Taxonomic composition and ploidy level among European water frogs (Anura: Ranidae: Pelophylax) in eastern Hungary
Abstract
The western Palaearctic water frogs in the genus Pelophylax comprise several distinct species and three hybridogenetic hybrid forms. In this study, we focus on the Pelophylax esculentus complex, which consists of two sexual species, Pelophylax ridibundus and Pelophylax lessonae, and their hybridogenetic hybrid, Pelophylax esculentus. Specifically, we investigated taxonomic composition and ploidy level of water frogs sampled in three different types of wetland habitats in the Hortobágy National Park (HNP), eastern Hungary. Using variation in serum albumin intron 1 (SAI-1) and 15 microsatellite loci, we detected the presence of all members of the P. esculentus complex in the studied localities. In one locality, all three taxa occurred syntopically, while in others water frog populations consisted of P. ridibundus and P. esculentus exclusively. The genomic composition of the 63 examined hybrid specimens analysed with microsatellites showed the occurrence of diploid genotypes only. We used a population genetic approach (allelic richness, gene diversity, multilocus genotypes and multilocus disequilibrium) to infer the breeding system of water frogs at HNP. Our data indicate that at least in two populations, hybrids form gametes with clonally transmitted P. ridibundus genome and produce a new hybrid generation by mating with P. lessonae.
Introduction
Western Palaearctic water frogs of the genus Pelophylax include fourteen distinct species and three hybrid forms reproducing by hybridogenesis (Graf and Polls-Pelaz 1989; Günther 1990; Plötner 2005; Lymberakis et al. 2007; Plötner et al. 2012). This special mode of reproduction resembles parthenogenesis in many aspects. While parthenogenetic females clonally produce diploid eggs, which give rise to a new generation of daughters, hybridogenetic individuals (hybrids) transmit clonally only one chromosome set; the other set of chromosomes is eliminated during gametogenesis and is restored in each generation by mating. Sexual and hybridogenetic taxa of water frogs form three different complexes in Europe. In central Europe, the Pelophylax esculentus complex is formed by two sexual species, the marsh frog, Pelophylax ridibundus (Pallas, 1771; genotype RR) and the pool frog, Pelophylax lessonae (Camerano, 1882; genotype LL). The interspecific mating of the two species produces the hybridogenetic edible frog, Pelophylax esculentus (Linnaeus, 1758; genotype LR). The mode of hybridogenesis in P. esculentus includes clonal transfer of either the genome of P. ridibundus (R genome) or the genome of P. lessonae (L genome), rarely simultaneous transfer of both genomes to gametes (Uzzell and Berger 1975; Berger et al. 1978; Polls-Pelaz 1994). Most P. esculentus in central Europe eliminate the L genome during gametogenesis, clonally transmit the R genome and backcross with syntopically living P. lessonae (LE system). Hybrid forms that eliminate the R genome form lessonae gametes and backcross with P. ridibundus (RE system) are less common and have been found only in eastern Germany, Poland and eastern Ukraine (Uzzell and Berger 1975; Berger 1977; Uzzell et al. 1980; Borkin et al. 2004; Biriuk et al. 2016). Besides hybrid forms living and mating with one or the other parental species, there are P. esculentus populations that are reproductively independent of the sexual species. Such all-hybrid populations (EE system) are found mostly in north-western, less in central and eastern Europe (Günther 1973; Ebendal 1979; Berger 1988; Fog 1994; Hoffmann et al. 2015). Persistence of all-hybrid populations is achieved by mating of individuals, which produce different types of gametes (e.g. Christiansen et al. 2005; Christiansen and Reyer 2009). In addition to diploid hybrids, these populations typically comprise a fraction of triploid individuals possessing either two L and one R genome (LLR) or two R and one L genome (LRR). Finally, syntopic occurrence of both parental species and hybrids (LER population) is recorded from central and eastern Europe (Günther et al. 1991; Gubányi 1992; Kotlík and Šůlová 1994; Mikulíček et al. 2015). However, these populations are rare and were not studied in details. In Slovak LER populations, hybrids form R gametes (as in the LE system) and P. ridibundus individuals likely do not contribute to perpetuation of the hybridogenetic lineages (Mikulíček et al. 2015).
