on an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, California, USA). The sequences were edited and assembled using Sequencher 5.1 (Gene Codes Corporation, Ann Arbor, Michigan, USA). Heterozygous positions (double peaks) were scored using the IUPAC ambiguity codes. Sequences generated for this study were deposited in the GenBank database (accession numbers MF038194-MF038764; for more details see Appendix 1).
2.4 Phylogenetic analyses of the mtDNA data sets
The phylogeny of Glauconycteris was initially analyzed using 116 specimens and three mitochondrial genes (CO1, Cytb, and 12S) in order to allow comparisons with all published mtDNA sequences of Glauconycteris. The 101 CO1, 99 Cytb, and 96 12S sequences newly generated in this study were compared to 10 CO1, 5 Cytb, and 7 12S sequences downloaded from GenBank (for more details see Appendix 1). To identify the sister-group of Glauconycteris, we included 20 outgroup taxa representing a large diversity of Vespertilionidae and, in particular, all genera found closely related to Glauconycteris in molecular studies (Hoofer & Van Den Bussche, 2003; Koubínová et al., 2013; Roehrs et al., 2011). DNA sequences were aligned automatically using MUSCLE (Edgar, 2004) and then manually on Seaview 4.4.0 (Gouy, Guindon, & Gascuel, 2010). The four data sets used for mtDNA analyses were CO1 (111 specimens and 705 nt), Cytb (104 specimens and 1,140 nt), 12S (103 specimens and 959 nt), and the concatenation of the three genes, named mtDNA-116T (116 specimens and 2,804 nt). All trees were rooted with the genus Myotis, in agreement with previous molecular studies (e.g., Hoofer & Van Den Bussche, 2003; Koubínová et al., 2013).
The Bayesian method was used to reconstruct phylogenetic relationships. The best-fitting model of sequence evolution was selected under jModelTest 2.1.7 (Darriba, Taboada, Doallo, & Posada, 2012) using the Akaike Information Criterion (AIC). Bayesian inferences were then conducted on MrBayes v3.2.1 (Ronquist et al., 2012) using the selected GTR+G+I model for CO1, Cytb, 12S, and mtDNA-116T data sets. To account for the combination of markers with contrasted molecular properties, we applied a partitioned approach using different models for 12S, and each of the three codon-positions of the two protein genes CO1 and Cytb. The posterior probabilities (PP) were calculated using four independent Markov chains run for 10,000,000 Metropolis-coupled MCMC generations, with trees sampled every 1,000 generations, and a burn-in of 25%.
The results obtained from the separate Bayesian analyses of the three mtDNA markers (Fig. S1) were also analyzed for congruence using the SuperTRI method (Ropiquet, Li, & Hassanin, 2009). The lists of bipartitions obtained from the three Bayesian analyses of mtDNA genes were transformed into a weighted binary matrix for supertree construction using SuperTRI v.57 (available at http://www.normalesup.org/~bli/Programs/programs.html). Each binary character corresponds to a node, which was weighted according to its frequency of occurrence in one of the three lists of bipartitions. In this manner, the SuperTRI method takes into account both principal and secondary signals, because all phylogenetic hypotheses found during the three separate analyses are represented in the weighted binary matrix used for supertree construction. The reliability of the nodes was assessed using three different measures. The first value is the Supertree Bootstrap Percentage (SBP), which was calculated under PAUP* v.4b10 (Swofford, 2003) after 1,000 BP replicates of the weighted binary matrix reconstructed with SuperTRI (2,038 characters; heuristic search). The second value is the “Mean Posterior Probability” (MPP) calculated using the lists of bipartitions obtained from Bayesian analyses of the three mtDNA data sets. The third value is the index of reproducibility (Rep), which is the ratio of the number of data sets supporting the node of interest to the total number of data sets. The MPP and Rep values were directly calculated on SuperTRI v.57. All SuperTRI values were reported on the Bayesian tree obtained from the analysis of the mtDNA-116T data set.
2.5 Phylogenetic analyses based on five independent data sets
The phylogeny of Glauconycteris was further investigated using a reduced sample of 69 individuals (including 54 Glauconycteris) sequenced for multiple loci, including four nuclear genes (HDAC2, RIOK3, ZFYVE27, and RAG2) and a concatenation of the three mtDNA genes CO1, Cytb, and 12S. It was not possible to include the species G. poensis in these analyses, because the quality of the DNA extracted from specimens DM 14186, DM 14187, and DM 14188 was not sufficient to obtain successful amplification of nuclear markers. The nuclear analyses were performed to test possible discordance between the phylogenetic signals extracted from independent markers, and to avoid misleading taxonomic interpretations due to either mtDNA introgression, lineage sorting, or different dispersal behavior between females and males (e.g., Hassanin et al., 2015; Nesi, Nakouné, Cruaud, & Hassanin, 2011). A few gaps were included in the alignments of the nuclear introns, but their positions were not found to be ambiguous. The indels shared by at least two individuals were coded as additional characters (“1”: insertion; “0”: deletion) and analyzed as a separate partition in the Bayesian analyses.
A total of seven data sets were analyzed: (i) supermatrix, combining all the seven genes (6,124 nt + 55 indels); (ii) nuDNA, combining all the four nuclear genes (3,320 nt + 55 indels); (iii) mtDNA-69T, combining the three mtDNA genes (2,804 nt); (iv) HDAC2 (888 nt + 20 indels); (v) RIOK3 (656 nt + 17 indels); (vi) ZFYVE27 (759 nt + 18 indels); and (vii) RAG2 (1,017 nt). The Bayesian analyses were performed as detailed above for mitochondrial data sets. The following models of nucleotide evolution were selected under jModelTest using ACI: GTR+G+I for mtDNA-69T and RAG2, GTR+G for HDAC2, and HKY+G for RIOK3 and ZFYVE27. The results obtained from the separate Bayesian analyses of the five independent data sets (mtDNA-69T and the four nuclear genes; Fig. S2) were also analyzed for congruence using the SuperTRI method (Ropiquet et al., 2009), and the reliability of the nodes was assessed using SBP (weighted binary matrix = 2,693 characters), MPP, and Rep values (see above for details). All SuperTRI values were reported on the Bayesian tree obtained from the nuDNA analysis.
2.6 Molecular dating
Divergence times were estimated with the Bayesian approach implemented in BEAST v.2.1.3 (Bouckaert et al., 2014). As no fossil record or biogeographic event is available for Glauconycteris, we employed two different strategies for molecular dating: The first one is based on Cytb haplotypes (77 sequences and 1,140 nt) and a priori assumptions on the rate of substitutions; the second one is based on RAG2 sequences from a large diversity of Vespertilionidae (126 sequences and 1,017 nt) and the use of several calibration points. In agreement with published data on the nucleotide substitution rates in the Cytb of mammals (Arbogast & Slowinski, 1998), we tested two substitution rates for estimating divergence times with the Cytb alignment: R1 = .02 ± .005 and R2 = .025 ± .005 per site per lineage per Myr. For the RAG2 alignment, we used three molecular calibration points extracted from the studies of Teeling et al. (2005) and Meredith et al. (2011): 10 ± 3 Mya for the most recent common ancestor (MRCA) of Antrozous and Rhogeessa; 20.5 ± 4.5 Mya for the MRCA of Myotis and Antrozous; and 46 ± 5 Mya for the MRCA of Myotis and Miniopterus. In agreement with Lack, Roehrs, Stanley, Ruedi, and Van Den Bussche (2010), the RAG2 tree of the family Vespertilionidae was rooted with Cistugo seabrae (Cistugonidae) and Miniopterus inflatus (Miniopteridae).
For all analyses, we applied a GTR+G+I model of evolution (based on jModelTest) for each of the three codon-positions of Cytb or RAG2 genes, and a relaxed-clock model with uncorrelated lognormal distribution for substitution rates. Node ages were estimated using a Yule speciation prior and 108 generations, with tree sampling every 1,000 generations, and a burn-in of 10%. Adequacy of chain mixing and MCMC chain convergence was assessed using the ESS values in Tracer v.1.6 (available in the BEAST package). The chronograms were generated with TreeAnnotator v.1.8.2 (available in the BEAST package) and visualized with FigTree v.1.4.1 (http://tree.bio.ed.ac.uk/software/).
3 RESULTS
3.1 Taxonomic identifications
In the DRC, we caught 53 individuals of Glauconycteris, including 29 females and 23 males. We identified seven species at different localities on the left bank [L] and/or right bank [R] of the Congo River: G. alboguttata (four individuals; Melume [R]), G. argentata (11 individuals; Yoko [L]; Melume [R], Mbiye [R], and Sukisa [R]), G. curryae (five individuals; Yaengo [L], Yatolema [L], and Yoko [L]), G. superba (11 individuals; Yoko [L] and Mbiye [R]), and three species within the beatrix/humeralis morpho-group (22 individuals; seven localities), that is, a first species identified as G. beatrix, which is represented by 16 dark sepia brown bats with faint white shoulder spots (Yatolema [L], and Yoko [L]; Bongandjolo, Mbiye [R], Melume [R], and Sukisa [R]); a second species identified as G. humeralis, characterized by smaller forearm and tibia lengths than females of G. beatrix (FA = 35.6 versus 36.9–39.5; TIB = 16.4 versus 18.8–20.1), a more reddish-brown color, and the presence of conspicuous white shoulder spots (rather than faint white shoulder spots in G. beatrix), which is represented by one female collected at Sukisa [R]; and a third species, which is described below, represented by five blackish brown bats without white markings (Yaengo [L], Yatolema [L], and Yoko [L]).
Our analyses also include 20 individuals of Glauconycteris collected in Dzanga-Sangha (southern CAR; code “R13”), representing five species, that is, G. alboguttata (eight individuals), G. curryae (one individual), G. egeria (six individuals), G. beatrix (three individuals with TIB > 19), and G. cf. humeralis (two females with TIB < 18 and a conspicuous white spot on each shoulder); one male of G. egeria from Mbaéré-Bodingué (southern CAR; MNHN 2016-2800); three individuals of G. poensis from Liberia (DM 14186 - DM 14188) (reported in Monadjem, Richards, & Denys, 2016); five individuals of G. variegata from Botswana (ECJS-43/2009), South Africa (TM 48494 - TM 48496), and Zambia (ECJS-92/2010); and one female from Cameroon (HNHM 23262) identified as G. beatrix (FA = 39.0; TIB = 20.1), which is characterized by the absence of white markings.
For all nine species identified in DRC and CAR, we found that females have longer forearms than males (Fig. S3). A female-biased size dimorphism was also observed for other measurements, such as the tibia length and weight (data not shown).
3.2 Systematic description
3.2.1 Glauconycteris atra sp. nov
Holotype
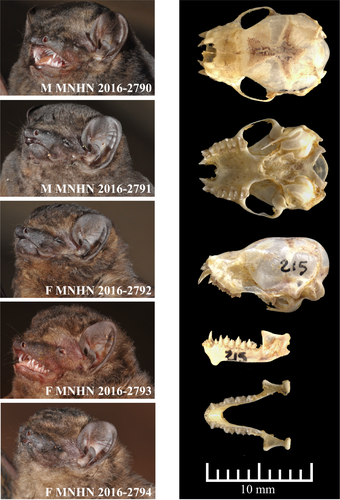
MNHN 2016-2792 (field number: K13-215), adult female, in alcohol, skull removed, collected on December 05, 2013 by AH, GCG, PMA, RC, TG, and VTT. The holotype was used in the molecular and morphological comparisons presented herein. W: 8.5 g; FA: 37.8; Tib: 17.9; T: 42.6; HB: 55.8; F1: 4.4; 2DM: 37.3; 3DM: 38.8; 3D1P: 13.8; 3D2P: 23.0; 4DM: 36.5; 5DM: 34.2; GLS: 12.63; ZW: 9.34; BCW: 7.49; IOW: 4.61; MB: 8.15; BCH: 7.66; LWP: 2.56; M3-M3: 4.05; C1-C1: 2.54; C-M3: 4.20; C-M3: 4.45; M3-M3: 5.87; C1-C1: 4.35; ML: 8.63; MW: 7.31 mm. Accession numbers of mitochondrial and nuclear sequences are MF038602 (CO1), MF038501 (Cytb), MF038702 (12S), MF038428 (HDAC2), MF038290 (RIOK3), MF038359 (RAG2), and MF038222 (ZFYVE27).
Type locality
Yaengo, Tshopo Province, Democratic Republic of the Congo: 0.34887°N; 24.48200°E; 400 m above sea level (a.s.l.).
Referred specimens
One additional individual from Yaengo (MNHN 2016-2791: adult male); two individuals from Yatolema (0.41319°N; 24.53908°E; 460 m a.s.l.; MNHN 2016-2790: adult male; MNHN 2016-2793: adult female); and one individual from Yoko (0.29410°N; 25.28895°E; 412 m a.s.l.; MNHN 2016-2794: adult female). Accession numbers of mitochondrial and nuclear sequences are detailed in Appendix 1.
Etymology
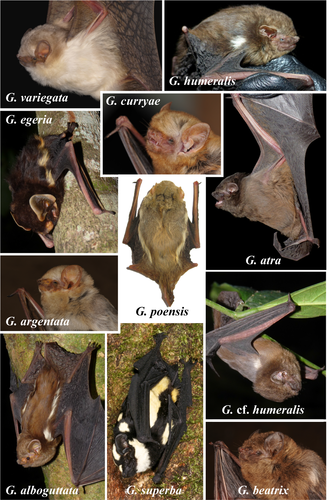
The specific epithet refers to the pelage color, which is dark and gloomy (blackish brown) without any white markings (Figure 2). We propose “Blackish Butterfly Bat” as the English common name and “Glauconyctère sombre” as the French common name.
Diagnosis
A small-sized species of Glauconycteris with a forearm length between 34.4 and 37.8 mm (n = 5), with some evidence of sexual dimorphism, as males tend to be slightly smaller than females (Fig. S3). It has a stocky build, with a weight range of 8.5–8.7 g for females (n = 3) and 6.9–7.7 g for males (n = 2), which is much heavier than other small-sized species of Glauconycteris collected in northeastern DRC, such as G. beatrix (3.9–6.1 g; n = 10), G. curryae (3.6–5.0 g; n = 5), and G. humeralis (4.7 g; n = 1). The dorsal pelage color of the living animal is blackish brown without any white markings. The ventral pelage is paler. Dorsally, the fur extends onto the plagiopatagium along the proximal half of the humerus and femur and onto the uropatagium along the proximal half of the tail. The two males are darker in color than the three females (Figure 2). The sexual dimorphism in pelage color, however, needs to be confirmed by observations of additional specimens. Dorsal hairs are 6–8 mm long and tricolored: dark brown at the base, beige in the middle, and blackish brown at the tip. The wings, interfemoral membrane, ears, and naked skin around muzzle and eyes are blackish brown. The inner margin of the tragus is straight or slightly curved, whereas the outer margin of the tragus is well rounded (Figure 2). Although similar in appearance to other species of Glauconycteris, the skull of G. atra sp. nov. is larger than in related species, such as G. beatrix, G. curryae, and G. humeralis, with little overlap in the different measurements (MW, ZW, BCW, MB). In addition, the braincase is relatively higher (BCH) (see in Table S1).
Description
Based on pelage coloration, G. atra sp. nov. cannot be confused with any other known species of Glauconycteris. It lacks the conspicuous body patterns of white spots, white stripes, or reticulated wings found in most other species of Glauconycteris. Its general color is much darker than the sepia- or reddish-brown colors of G. beatrix, G. curryae, and G. humeralis. The forearm length of G. atra sp. nov. ranges from 36.1 to 37.8 mm in females (n = 3) and from 34.5 to 36.8 mm in males (n = 2), which is similar to G. curryae and G. humeralis, but slightly smaller than G. beatrix (Fig. S3). The tibia length ranges from 17.2 to 17.9 mm in females (n = 3) and from 16.3 to 17.2 mm in males (n = 2), which is similar to G. curryae, but slightly smaller than G. beatrix, and slightly larger than G. humeralis. The tail is shorter than HB. The ears are separated, short (10–11 mm) and rounded. In lateral view, the skull profile of the forehead region is moderately concave in G. atra sp. nov., as observed in G. humeralis and some individuals of G. beatrix. By contrast, the profile is strongly concave in G. curryae. The dental formula of G. atra sp. nov. is I 2/3 C 1/1 P 1/2 M 3/3 = 32 teeth, which is identical to that of other Glauconycteris species. The inner upper incisors are strongly bicuspid with the outer cusp smaller than the inner cusp, as observed in G. beatrix, G. curryae, and G. humeralis. The inner lower incisors have three cusps, whereas the two outer incisors have four cusps.
Distribution and natural history
All five specimens of G. atra sp. nov. were caught in riparian zones on the left bank of the Congo River, at three localities of the Tshopo Province of the Democratic Republic of the Congo: Yaengo, Yatolema, and Yoko. The new species was found in sympatry with G.argentata (Yoko), G. beatrix (Yatolema and Yoko), G. curryae (Yaengo and Yatolema), and G. superba (Yoko).
3.3 Morphological analyses
We examined 45 adult specimens, representing 11 species, for which we measured 26 quantitative variables and coded 11 qualitative variables (Table S1). Using the FAMD method, the dimensions 1, 2, 3, 4, and 5 account for 46.8%, 11.5%, 8.8%, 7.2%, and 6.5% of total variance, respectively. The plot of Dimension 1 against Dimension 2 (Figure 3) shows the existence of several clusters corresponding to G. alboguttata, G. argentata, G. beatrix, G. poensis, G. superba, and G. variegata. By contrast, these two dimensions cannot be used to distinguish individuals of G. atra sp. nov. from G. curryae, G. cf. beatrix, and G. egeria. Several specimens housed in the ROM identified as G. beatrix are hereafter referred to as. G. cf. beatrix as they do not plot with other G. beatrix, and overlap instead with individuals of G. curryae. Most variables contribute to the construction of Dimension 1, whereas Dimension 2 is mainly explained by the quantitative variable 18 (tail length) and the qualitative variable q11 (skull profile).
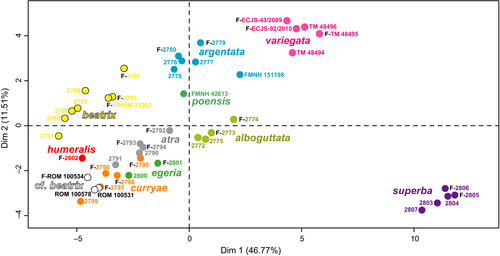
The morphological analysis based only on similar smaller forms (i.e., discarding the obviously different species: G. alboguttata, G. argentata, G. poensis, G. superba, and G. variegata) did not improve the discrimination between these closely related species (data not shown).