Due to the high rate of hybridization and overlapping morphological characters, there is no easy way to distinguish between hybrids of different ploidy levels and parental species. Various methods give reasonable results including erythrocyte size measurements (Günther 1977; Polls-Pelaz and Graf 1988; Ogielska et al. 2001; Mezhzherin et al. 2010) combined with morphometry (Hotz and Uzzell 1982; Gubányi and Korsós 1992; Günther and Plötner 1994; Tognarelli et al. 2014), electrophoresis of allozymes (Plötner and Klinkhardt 1992; Hotz et al. 2001; Mikulíček and Kotlík 2001; Mezhzherin et al. 2010) or albumin (Lőw et al. 1989), DNA flow cytometry (Vinogradov et al. 1990; Borkin et al. 2004; Arioli et al. 2010; Mikulíček et al. 2015) and microsatellites (Christiansen 2005; Arioli 2007; Christiansen and Reyer 2009; Pruvost et al. 2013).
Our understanding of the taxonomic and genotypic composition in Hungarian water frog populations is limited: few studies have been published and many of them are not based on genetic data. Several publications from the early and mid-20th century did not distinguish between P. lessonae and P. esculentus individuals (Bolkay 1909; Fejérváry 1921; Fejérváry-Lángh 1943; Dely 1953). In Hungary, studies based on both morphology and genetic data include those conducted in western (Gubányi 1988, 1990; Lőw et al. 1989; Gubányi and Pekli 1991; Tunner and Heppich-Tunner 1992), north-eastern (Berger et al. 1988) and eastern parts of the country (Mészáros 1973; Mészáros and Bartos 1978; Gubányi and Korsós 1992). Eastern Hungarian populations have not been well represented, and only the papers published by Mészáros and Bartos (1978) and Gubányi and Korsós (1992) examined the genetic structure of water frogs collected in Hortobágy National Park (hereafter HNP). Except from one population in western Hungary (Tunner and Heppich-Tunner 1992), we have no information about the type of breeding system of water frogs. Furthermore, our study is the first which attempts to assess breeding system of water frogs without the implementation of crossing experiments. Our intention was to apply population genetic approach to determine the type of gametes produced by hybrids and to infer the breeding system of water frogs.
The aims of the present study were to (1) determine the taxonomic composition of the P. esculentus complex in HNP using molecular markers, (2) specify the ploidy levels of hybrid individuals and (3) examine the effect of population structure on genetic diversity and the rate of clonal inheritance in water frogs.
Materials and Methods
Study area
This study was conducted in HNP, the largest continuous alkaline steppe in Europe covering 80 000 hectares. Founded in 1973, the area is the largest and oldest National Park in Hungary, abundant in wetland habitats like alkaline marshes, fishponds, wet grasslands and wet meadows. The expanded protected area also includes 75 fishponds covering a total of 6000 ha and ranging between 1 and 790 ha each (Ecsedi 2004).
Sampling
We collected 164 water frogs during the years 2012–2014 from three different types of wetland habitats. Fieldwork was carried out between April and October each year, respectively. Each site was visited at least three times during the years (in total, thirteen times). Habitats comprise slowly flowing water at the Nádudvar-Kösély canal (hereafter NKC), a fish pond system at Hortobágy fish ponds (HFP) and a marshland system at Egyek-Pusztakócs (EPMS). There are no direct connections between these wetland habitats which can provide easy passage for water frog migration. The straight geographic distance between the localities EPMS and NKC is more than 27 km, 15 km between EPMS and HFP and 21 km between HFP and NKC (Fig. 1). Sampling sites were visualized with Quantum GIS 2.8 (QGIS Development Team, 2015). A brief description of localities, sample sizes and GPS coordinates is given in Table 1. Frogs were captured at night manually and with dip nets. Additional individuals were collected from fresh road kills. DNA from water frogs was extracted from buccal swabs (Tubed Sterile Dryswab™, MWE) using DNeasy Blood and Tissue Kit spin columns (QIAGEN Inc., Valencia, CA, USA) and from toe clips of road-killed individuals. Voucher specimens of water frogs were deposited in the Herpetological Collection of the Hungarian Natural History Museum (Table S1).