3.4 Mitochondrial phylogeny
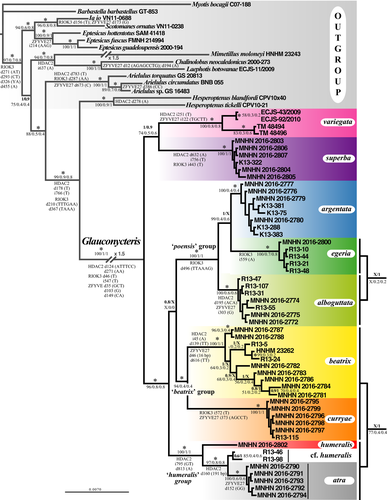
The Bayesian tree obtained from the concatenation of the three mitochondrial genes (CO1, Cytb and 12S; 116 specimens and 2,804 nt) is shown in Figure 4. In the total alignment, the missing data represent 9.5%, because we included 13 additional individuals of Glauconycteris from GenBank, for which no sequence was available for one or two mitochondrial genes (see Appendix 1 for details). Most of the nodes are supported by high values of posterior probabilities (PP = 0.9–1) and high SuperTRI values (SBP ≥ 80, MPP ≥ .5, Rep ≥ .7). As expected, the nodes containing one or several specimens with missing data have lower values of SBP. For instance, the sister-group relationship between individuals of G. egeria (MNHN 2016-2800 and AMNH 268381) was supported by SBP = 40, because only the 12S gene was sequenced for the AMNH specimen.
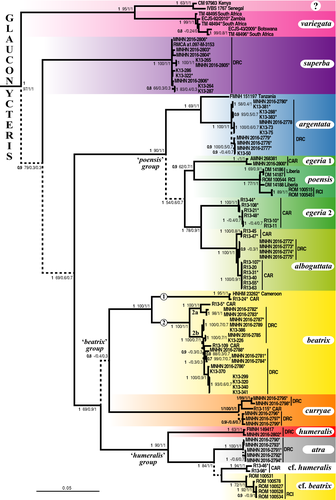
The monophyly of the genus Glauconycteris was strongly supported by the analyses, as well as that of the three other genera of Vespertilionidae for which at least two species were included in the data set, that is, Arielulus, Eptesicus, and Hesperoptenus (PP = 1; SBP = 87–100; MPP = 0.7–1; Rep = 0.7–1). Four intergeneric relationships were supported by PP = 1 and by at least two mitochondrial genes (Rep ≥ .7): (i) Glauconycteris + Hesperoptenus; (ii) Ia + Scotomanes; (iii) Chalinolobus + Laephotis, and (iv) their association with Mimetillus.
Within Glauconycteris, our analyses revealed the existence of three major clades: (i) G. variegata; (ii) G. superba; and (iii) a clade uniting all other species, which can be further divided into four robust groups: (i) the “poensis” group, which includes G. poensis, G. alboguttata, G. argentata, and G. egeria 1 and 2; (ii) G. beatrix from Central Africa (Cameroon, CAR, and DRC); (iii) G. curryae; and (iv) the “humeralis” group, which contains G. humeralis and G. atra sp. nov. in DRC, G. cf. humeralis in CAR, and G. cf. beatrix in Republic of Côte d'Ivoire (RCI).
Most species were found to be monophyletic with PP = 1 and Rep = 1 (i.e., supported by the three mtDNA markers), with the exception of G. egeria, which splits into two unrelated haplogroups. The first haplogroup, named G. egeria 1, contains two individuals from CAR (AMNH 268381 from Dzanga-Sangha; MNHN 2016-2800 from Mbaéré Bodingué National Park) (PP = 1; SBP = 40; MPP/Rep = 1). It appears as the sister-group of G. poensis (PP = 0.9; SBP = 62; MPP = 0.7; Rep = 1), from which it differs by 6.5%–8.0% in CO1 sequences and 10% in Cytb sequences. The second haplogroup, named G. egeria 2, is composed of all other individuals collected in Dzanga-Sangha (PP = 1; SBP = 100; MPP/Rep = 1). It appears as the sister-group of G. alboguttata (PP = 1; SBP = 78; MPP = .9; Rep = 1), from which it differs by 5.8%–6.2% in CO1 sequences, 5.8%–6.6% in Cytb sequences, and 2.3%–2.6% in 12S sequences.
Highly divergent mitochondrial haplogroups were also detected in several other species. In G. variegata, there are two geographic haplogroups separated by more than 10.3% in Cytb and 4.2% in 12S. The first haplogroup includes the individuals collected in southern Africa (Botswana, South Africa and Zambia), and the second contains two individuals from two widely spaced localities in East and West Africa (CM 97983: Kenya; IVBS 1767: Senegal). In G. argentata, the 12S sequence from an individual from Tanzania (FMNH 151197) was found to be >2.2% distant from that of individuals collected in DRC. In G. beatrix sensu stricto (s.s.), that is, from Central Africa, there are two highly divergent haplogroups (CO1: >9.4%; Cytb: >8.7%; 12S: >2.8%): The first is only represented by two individuals, that is, one from Cameroon (HNHM 23262) and another one from CAR (R13-24); and the second includes a few individuals from CAR and all the individuals collected in eastern equatorial Africa (DRC). The second haplogroup of G. beatrix s.s. can be subdivided into two subgroups, named 2a and 2b in Figure 4 (CO1: >2.6%; Cytb: >2.3%; 12S: >1.2%).
In the “humeralis” group, there are three geographic haplogroups corresponding to G. atra sp. nov. (DRC), G. cf. beatrix (RCI), and G. cf. humeralis (CAR) (CO1: >4.2%; Cytb: >4.6%; 12S: >3.1%), which are highly distinct from the two specimens from DRC assigned to the species G. humeralis (MNHN 2016-2802 and FMNH 149417, highlighted in red in Figure 4) (CO1: >9.2%; Cytb: >11.7%; 12S: >4.6%).
3.5 Nuclear, Supermatrix, and SuperTRI analyses
The Bayesian tree obtained from the concatenation of the four nuclear genes HDAC2, RAG2, RIOK3, and ZFYVE27 (nuDNA; 69 specimens and 3,375 characters; missing data = 1.8%) is shown in Figure 5, while the Bayesian tree reconstructed with the supermatrix (all mitochondrial and nuclear genes; 69 specimens and 6,179 characters; missing data = 1.3%) is provided in Fig. S4. The topologies are very similar to that inferred from the mtDNA data set (Figure 4). Most nodes of the mtDNA tree were recovered with maximal support values in the nuDNA and supermatrix (mtDNA+nuDNA) trees (PP = 1): Vespertilioninae; the monophyly of the genera Glauconycteris, Arielulus, Eptesicus, and Hesperoptenus; Ia + Scotomanes; Chalinolobus + Laephotis, and their association with Mimetillus; the sister-group relationship between E. fuscus and E. guadeloupensis; the monophyly of the species G. alboguttata, G. argentata, G. beatrix s.s., G. curryae, G. variegata, and G. superba; the monophyly of the “beatrix” group (G. beatrix s.s. + G. curryae), “humeralis” group (G. humeralis + G. atra sp. nov. + G. cf. humeralis + G. cf. beatrix), and “poensis” group (as represented by G. alboguttata, G. argentata, and G. egeria). Most of these nodes were supported by high values in the SuperTRI analyses of the five independent markers (mtDNA and four nuclear genes): SBP ≥ 94; MPP ≥ .7; and Rep ≥ .8. Two nodes were moderately supported: the species G. alboguttata (SBP = 100; MPP/Rep = .6), which is recovered monophyletic in the separate analyses of three independent markers (mtDNA, HDAC2, ZFYVE27; Fig. S2); and the “beatrix” group (SBP = 94; MPP/Rep = .4), which was found monophyletic with only two markers (mtDNA and RAG2; Fig. S2).
A few nodes of the mtDNA tree are clearly discordant with the nuDNA topology. The species G. egeria, which appeared polyphyletic in the mitochondrial tree (PP = 1), is monophyletic in the nuclear tree (PP = 1) and in all separate analyses of the four nuclear genes (Fig. S2). The species G. egeria can be also diagnosed by an insertion of A nucleotide in position 559 of the RIOK3 alignment. Three nuclear genes indicate that it is closely related to G. argentata (HDAC2, RAG2, and ZFYVE27). Within G. beatrix s.s., the nuDNA tree does not corroborate the existence of three divergent haplogroups, as shown in the mitochondrial tree (1, 2a and 2b in Figure 4). Most of the poorly supported nodes of the mitochondrial tree (PP ≤ .9) were not recovered in the nuclear tree. The species G. variegata, which occupied a basal position within Glauconycteris in the mitochondrial tree (PP = .9), is the sister-species of G. superba in the nuclear tree (PP = 1), a result supported by the separate analyses of HDAC2, RAG2, and ZFYVE27 genes (Fig. S2). The genus Arielulus, which was grouped with Eptesicus, Ia and Scotomanes in the mtDNA tree (PP = .8), is allied to Glauconycteris and Hesperoptenus in the nuclear tree (PP = 1), a result recovered in the separate analyses of RAG2 and ZFYVE27 genes (Fig. S2).
The tree reconstructed from the supermatrix (Fig. S4) is in agreement with the nuclear tree for the deepest nodes (i.e., position of Arielulus, G. variegata + G. superba), but it is more similar to the mitochondrial tree for the most recent nodes (e.g., G. egeria +G. alboguttata, the three haplogroups within G. beatrix s.s.). Most topological conflicts between nuclear and supermatrix trees can be resolved using the results from SuperTRI analyses. The topology of the SuperTRI Bootstrap 50% majority-rule consensus tree (data not shown) is identical to the nuclear tree, except some unsupported nodes at the intra-specific levels. The SuperTRI analyses also support the placement of Arielulus as sister-group of the clade uniting Glauconycteris and Hesperoptenus (SBP = 88; MPP = .5; Rep = .4), and the sister-group relationship between G. superba and G. variegata (SBP = 74; MPP = .5; Rep = .6). In agreement with the nuclear tree, the SuperTRI analyses favor the association between G. egeria and G. argentata (SBP = 99; MPP = .4; Rep = .6). In addition, they do not confirm the existence of three haplogroups within G. beatrix s.s., as most intra-specific relationships are associated to low MPP and Rep values (≤.4).
3.6 Molecular dating of Glauconycteris
Two data sets were used for molecular dating: an alignment of 126 RAG2 sequences, which is more appropriate to estimate the deeper nodes of our phylogeny; and an alignment of 77 Cytb haplotypes, which contains a large diversity of Glauconycteris sequences, and which is therefore more accurate to estimate the most recent divergence times. The Cytb alignment was analyzed with two different rates of substitution, R1 = .02 ± .005 and R2 = .025 ± .005 per site per lineage per Myr. The three chronograms RAG2, Cytb R1, and Cytb R2 are available in Fig. S5. For nodes in common, we found that the RAG2 ages estimated with three calibration points were systematically younger than the Cytb ages estimated using either R1 or R2 substitution rates. These comparisons suggest that the faster Cytb R2 rate was more reliable than the slower Cytb R1 rate. In Table 1, we therefore only show the ages estimated with RAG2 and Cytb R2. The 95% intervals were found to be generally larger for RAG2 ages, because the RAG2 data set was analyzed using three calibration points (10, 20.5 and 46 Mya) older than the MRCA of the ingroup and with large standard deviations (between 3 and 5 Mya).
| Taxa | RAG2 | CytbR2 | Epochs |
|---|---|---|---|
| Interspecific relationships | |||
| Glauconycteris + Hesperoptenus + Arielulus | 12.61 (16.7–8.7) | 10.98 (14.4–8.0) | Middle Late Miocene |
| Glauconycteris + Hesperoptenus | 10.93 (14.9–7.1) | 10.25 (13.6–7.4) | Middle Late Miocene |
| Genus Arielulus | 4.48 (8.4–1.3) | 6.36 (8.8–4.1) | Miocene/Pliocene |
| Genus Hesperoptenus | 7.82 (11.4–4.4) | 7.02 (10.0–4.3) | Late Miocene |
| Genus Glauconycteris | 5.70 (8.4–3.3) | 6.32 (8.2–4.6) | Late Miocene |
| G. superba + G. variegata | 3.91 (6.6–1.4) | 5.60 (7.6–3.8) | Pliocene |
| G. superba | NA | 0.19 (0.3–0.1) | Middle/Late Pleistocene |
| G. variegata (southern Africa + Senegal) | NA | 2.44 (3.6–1.4) | Early Pleistocene |
| G. variegata (only southern Africa) | 0.36 (1.2–0.0) | 0.34 (0.5–0.2) | Middle Pleistocene |
| Groups “poensis” + “beatrix” + “humeralis” | 4.19 (6.3–2.3) | 5.17 (6.8–3.8) | Pliocene |
| Groups “beatrix” + “humeralis” | 3.48 (6.3–2.3) | 4.45 (5.8–3.2) | Pliocene |
| Group “beatrix” | 2.87 (4.5–1.4) | 3.93 (5.3–2.7) | Pliocene |
| G. beatrix (Central Africa) | 1.89 (3.2–0.7) | 2.35 (3.4–1.4) | Early Pleistocene |
| G. beatrix but HNHM 23262 (+ R13-24) | 0.85 (1.9–0.0) | 0.67 (1.0–0.4) | Middle Pleistocene |
| G. curryae | 0.28 (0.9–0.0) | 0.41 (0.6–0.2) | Middle Pleistocene |
| Group “humeralis” | 1.47 (3.0–0.3) | 2.61 (3.7–1.7) | Pliocene/Pleistocene |
| G. atra + G. cf. humeralis | 0.31 (1.0–0.0) | 1.02 (1.5–0.6) | Early/Middle Pleistocene |
| Group “poensis” | 1.67 (3.3–0.4) | 3.26 (4.4–2.3) | Pliocene/Pleistocene |
| G. poensis + G. egeria + G. alboguttata | NA | 2.53 (3.4–1.7) | Pliocene/Pleistocene |
| G. argentata + G. egeria | 0.87 (1.9–0.1) | NA | Pleistocene |
| G. poensis + G. egeria1 | NA | 2.21 (3.1–1.4) | Early Pleistocene |
| G. alboguttata + G. egeria2 | NA | 1.49 (2.2–0.9) | Early Pleistocene |
| G. poensis | NA | 0.15 (0.3–0.0) | Middle/Late Pleistocene |
| G. alboguttata | NA | 0.19 (0.3–0.1) | Middle/Late Pleistocene |
| G. argentata | NA | 0.20 (0.3–0.1) | Middle/Late Pleistocene |
| G. egeria | 0.19 (0.6–0.0) | NA | Middle/Late Pleistocene |
- R2 = .025 ± .005 per lineage/Myr; NA, not applicable
Our results suggest that Glauconycteris diverged from Hesperoptenus during the Middle/Late Miocene at around 10 ± 3 Mya, and then it diversified into three main lineages (G. superba, G. variegata, and the clade uniting all other species) at the end of the Miocene at around 6 ± 2 Mya. All other speciation events occurred during the Pliocene and up until the Early Pleistocene. Divergence times at the species level were estimated between the Middle and Late Pleistocene, with the exception of G. beatrix s.s. (Early Pleistocene), and the separation between northern and southern groups of G. variegata (Early Pleistocene).
4 DISCUSSION
4.1 Is Glauconycteris monophyletic?
The monophyly of Glauconycteris was supported in the molecular studies of Hoofer and Van Den Bussche (2003), Roehrs, Lack, and Van Den Bussche (2010), and Roehrs et al. (2011) based on DNA sequences from four species of the genus. More recently, however, Glauconycteris appeared paraphyletic in the molecular tree of Koubínová et al. (2013), the species G. variegata being separated from the clade comprising G. argentata, G. beatrix, G. egeria, and Arielulus cuprosus (a species endemic to Borneo). We suspected that this result was an artifact due to the high percentages of missing data in the supermatrix used in Koubínová et al. (2013): 59% in the DNA sequences of Glauconycteris and 93% in those of A. cuprosus (only a partial Cytb sequence was included in the data set). In agreement with that hypothesis, our phylogenetic analyses showed high support for the monophyly of both genera Glauconycteris and Arielulus (Figures 4 and 5), even when the available GenBank sequences of A. cuprosus were included in the Cytb, 12S, and RAG2 alignments (see results in Fig. S6).
Only four species of Glauconycteris were included in previous molecular studies (Hoofer & Van Den Bussche, 2003; Koubínová et al., 2013; Roehrs et al., 2010, 2011) of the 12 species that are currently recognized in the genus (ACR 2016; Happold & Happold, 2013). In particular, the rarely collected species G. superba has not been sequenced prior to our study. Reeder et al. (2013) based their conclusion to place this species in its own genus Niumbaha solely on morphological characters. Our morphological analysis using both quantitative and qualitative variables (Figure 3) corroborates their finding that G. superba is readily distinguishable from other species of Glauconycteris, mainly based on its larger body size and unique pelage pattern. Our mtDNA and nuDNA trees (Figures 4 and 5) showed that G. superba is highly divergent from two other lineages, that is, G. variegata and a large clade uniting all other species of Glauconycteris. However, our nuclear analyses provided strong support for a sister-group relationship between G. superba and G. variegata (Figure 5), which invalidates the taxonomic conclusion of Reeder et al. (2013). Hence, we recommend that the species G. superba should be retained in the genus Glauconycteris.
4.2 On the origin and diversification of butterfly bats
Based on a mitochondrial alignment covering both 12S and 16S rRNA genes, Hoofer and Van Den Bussche (2003) have concluded that the genus Glauconycteris belongs to the tribe Nycticeiini, which also contains Nycticeius, Lasionycteris, and the clade uniting Eptesicus and Scotomanes. Using both mitochondrial and nuclear genes, Roehrs et al. (2010, 2011) and Koubínová et al. (2013) have suggested that the tribe also includes the three Asian genera Arielulus, Hesperoptenus and Ia. However, none of the previous studies provided strong support for either the monophyly of the tribe Nycticeiini or the position of Glauconycteris.