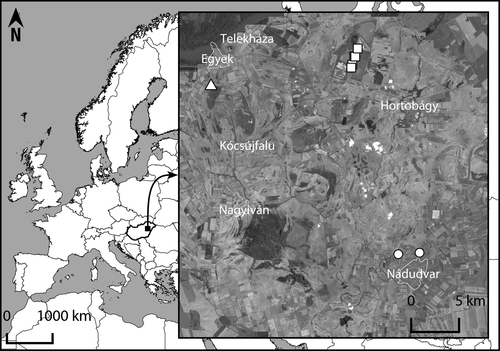
| Locality | Abbreviation | Longitude | Latitude | Sample size | Habitat |
|---|---|---|---|---|---|
| Nádudvar-Kösély canal | NKC | 47°26′43 | 21°10′11 | 30 | Irrigation canal with moderate reed belt |
| 47°26′38 | 21°8′25 | 32 | Irrigation canal with moderate reed belt | ||
| Hortobágy fish ponds | HFP | 47°37′15 | 21°4′36 | 13 | Small ‘pool’ near bigger fishpond |
| 47°37′43 | 21°4′46 | 12 | Bank of bigger fishpond with moderate reed belt | ||
| 47°37′42 | 21°4′51 | 12 | Bank of bigger fishpond with no reed belt | ||
| 47°38′9 | 21°5′3 | 20 | Bank of bigger fishpond with moderate reed belt | ||
| 47°37′5 | 21°4′31 | 24 | Canal alongside the railway with poor vegetation | ||
| Egyek-Pusztakócs marsh system | EPMS | 47°36′3 | 20°52′53 | 21 | Marshland with temporary water level |
Species and ploidy level identification
We used molecular markers to distinguish, first, among the taxa P. ridibundus, P. lessonae and P. esculentus, and second, between the diploid and triploid individuals of hybrids. Taxon identification was determined using the technique described by Hauswaldt et al. (2012) and is based on allele size polymorphism in intron 1 of the serum albumin gene (SAI-1; Plötner et al. 2009), with a slight modification in PCR protocol. Specifically, we used a PCR mix of a total volume of 10 μl containing 1 μl 10X Taq buffer with (NH4)2SO4 (ThermoScientific, Waltham, MA, USA), 0.48 μl of both forward (5′-TCCATACAAATGTGCTAAGTAGGTT-3′) and reverse (5′-GACGGTAAGGGGACATAATTCA-3′) primers (10 μM), 1 μl dNTPs (10 μM), 0.6 μl MgCl2 (25 mM), 0.08 μl Taq DNA polymerase (5 U μl−1) (ThermoScientific, Waltham, MA, USA), 5.56 μl nuclease free water and 0.8 μl of DNA. The PCR profile comprised an initial denaturation at 94°C for 90 sec, followed by 35 cycles of denaturation (30 sec at 94°C), annealing (40 sec at 59°C) and elongation (100 sec at 72°C), and a final elongation step at 72°C for 10 min. Three μL of PCR product was run on 1.8% of agarose gels. To verify SAI-1 fragments, we sequenced representative alleles on a Hitachi 3130 Genetic Analyzer (Applied Biosystems, UK). Consensus sequences were compiled using BioEdit version 7.0.9.0 (Hall 1999) and aligned manually. The sequences were then compared with those deposited in GenBank.
Ploidy determination and genotype composition of hybrids (diploid RL, triploid LLR and LRR) was based on analysis of species-specific polymorphism in selected microsatellite loci (Table 2). Besides Hungarian samples, we added three triploid LLR frogs from Kozí Chrbát, Slovakia, whose ploidy and genotypic composition was confirmed in the study of Pruvost et al. (2015). Two multiplex PCR amplification sets with forward primers fluorescently labelled with FAM, VIC, NED and PET were performed as follows. Reactions for Multiplex 1 were 6 μl total volume and contained 1 μl template DNA, 3 μl 2× QIAGEN Multiplex PCR kit (QIAGEN Inc., Valencia, CA, USA), 1.58 μl nuclease free water and 0.06 μl (10 μM) of both forward and reverse primers. Reactions for Multiplex 2 were the same except that we used 0.12 μl from Res14 and 0.18 μl from Re2Caga3 forward and reverse primers. In order to achieve equivalent allele intensity (i.e. those concentration of primers to all alleles amplify in similar amount), different primer volume was used. Multiplex PCR was performed using the following conditions: 15 min at 95°C, followed by 30 cycles of 30 sec at 94°C, 90 sec at 60°C and 60 sec at 72°C, followed by the final step 30 min at 60°C. PCR products were run on an ABI 3130 Genetic Analyzer (Applied Biosystems, UK) with a LIZ-500 size standard. The alleles (relative peak heights) were scored with GeneMarker software (Softgenetics, State College, PA, USA) manually by a single observer.
| Locus | Genome specificity (alleles) | References | Tag |
|---|---|---|---|
| Multiplex 1 | |||
| RICA1b6 | L (73, 79, 83, 85) R (73, 85, 90, 97, 99) | Arioli (2007) | PET |
| RICA1b5 | L (120, 122) R (135) | Garner et al. (2000) | NED |
| Ga1a19 | L (99) R (105, 108, 118, 120, 134, 148) | Arioli (2007) | FAM |
| Rrid064A | R (223, 225, 227, 228, 230, 232, 236) | Christiansen and Reyer (2009) | NED |
| RICA5 | L (248, 258, 260, 264) | Garner et al. (2000) | VIC |
| RICA2a34 | L (110, 131, 137, 142, 144) | Christiansen and Reyer (2009) | VIC |
| Rrid013A | L (288) R (289, 292, 295, 298, 301, 304) | Hotz et al. (2001) | PET |
| Multiplex 2 | |||
| Res14 | L (139) R (139, 145, 150) | Zeisset et al. (2000) | FAM |
| RICA18 | L (178, 180, 182, 184, 206) | Garner et al. (2000) | NED |
| Res22 | R (84, 86, 110, 112) | Zeisset et al. (2000) | NED |
| Rrid059A | L (101) R (113, 115, 133, 136, 138) | Hotz et al. (2001) | VIC |
| Rrid169A | R (184, 188, 192, 194, 196, 210, 212, 215) | Christiansen and Reyer (2009) | VIC |
| Re1Caga10 | L (96, 98) R (102, 106, 108, 110, 114, 116, 118) | Arioli (2007) | FAM |
| Res20 | a | Zeisset et al. (2000) | PET |
| Re2Caga3 | R (173, 189, 204, 209, 213, 218, 231, 239, 244, 248) | Arioli (2007) | PET |
- a Neither L nor R genome was amplified by the primer.
Estimation of null alleles, genetic diversity and clonality
Null alleles in microsatellite loci refer to alleles present in an individual that are not amplified because of mutation(s) in priming sites. An individual heterozygous in a specific locus, but having a null allele, is thus scored as a homozygote, because the null allele is not amplified during PCR. In hybridogenetic water frogs, which behave genetically as F1 hybrids, all individuals should be heterozygous in loci that amplify in both parental genomes. A homozygous genotype must be due to either a null allele or an allele, which is not species-specific, but occurs in both genomes. For loci that amplify only in one parental genome, all null alleles are ‘visible’ as non-amplification. We estimated frequency of null alleles in microsatellite loci as follows: first, we assigned all alleles to specific parental genomes (R or L) according to microsatellite data from the nearest genetically investigated populations in Slovakia (Mikulíček and Pišút 2012; Pruvost et al. 2013, 2015; Hoffmann et al. 2015). In loci amplifying in both genomes, alleles were either genome specific (e.g. allele 135 was present in R genome and alleles 120 and 122 in L genome in the locus RICA1b5; Table 2) or revealed significant differences in frequency between the genomes (e.g. allele 139 in the locus Res14 was fixed in L genome, but occurred in low frequency also in R genome). When an allele was not species-specific, it was assigned to the R or L genome according to its frequency observed in parental gene pools. After assigning the alleles to specific parental genomes, we counted the number of missing alleles and expected they were not amplified because they were null alleles. Finally, we calculated the frequency of null alleles locus by locus.
Because R and L genomes rarely or never recombine, all population genetic parameters were calculated for each genome separately. To estimate genetic diversity within the P. esculentus populations, we used allelic richness (AR) and gene diversity (He). AR and He were calculated in the program SPAGeDi 1.5, allowing simultaneous analysis of haploid and diploid data (Hardy and Vekemans 2002). The level of genome clonality was determined, based on the number of multilocus genotypes (MLG) and multilocus disequilibrium (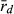 ). The number of MLGs was calculated using the program GenAlEx 6.4 (Peakall and Smouse 2006). MLG is the number of identical combinations of alleles found in microsatellite datasets; few combinations and high frequencies of these combinations may indicate clonal reproduction. In the absence of recombination, frequent MLGs here defined as four or more encounters (in agreement with Pruvost et al. 2015) would represent hemiclones (i.e. clonally transmitted haploid genomes sensu Vrijenhoek 1979). Multilocus disequilibrium is a score of non-random association of alleles at microsatellite loci and varies on a scale from ca. zero to one. Low
). The number of MLGs was calculated using the program GenAlEx 6.4 (Peakall and Smouse 2006). MLG is the number of identical combinations of alleles found in microsatellite datasets; few combinations and high frequencies of these combinations may indicate clonal reproduction. In the absence of recombination, frequent MLGs here defined as four or more encounters (in agreement with Pruvost et al. 2015) would represent hemiclones (i.e. clonally transmitted haploid genomes sensu Vrijenhoek 1979). Multilocus disequilibrium is a score of non-random association of alleles at microsatellite loci and varies on a scale from ca. zero to one. Low 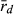 values are related to high recombination rate and thus would indicate low levels of clonality. Contrarily, high
values are related to high recombination rate and thus would indicate low levels of clonality. Contrarily, high 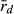 values might be associated with clonal inheritance, as clonal genomes are inherited en bloc, without recombination. Multilocus disequilibrium was calculated in the program Multilocus 1.3 (Agapow and Burt 2001).