Due to the lack of nuclear data for Nycticeius, our study is not really appropriate for testing the monophyly of Nycticeiini. However, it should be noted that none of our analyses supported the existence of this tribe. By contrast, all our mtDNA and nuDNA analyses (Figures 4 and 5) showed a strong support for a sister-group relationship between the African genus Glauconycteris and the Asian genus Hesperoptenus. In addition, these two genera share two diagnostic indels (insertion or deletion) in HDAC2 and two others in RIOK3 (Figure 5). Our nuDNA and supermatrix analyses suggested that the Asian genus Arielulus is the sister-group of the clade composed of Glauconycteris and Hesperoptenus (Figures 5 and S4). Since all species of Arielulus and Hesperoptenus are found in Southeast Asia, except A. torquatus, which is endemic to Taiwan (IUCN 2016), we can propose that the Asian ancestor of Glauconycteris originated from this region. Our molecular dating estimates suggested that the ancestor of Glauconycteris diverged from that of Hesperoptenus at around 10/11 Mya and diversified in Africa at around 6 Mya (Table 1). The ancestor of Glauconycteris dispersed from Asia into Africa probably during the Tortonian age of the Late Miocene (11.6–7.2 Mya), when the climate of northeastern Africa and Arabian Peninsula was less arid than today (Kürschner, 1998; Pound et al., 2011).
At the end of the Miocene, around 6 ± 2 Mya, Glauconycteris diversified in Africa into three main groups, which today occupy different habitats: The lineage corresponding to G. variegata (which may also contain G. machadoi) is predominantly associated with savannah, woodland, and bushveld habitats (Monadjem, Taylor, Cotterill, & Schoeman, 2010; Rambaldini, 2010); in the tropical rainforests, G. superba seems to be an open space forager, that is, concentrating its activity above the canopy (Ing et al., 2016), whereas all other species of Glauconycteris, which have a smaller size and a less conspicuous color pattern, are expected to be edge foragers, that is, exploiting the spaces immediately below the canopy, as do the majority of bat species living in Neotropical rainforests (Tiago Marques, Ramos Pereira, & Palmeirim, 2016). The basal diversification of Glauconycteris occurred when a brief event of aridity, between 6.5 and 6 Mya, was followed by more humid conditions (Bonnefille, 2010). It also coincides with the first phase of diversification in the two fruit bat tribes that are endemic to Africa, that is, Epomophorini and Scotonycterini (Hassanin et al., 2015, 2016; Nesi et al., 2013). As in Glauconycteris, fruit bats diversified into taxa characterized by different body sizes and different habitat types, suggesting that climatic changes led to the exploitation of new ecological niches.
Our molecular dating estimates suggest that the three groups of Glauconycteris found in tropical rainforests, that is, “beatrix,” “humeralis,” and “poensis,” appeared during the Early Pliocene (5.3–3.6 Mya). At that time, the warmer and wetter climate led to the expansion of tropical rainforests in Africa (Bonnefille, 2010; Salzmann et al., 2011), which may have offered more favorable conditions for forest-adapted bats. This was followed by the majority of speciation events during the Late Pliocene and Early Pleistocene epochs, suggesting that most of them were driven by allopatric isolation in Pleistocene forest refugia, as it has previously been found in fruit bats of the tribe Scotonycterini (Hassanin et al., 2015). However, Glauconycteris species need to be sampled from additional geographic localities in Cameroon, Gabon, and West Africa, to determine the real influence of forest refugia on their evolution.
4.3 Mito-nuclear discordance for the monophyly of G. egeria
The species G. egeria is poorly represented in museum collections (Happold & Happold, 2013): four from Cameroon, four from Uganda, and five from CAR, including two new specimens collected for our study. Morphologically, it can be recognized on the basis of its pelage, which is dark brown or almost black with conspicuous whitish dorsal flank stripe, and its subquadrangular ears, which are dark brown with a conspicuous pale rim (Happold & Happold, 2013; Figure 6). The eight individuals from southern CAR included in our molecular analyses fall into two divergent and unrelated mitochondrial haplogroups (Figure 4): The first haplogroup is the sister-group of G. poensis; whereas the second haplogroup is more closely related to G. alboguttata. However, the monophyly of G. egeria was supported in all our nuclear analyses (Figure 5), and by an insertion of a nucleotide A in position 559 of the RIOK3 alignment. All these results indicate therefore that the signal provided by the mitochondrial genome is misleading.
Three non-exclusive hypotheses can be advanced to explain such a mito-nuclear discordance: different sexual dispersal behaviors, incomplete lineage sorting of ancestral mitochondrial haplotypes, and mitochondrial introgression (e.g., Hassanin et al., 2013, 2015; Nesi et al., 2011; Ropiquet & Hassanin, 2006). Our analyses suggest that the southern CAR region is a key geographic area for bat species endemic to African rainforests. Indeed, divergent mitochondrial haplogroups were also detected in this region for two other forest species: G. beatrix (Figure 4) and the fruit bat species Casinycteris argynnis (Hassanin et al., 2015). In the two latter species, the two haplogroups represent two geographic regions: western Equatorial Africa (Gabon, Republic of the Congo, and/or southeastern Cameroon) and eastern Equatorial Africa (eastern DRC). A similar geographic division was also detected for Scotonycteris bergmansi, another fruit bat species endemic to the forests of Equatorial Africa (Hassanin et al., 2015). All these data suggest that southern CAR may therefore be a potential hybrid zone between bat populations isolated into distant forest refugia during glacial periods of the Pleistocene epoch, located, respectively, in western and eastern Equatorial Africa. In this context, female philopatry may explain the divergence of two mitochondrial haplogroups in distant western and eastern Pleistocene forest refugia, whereas male-biased dispersal may have maintained nuclear gene flow during interglacial periods. In G. egeria, however, it is impossible to know if the two mitochondrial haplogroups have any geographical significance, because all of our specimens were collected in CAR. Another issue that remains to be resolved is that the two mitochondrial haplogroups identified in G. egeria are related to two different species: the haplogroup 1 diverged from G. poensis at 2.2 ± 0.8 Mya, whereas the haplogroup 2 diverged from G. alboguttata at 1.5 ± 0.6 Mya (Table 1). In the absence of nuclear sequences for G. poensis, it is difficult to provide a reliable interpretation. However, our nuclear analyses showed that G. egeria is more closely related to G. argentata than to G. alboguttata (Figure 5). This phylogenetic result suggests that the mitochondrial genome of G. alboguttata was transferred into one population of G. egeria at around 1.5 Mya. To provide further support for this scenario, and to better understand the potential role of female philopatry and Pleistocene forest refugia, it will be necessary to sequence mitochondrial and nuclear markers from several other specimens of G. egeria and G. poensis collected in both western and eastern Equatorial Africa.
4.4 How many species exist within the “beatrix” and “humeralis” groups?
Although our molecular analyses showed that specimens identified as G. beatrix and G. humeralis belong to two distinct groups, the morphological identification of individuals to either G. beatrix or G. humeralis remains problematic (Figure 3).
The species G. beatrix was described by Thomas (1901) based on a single specimen (BMNH 1898.5.4.19) collected at the Benito River in Equatorial Guinea. As pointed out by Rosevear (1965), the description of Thomas (1901) “was misleading as to skull length, pattern, and possibly color.” Thomas (1901) described G. beatrix as “General color above and below uniform blackish brown without lighter markings,” but Rosevear (1965) noted that the color of the type specimen was “pale red brown… with white tufts on the shoulders,” as described by Sanborn (1953) in several specimens from Gabon. All 16 specimens of G. beatrix we collected in DRC also have white shoulder spots, and the three specimens from CAR share the same pattern. By contrast, the adult female from Cameroon (HNHM 23262) does not have white shoulder spots. The holotype of G. beatrix is a female characterized by the following measurements (data from Rosevear, 1965): FA = 39, HB = 45, TIB = 19.5, 3DM = 38, GLS = 11.6, ZW = 8.5, BCW = 7.1, and MB = 7.5. Our specimens identified as G. beatrix from Cameroon, CAR and DRC fit all these characteristics (mean values for ♀ [n = 3], FA = 38.5, TIB = 19.7, 3DM = 39.0; ♂ [n = 7]: FA = 36.0, TIB = 18.5, 3DM = 36.6; see details in Table S1). By contrast, the three specimens from RCI formerly identified as G. beatrix in the ROM collection have a smaller body size (♀ [n = 1], FA = 35, TIB = 17.5, 3DM = 34.1; ♂ [n = 2]: FA = 34.5, TIB = 16.5, 3DM = 34.6). In the mitochondrial tree (Figure 4), the species G. beatrix was found to be polyphyletic: the individuals from Central Africa constitute the sister-group of G. curryae, whereas the five G. beatrix specimens from RCI are closely related to the “humeralis” group. As a consequence, both morphological and molecular results suggest that the five specimens housed at the ROM (under N° 100527, 100528, 100531, 100534, and 100578) belong to a species different from G. beatrix, here referred to as G. cf. beatrix.
Various authors have expressed doubts about the specific status of G. humeralis. Koopman (1971, 1994), Peterson and Smith (1973), Eger and Schlitter (2001) and Monadjem et al. (2010) all considered it as a subspecies of G. beatrix. Despite their apparent morphological similarities, our molecular analyses indicate that G. beatrix and G. humeralis are not closely related. The species G. humeralis was described by Allen (1917) on the basis of five specimens collected in DRC: a holotype and three topotypes from Medje, and another individual from Avakubi. The measurements of the holotype (AMNH 49013, female) published by Allen (1917) are: FA = 36.8, HB = 42, T = 40, TIB = 16.8,1 3DM = 35.8, GLS = 11.3, ZW = 8.2, MB = 7.3, C-M3 = 3.6, ML = 7.9, and C-M3 = 3.9. These measurements fit with those taken from a female, MNHN 2016-2802, that was also used in our molecular study, which was collected <250 km from the type locality at Medje (Table S1). In addition, the forearm lengths of the two female topotypes are also similar to that of MNHN 2016-2802 (35.8 and 35.3 versus 35.6 mm). By contrast, the FA is significantly larger in a female, AMNH 49014, collected at Avakubi (38.8 mm). As pointed out by Allen (1917) “The pure white shoulder tuft is a conspicuous feature in the type and topotypes; it is present in the Avakubi specimen, but only the tips of the hairs are white (yellowish white instead of pure white).” We therefore suggest that the Avakubi specimen (AMNH 49014) belongs instead to G. beatrix. This point may explain previous taxonomic confusions between G. beatrix and G. humeralis. In the DRC, we collected two species of the “beatrix” group, that is, G. beatrix and G. curryae, and two species of the “humeralis” group, G. humeralis and G. atra sp. nov.. Whereas G. curryae and G. atra sp. nov. can be easily identified by phenotypic characteristics, such as the fur color, G. beatrix and G. humeralis have very similar external appearances. In light of our study, we consider, however, that three characters can be used to distinguish the two species: The shoulder spots are more conspicuous in G. humeralis; and FA and TIB lengths are significantly smaller in G. humeralis than in G. beatrix (Table S1).
Within the “humeralis” group, our analyses suggest two potentially new species. According to our mitochondrial and nuclear analyses, the taxon identified as G. cf. humeralis may belong to a new species. Unfortunately, the individuals of G. cf. humeralis, R13-46 and R13-98, that were captured in southern CAR were released after being examined and punched for DNA sequencing. Without type specimen(s) deposited in museum collection, we refrain to describe the new species herein. According to our morphological and mitochondrial analyses, the taxon here referred to as G. cf. beatrix may also be recognized as a putative new species. However, nuclear markers need to be sequenced on the five specimens housed at the ROM in order to test possible gene flow with G. atra sp. nov. and G. cf. humeralis.
4.5 Evolution of color pattern within the genus Glauconycteris
The species of butterfly bats show a wide a variety of colors (piebald, blackish brown, dark brown, sepia brown, reddish-brown, rusty brown with yellowish tints, grayish brown, golden-fawn, creamy-fawn, and pale-brown; Figure 6). In addition, most species exhibit specific body patterns. Three species have reticulated wings (dark lines on pale background): G. gleni, G. machadoi, and G. variegata. The species G. kenyacola is characterized by whitish facial markings on nose and at base of ears (Happold & Happold, 2013). All species of the “poensis” group have white or whitish stripes along the flank above the wing (G. poensis, G. alboguttata, G. argentata, and G. egeria,), suggesting that this character probably constitutes a good synapomorphy. In G. superba, there are two pairs of white stripes that are located more dorsally on the back. Five species have generally one spot on each shoulder: G. alboguttata, G. beatrix, G. cf. beatrix, G. cf. humeralis, and G. humeralis. In G. egeria, we observed that the shoulder spot and dorsal flank stripe are confluent. In G. superba, there are two or rarely three white spots on each shoulder (Ing et al., 2016). In addition, two species are without body patterns or reticulated wings: G. atra sp. nov. and G. curryae.
Hayman and Jones (1950) described a remarkably wide variation in the pattern of white shoulder spots and flank stripes for several individuals of G. poensis collected in Sierra Leone: “Of the 40 specimens… 20 have the shoulder spots and flank stripes distinct on both sides, while six have no markings of any kind. The remaining 14 show every type of variation. Several are asymmetrical, having shoulder spots on one side only, with or without a flank stripe.” As a consequence, Hayman and Jones (1950) raised the question whether these white markings are in themselves sufficiently constant for use in specific diagnoses. In all species examined in our study, however, we found no variation in body pattern, that is, spots and/or flank stripes are consistently present or absent in all individuals of each species. The sole exception is G. beatrix, in which individuals from CAR and DRC have faint white shoulder spots, whereas the individual from Cameroon does not exhibit any white markings. In addition, we never observed individuals with asymmetrical markings, that is, with a spot on one shoulder only. More importantly, all species of the “poensis” group share the presence of dorsal flank stripes. In this context, the observations published by Hayman and Jones (1950) for G. poensis should be regarded with caution, and all of their specimens from Sierra Leone need to be re-examined and sequenced to provide definitive conclusions on their taxonomic status and the levels of intraspecific variation in G. poensis.
ACKNOWLEDGEMENTS
We thank Aimée Francis Bingo, Laurent Daudet, Jean-François Julien, Emmanuel Nakouné, Nicolas Nesi, and Carine Ngoagouni for assistance with field studies. In DRC, we are very grateful to Faustin Toengaho Lokundo, the Rector of the University of Kisangani, for his invitation, and Benjamin Dudu, Hilde Keunen, Erik Verheyen, and all other members of the Centre de Surveillance de la Biodiversité, who provided administrative and logistic support. In CAR, we would like to acknowledge Pr. Alain Le Faou, the director of the Institut Pasteur de Bangui, for his invitation in 2008, and Philippe Annoyer, who organized the expedition “Sangha 2012, Biodiversité en terre pygmée,” the Ministries of Environment and Forest, Pr. Georgette Florence Koyt Deballé, the Rector of the University of Bangui, Dr. Bolevane Ouantinam Serge Florent and Dr. Yongo Olga. We are very grateful to collection managers and curators who loaned museum specimens or provided measurements, pictures, or samples for DNA analyses: Leigh Richards from the DNSM; Steve Goodman and Bruce Patterson from the FMNH; Burton Lim and Jacqueline Miller from the ROM; and Nguyen Truong Son from the IEBR (Hanoi, Vietnam). We also acknowledge Alexandre Hanquet and Didier Geffard-Kuriyama (UMS 2700 MNHN) for skull images, Ernest Seamark (AfricanBats) for the image of G. variegata, and Wilderness Safaris for supporting his fieldwork in Botswana and Zambia. AH would like to thank Meredith Happold for measurements on Glauconycteris species, Darina Koubínová for the Nexus file, Dos Santos Martins Carlos, Kevin Racine, and the World Bat Library (Geneva, Switzerland) for bibliography, and Céline Bonillo and Stéphanie Varizat, who kindly helped in the laboratory work. We also acknowledge the reviewers for their helpful comments on the first version of the manuscript. This study was supported by the MNHN, CNRS, LabEx BCDiv 2012-2013, the Institut Langevin, CNRS, ESPCI, Labex WIFI 2012-2013, and the “PPF Biodiversité actuelle et fossile.”
Note
APPENDIX 1: SPECIMENS ANALYZED IN THIS STUDY
| Species | Sample code | Sex | Locality | CO1 | Cytb | 12S rRNA | HDAC2 | RIOK3 | ZFYVE | RAG2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Glauconycteris alboguttata | MNHN 2016-2774 (K13-102) | F | Melume, DRC | MF038612 | MF038511 | MF038712 | MF038437 | MF038299 | MF038231 | MF038368 |
| Glauconycteris alboguttata | MNHN 2016-2772 (K13-98) | M | Melume, DRC | MF038614 | MF038513 | MF038714 | MF038439 | MF038301 | MF038233 | MF038370 |
| Glauconycteris alboguttata | MNHN 2016-2773 (K13-101) | F | Melume, DRC | MF038611 | MF038510 | MF038711 | NA | NA | NA | NA |
| Glauconycteris alboguttata | MNHN 2016-2775 (K13-118) | M | Melume, DRC | MF038613 | MF038512 | MF038713 | MF038438 | MF038300 | MF038232 | MF038369 |
| Glauconycteris alboguttata | R13-107 | F | Dzanga-Sangha, CAR | MF038615 | MF038514 | MF038715 | MF038440 | MF038302 | MF038234 | MF038371 |
| Glauconycteris alboguttata | R13-20 | F | Dzanga-Sangha, CAR | MF038616 | MF038515 | MF038716 | NA | NA | NA | NA |
| Glauconycteris alboguttata | R13-31 | M | Dzanga-Sangha, CAR | MF038617 | MF038516 | MF038717 | MF038441 | MF038303 | MF038235 | MF038372 |
| Glauconycteris alboguttata | R13-40 | F | Dzanga-Sangha, CAR | MF038618 | MF038517 | MF038718 | NA | NA | NA | NA |
| Glauconycteris alboguttata | R13-45 | F | Dzanga-Sangha, CAR | MF038619 | MF038518 | MF038719 | NA | NA | NA | NA |
| Glauconycteris alboguttata | R13-47 | F | Dzanga-Sangha, CAR | MF038620 | MF038519 | MF038720 | MF038442 | MF038304 | MF038236 | MF038373 |
| Glauconycteris alboguttata | R13-55 | M | Dzanga-Sangha, CAR | MF038621 | MF038520 | MF038721 | MF038443 | MF038305 | MF038237 | MF038374 |
| Glauconycteris alboguttata | R13-63 | F | Dzanga-Sangha, CAR | MF038622 | MF038521 | MF038722 | NA | NA | NA | NA |
| Glauconycteris argentata | FMNH 151197 | NA | Gonja FR, Tanzania | NA | NA | AY495468 | NA | NA | NA | NA |
| Glauconycteris argentata | FMNH 151198 | M | Gonja FR, Tanzania | NA | NA | NA | NA | NA | NA | NA |
| Glauconycteris argentata | K13-288 | F | Mbiye, DRC | MF038588 | MF038487 | MF038688 | MF038417 | MF038279 | MF038211 | MF038348 |
| Glauconycteris argentata | K13-381 | F | Yoko, DRC | MF038589 | MF038488 | MF038689 | MF038418 | MF038280 | MF038212 | MF038349 |
| Glauconycteris argentata | K13-383 | F | Yoko, DRC | MF038590 | MF038489 | MF038690 | MF038419 | MF038281 | MF038213 | MF038350 |
| Glauconycteris argentata | K13-50 | F | Sukisa, DRC | MF038593 | MF038492 | MF038693 | NA | NA | NA | NA |
| Glauconycteris argentata | K13-73 | F | Sukisa, DRC | MF038595 | MF038494 | MF038695 | NA | NA | NA | NA |
| Glauconycteris argentata | K13-75 | F | Sukisa, DRC | MF038596 | MF038495 | MF038696 | MF038422 | MF038284 | MF038216 | MF038353 |
| Glauconycteris argentata | MNHN 2016-2776 (K13-45) | M | Sukisa, DRC | MF038591 | MF038490 | MF038691 | MF038420 | MF038282 | MF038214 | MF038351 |
| Glauconycteris argentata | MNHN 2016-2777 (K13-46) | M | Sukisa, DRC | MF038592 | MF038491 | MF038692 | MF038421 | MF038283 | MF038215 | MF038352 |
| Glauconycteris argentata | MNHN 2016-2778 (K13-59) | M | Sukisa, DRC | MF038594 | MF038493 | MF038694 | NA | NA | NA | NA |
| Glauconycteris argentata | MNHN 2016-2779 (K13-103) | F | Melume, DRC | MF038586 | MF038485 | MF038686 | MF038415 | MF038277 | MF038209 | MF038346 |
| Glauconycteris argentata | MNHN 2016-2780 (K13-272) | F | Mbiye, DRC | MF038587 | MF038486 | MF038687 | MF038416 | MF038278 | MF038210 | MF038347 |
| Glauconycteris atra sp. nov. | MNHN 2016-2790 (K13-206) | M | Yatolema, DRC | MF038600 | MF038499 | MF038700 | MF038426 | MF038288 | MF038220 | MF038357 |
| Glauconycteris atra sp. nov. | MNHN 2016-2791 (K13-210) | M | Yaengo, DRC | MF038601 | MF038500 | MF038701 | MF038427 | MF038289 | MF038221 | MF038358 |
| Glauconycteris atra sp. nov. | MNHN 2016-2792 (K13-215) | F | Yaengo, DRC | MF038602 | MF038501 | MF038702 | MF038428 | MF038290 | MF038222 | MF038359 |
| Glauconycteris atra sp. nov. | MNHN 2016-2793 (K13-224) | F | Yatolema, DRC | MF038603 | MF038502 | MF038703 | MF038429 | MF038291 | MF038223 | MF038360 |
| Glauconycteris atra sp. nov. | MNHN 2016-2794 (K13-380) | F | Yoko, DRC | MF038604 | MF038503 | MF038704 | MF038430 | MF038292 | MF038224 | MF038361 |
| Glauconycteris beatrix | HNHM 23262 | F | Nki NP, Cameroon | MF038641 | MF038540 | MF038741 | MF038452 | MF038314 | MF038246 | MF038383 |
| Glauconycteris beatrix | K13-226 | F | Yatolema, DRC | MF038626 | MF038525 | MF038726 | NA | NA | NA | NA |
| Glauconycteris beatrix | K13-299 | F | Yoko, DRC | MF038628 | MF038527 | MF038728 | NA | NA | NA | NA |
| Glauconycteris beatrix | K13-320 | M | Yoko, DRC | MF038629 | MF038528 | MF038729 | NA | NA | NA | NA |
| Glauconycteris beatrix | K13-340 | F | Yoko, DRC | MF038631 | MF038530 | MF038731 | NA | NA | NA | NA |
| Glauconycteris beatrix | K13-341 | F | Yoko, DRC | MF038632 | MF038531 | MF038732 | NA | NA | NA | NA |
| Glauconycteris beatrix | K13-370 | F | Yoko, DRC | MF038634 | MF038533 | MF038734 | NA | NA | NA | NA |
| Glauconycteris beatrix | K13-386 | M | Yoko, DRC | MF038636 | MF038535 | MF038736 | NA | NA | NA | NA |
| Glauconycteris beatrix | MNHN 2016-2781 (K13-19) | F | Bongandjola, DRC | MF038624 | MF038523 | MF038724 | MF038445 | MF038307 | MF038239 | MF038376 |
| Glauconycteris beatrix | MNHN 2016-2782 (K13-99) | M | Melume, DRC | MF038639 | MF038538 | MF038739 | MF038450 | MF038312 | MF038244 | MF038381 |
| Glauconycteris beatrix | MNHN 2016-2783 (K13-100) | M | Melume, DRC | MF038638 | MF038537 | MF038738 | MF038449 | MF038311 | MF038243 | MF038380 |
| Glauconycteris beatrix | MNHN 2016-2784 (K13-156) | M | Yatolema, DRC | MF038623 | MF038522 | MF038723 | MF038444 | MF038306 | MF038238 | MF038375 |
| Glauconycteris beatrix | MNHN 2016-2785 (K13-225) | M | Yatolema, DRC | MF038625 | MF038524 | MF038725 | NA | NA | NA | NA |
| Glauconycteris beatrix | MNHN 2016-2786 (K13-256) | F | Mbiye, DRC | MF038627 | MF038526 | MF038727 | MF038446 | MF038308 | MF038240 | MF038377 |
| Glauconycteris beatrix | MNHN 2016-2787 (K13-339) | M | Yoko, DRC | MF038630 | MF038529 | MF038730 | MF038447 | MF038309 | MF038241 | MF038378 |
| Glauconycteris beatrix | MNHN 2016-2788 (K13-347) | M | Yoko, DRC | MF038633 | MF038532 | MF038733 | MF038448 | MF038310 | MF038242 | MF038379 |
| Glauconycteris beatrix | MNHN 2016-2789 (K13-385) | M | Yoko, DRC | MF038635 | MF038534 | MF038735 | NA | NA | NA | NA |
| Glauconycteris beatrix | R13-109 | F | Dzanga-Sangha, CAR | MF038637 | MF038536 | MF038737 | NA | NA | NA | NA |
| Glauconycteris beatrix | R13-24 | F | Dzanga-Sangha, CAR | MF038642 | MF038541 | MF038742 | MF038453 | MF038315 | MF038247 | MF038384 |
| Glauconycteris beatrix | R13-5 | M | Dzanga-Sangha, CAR | MF038640 | MF038539 | MF038740 | MF038451 | MF038313 | MF038245 | MF038382 |
| Glauconycteris cf. beatrix | ROM 100527 | NA | Mont Peko NP, RCI | JF444140 | NA | NA | NA | NA | NA | NA |
| Glauconycteris cf. beatrix | ROM 100528 | NA | Mont Peko NP, RCI | JF444141 | NA | NA | NA | NA | NA | NA |
| Glauconycteris cf. beatrix | ROM 100531 | M | Mont Peko NP, RCI | JF444142 | NA | NA | NA | NA | NA | NA |
| Glauconycteris cf. beatrix | ROM 100534 | F | Mont Peko NP, RCI | JF444143 | NA | NA | NA | NA | NA | NA |
| Glauconycteris cf. beatrix | ROM 100578 | M | Mont Peko NP, RCI | JF444144 | NA | NA | NA | NA | NA | NA |
| Glauconycteris cf. humeralis | R13-46 | F | Dzanga-Sangha, CAR | MF038598 | MF038497 | MF038698 | MF038424 | MF038286 | MF038218 | MF038355 |
| Glauconycteris cf. humeralis | R13-98 | F | Dzanga-Sangha, CAR | MF038599 | MF038498 | MF038699 | MF038425 | MF038287 | MF038219 | MF038356 |
| Glauconycteris curryae | MNHN 2016-2795 (K13-15) | F | Bongandjola, DRC | MF038605 | MF038504 | MF038705 | MF038431 | MF038293 | MF038225 | MF038362 |
| Glauconycteris curryae | MNHN 2016-2796 (K13-213) | F | Yaengo, DRC | MF038606 | MF038505 | MF038706 | MF038432 | MF038294 | MF038226 | MF038363 |
| Glauconycteris curryae | MNHN 2016-2797 (K13-241) | F | Yatolema, DRC | MF038607 | MF038506 | MF038707 | MF038433 | MF038295 | MF038227 | MF038364 |
| Glauconycteris curryae | MNHN 2016-2798 (K13-284) | F | Mbiye, DRC | MF038608 | MF038507 | MF038708 | MF038434 | MF038296 | MF038228 | MF038365 |
| Glauconycteris curryae | MNHN 2016-2799 (K13-310) | M | Yoko, DRC | MF038609 | MF038508 | MF038709 | MF038435 | MF038297 | MF038229 | MF038366 |
| Glauconycteris curryae | R13-115 | NA | Dzanga-Sangha, CAR | MF038610 | MF038509 | MF038710 | MF038436 | MF038298 | MF038230 | MF038367 |
| Glauconycteris egeria | AMNH 268381 | NA | Dzanga-Sangha, CAR | NA | NA | AY495470 | NA | NA | NA | NA |
| Glauconycteris egeria | MNHN 2016-2800 (R08-61) | M | Mbaéré-Bodingué, CAR | MF038649 | MF038548 | MF038749 | MF038458 | MF038320 | MF038252 | MF038389 |
| Glauconycteris egeria | MNHN 2016-2801 (R13-02) | F | Dzanga-Sangha, CAR | NA | NA | NA | NA | NA | NA | NA |
| Glauconycteris egeria | R13-10 | NA | Dzanga-Sangha, CAR | MF038643 | MF038542 | MF038743 | MF038454 | MF038316 | MF038248 | MF038385 |
| Glauconycteris egeria | R13-108 | M | Dzanga-Sangha, CAR | MF038644 | MF038543 | MF038744 | NA | NA | NA | NA |
| Glauconycteris egeria | R13-11 | NA | Dzanga-Sangha, CAR | MF038645 | MF038544 | MF038745 | NA | NA | NA | NA |
| Glauconycteris egeria | R13-21 | M | Dzanga-Sangha, CAR | MF038646 | MF038545 | MF038746 | MF038455 | MF038317 | MF038249 | MF038386 |
| Glauconycteris egeria | R13-44 | F | Dzanga-Sangha, CAR | MF038647 | MF038546 | MF038747 | MF038456 | MF038318 | MF038250 | MF038387 |
| Glauconycteris egeria | R13-48 | F | Dzanga-Sangha, CAR | MF038648 | MF038547 | MF038748 | MF038457 | MF038319 | MF038251 | MF038388 |
| Glauconycteris humeralis | FMNH 149417 | M | Epulu, D.R. Congo | NA | NA | AY495469 | NA | NA | NA | NA |
| Glauconycteris humeralis | MNHN 2016-2802 (K13-47) | F | Sukisa, DRC | MF038597 | MF038496 | MF038697 | MF038423 | MF038285 | MF038217 | MF038354 |
| Glauconycteris poensis | DM 14186 | NA | Mt Nimba, Liberia | MF038650 | MF038549 | NA | NA | NA | NA | NA |
| Glauconycteris poensis | DM 14187 | NA | Mt Nimba, Liberia | MF038651 | MF038550 | NA | NA | NA | NA | NA |
| Glauconycteris poensis | DM 14188 | NA | Mt Nimba, Liberia | MF038652 | MF038551 | NA | NA | NA | NA | NA |
| Glauconycteris poensis | FMNH 42613 | M | Ifon, Nigeria | NA | NA | NA | NA | NA | NA | NA |
| Glauconycteris poensis | ROM 100515 | NA | Mont Peko NP, RCI | JF444145 | NA | NA | NA | NA | NA | NA |
| Glauconycteris poensis | ROM 100544 | NA | Mont Peko NP, RCI | JF444146 | NA | NA | NA | NA | NA | NA |
| Glauconycteris poensis | ROM 100545 | NA | Mont Peko NP, RCI | JF444147 | NA | NA | NA | NA | NA | NA |
| Glauconycteris superba | K13-264 | M | Mbiye, DRC | MF038655 | MF038554 | MF038752 | NA | NA | NA | NA |
| Glauconycteris superba | K13-265 | M | Mbiye, DRC | MF038656 | MF038555 | MF038753 | NA | NA | NA | NA |
| Glauconycteris superba | K13-286 | M | Mbiye, DRC | MF038658 | MF038557 | MF038755 | NA | NA | NA | NA |
| Glauconycteris superba | K13-287 | F | Mbiye, DRC | MF038659 | MF038558 | MF038756 | NA | NA | NA | NA |
| Glauconycteris superba | K13-322 | F | Yoko, DRC | MF038661 | MF038560 | MF038758 | MF038463 | MF038325 | MF038257 | MF038394 |
| Glauconycteris superba | MNHN 2016-2803 (K13-262) | M | Mbiye, DRC | MF038653 | MF038552 | MF038750 | MF038459 | MF038321 | MF038253 | MF038390 |
| Glauconycteris superba | MNHN 2016-2804 (K13-263) | M | Mbiye, DRC | MF038654 | MF038553 | MF038751 | MF038460 | MF038322 | MF038254 | MF038391 |
| Glauconycteris superba | MNHN 2016-2805 (K13-266) | F | Mbiye, DRC | MF038657 | MF038556 | MF038754 | MF038461 | MF038323 | MF038255 | MF038392 |
| Glauconycteris superba | MNHN 2016-2806 (K13-289) | F | Mbiye, DRC | MF038660 | MF038559 | MF038757 | MF038462 | MF038324 | MF038256 | MF038393 |
| Glauconycteris superba | MNHN 2016-2807 (K13-351) | M | Yoko, DRC | MF038662 | MF038561 | MF038759 | MF038464 | MF038326 | MF038258 | MF038395 |
| Glauconycteris superba | RMCA a1.097-M-3153 | M | Mbiye, DRC | MF038663 | MF038562 | NA | NA | NA | NA | NA |
| Glauconycteris variegata | CM 97983 | NA | Western province, Kenya | NA | NA | AY495471 | NA | NA | NA | NA |
| Glauconycteris variegata | ECJS-43/2009 | F | Chitabe, Botswana | MF038664 | MF038563 | MF038760 | MF038465 | MF038327 | MF038259 | MF038396 |
| Glauconycteris variegata | ECJS-92/2010 | F | Kafue NP, Zambia | MF038665 | MF038564 | MF038761 | MF038466 | MF038328 | MF038260 | MF038397 |
| Glauconycteris variegata | IVBS 1767 | NA | Niokolo-Koba NP, Senegal | NA | JX276108 | JX276363 | NA | NA | NA | NA |
| Glauconycteris variegata | TM 48494 | M | Kruger NP, South Africa | MF038666 | MF038565 | MF038762 | MF038467 | MF038329 | MF038261 | MF038398 |
| Glauconycteris variegata | TM 48495 | NA | Kruger NP, South Africa | MF038667 | MF038566 | MF038763 | NA | NA | NA | NA |
| Glauconycteris variegata | TM 48496 | M | Kruger NP, South Africa | MF038668 | MF038567 | MF038764 | MF038468 | MF038330 | MF038262 | MF038399 |
| Outgroup | ||||||||||
| Arielulus circumdatus | BNB 055 | M | MF038573 | MF038474 | MF038674 | MF038405 | MF038267 | MF038199 | MF038336 | |
| Arielulus circumdatus | BNB 097 | F | MF038574 | MF038475 | MF038675 | NA | NA | NA | NA | |
| Arielulus sp. | GS 16483 | M | MF038575 | MF038476 | NA | MF038406 | MF038268 | MF038200 | MF038337 | |
| Arielulus torquatus | GS 20811 | F | MF038576 | MF038477 | MF038676 | NA | NA | NA | NA | |
| Arielulus torquatus | GS 20813 | M | MF038577 | MF038478 | MF038677 | MF038407 | MF038269 | MF038201 | MF038338 | |
| Barbastella barbastellus | GT-853 | F | MF038569 | MF038470 | MF038670 | MF038401 | MF038264 | MF038195 | MF038332 | |
| Chalinolobus neocaledonicus | 2000-273 | NA | MF038571 | MF038472 | MF038672 | MF038403 | NA | MF038197 | MF038334 | |
| Eptesicus fuscus | FMNH 214994 | NA | MF038578 | MF038479 | MF038678 | MF038408 | MF038270 | MF038202 | MF038339 | |
| Eptesicus guadeloupensis | 2000-194 | NA | MF038579 | MF038480 | MF038679 | MF038409 | MF038271 | MF038203 | MF038340 | |
| Eptesicus hottentotus | SAM 41418 | NA | MF038580 | AJ841963 | MF038680 | MF038410 | MF038272 | MF038204 | MF038341 | |
| Eptesicus serotinus | 124203 | M | MF038581 | MF038481 | MF038681 | NA | NA | NA | NA | |
| Hesperoptenus blandfordi | CPV10-40 | M | MF038582 | MF038482 | MF038682 | MF038411 | MF038273 | MF038205 | MF038342 | |
| Hesperoptenus tickelli | CPV10-21 | M | MF038583 | MF038483 | MF038683 | MF038412 | MF038274 | MF038206 | MF038343 | |
| Ia io | VN11-0688 | M | MF038584 | JX465365 | MF038684 | MF038413 | MF038275 | MF038207 | MF038344 | |
| Laephotis botswanae | ECJS-11/2009 | NA | MF038572 | MF038473 | MF038673 | MF038404 | MF038266 | MF038198 | MF038335 | |
| Lasionycteris noctivagans | NA | NA | GU722942 | KC747682 | AF326095 | NA | NA | NA | NA | |
| Mimetillus moloneyi | HNHM 23243 | F | MF038570 | MF038471 | MF038671 | MF038402 | MF038265 | MF038196 | MF038333 | |
| Myotis bocagii | C07-188 | M | MF038568 | MF038469 | MF038669 | MF038400 | MF038263 | MF038194 | MF038331 | |
| Nycticeius humeralis | NA | NA | GU723163 | KC747697 | AF326102 | NA | NA | NA | NA | |
| Scotomanes ornatus | VN11-0238 | M | MF038585 | MF038484 | MF038685 | MF038414 | MF038276 | MF038208 | MF038345 | |
- NA, NOT APPLICABLE.