values might be associated with clonal inheritance, as clonal genomes are inherited en bloc, without recombination. Multilocus disequilibrium was calculated in the program Multilocus 1.3 (Agapow and Burt 2001).
Results
Taxonomic composition
SAI-1 gene fragment and microsatellites data identified all members of the P. esculentus complex in the studied localities. The 164 frogs examined comprised 100 P. ridibundus, 1 P. lessonae and 63 P. esculentus individuals. In P. ridibundus, SAI-1 amplification resulted in a single band on agarose gels at approximately 850 bp, while in several individuals there was a second band at approximately 700 bp. In P. lessonae, the PCR produced one band at approximately 300 bp. In hybrids, a 300-bp-long fragment was always in combination with either 850-bp or 700-bp fragment. We compared our sequences with GenBank entries and found that the 300-bp PCR band generated from P. lessonae and P. esculentus (GenBank accession number KX838289) individuals was identical to P. lessonae GenBank entries FN432385 from Germany and FN432383 from P. lessonae in Italy. The sequence obtained from the 850-bp band of P. ridibundus (GenBank accession number KX838291) and P. esculentus matched Genbank entry JQ965524 from P. ridibundus in Latvia. The 700-bp band derived from both P. ridibundus and P. esculentus (GenBank accession number KX838290) matched GenBank entries JQ965529 and JQ965528 sampled in Poland. Both GenBank sequences are assigned to Pelophylax kurtmuelleri (Gayda, 1940), a disputable taxon distributed in the Balkans, whose allele in the SAI-1 gene was found in central European P. ridibundus (Hauswaldt et al. 2012). Table 3 summarizes the species composition ratio between different habitats and populations. Pelophylax ridibundus was the most frequent species (10–69% per each locality, 61% of all individuals), whereas P. esculentus occurred in moderate frequencies (31–86% per each locality, 38% of all individuals), and P. lessonae was represented by a single individual (0–5% per each locality, 0.6% of all individuals).
| Locality | Taxon | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. ridibundus | P. lessonae | P. esculentus | |||||||||||||
| N | % | ♀ | ♂ | ASR | N | % | ♀ | ♂ | ASR | N | % | ♀ | ♂ | ASR | |
| NKC | 42 | 68 | 17 | 18 | 0.51 | 0 | 0 | 0 | 0 | – | 20 | 32 | 18 | 1 | 0.05 |
| HFP | 56 | 69 | 12 | 24 | 0.66 | 0 | 0 | 0 | 0 | – | 25 | 31 | 11 | 7 | 0.38 |
| EPMS | 2 | 10 | 1 | 0 | 0 | 1 | 5 | 0 | 0 | – | 18 | 86 | 5 | 0 | 0 |
| Total | 100 | 61 | 30 | 42 | 0.58 | 1 | 0.6 | 0 | 0 | – | 63 | 38 | 34 | 8 | 0.19 |
- ASR, adult sex ratio is expressed by convention as the proportion of males in the adult population.
We recorded two types of populations occurring in HNP. In NKC and HFP, we found P. ridibundus and P. esculentus individuals (RE population) and a population with all three taxa, but dominated by P. esculentus occurred at EPMS (LER population; Table 3). In NKC and HFP, both sexes of P. ridibundus co-occurred with females and males of P. esculentus. In NKC, 95% of the hybrids were females, while in HFP, the proportion was 61% compared to males.
Ploidy level of hybridogenetic hybrids
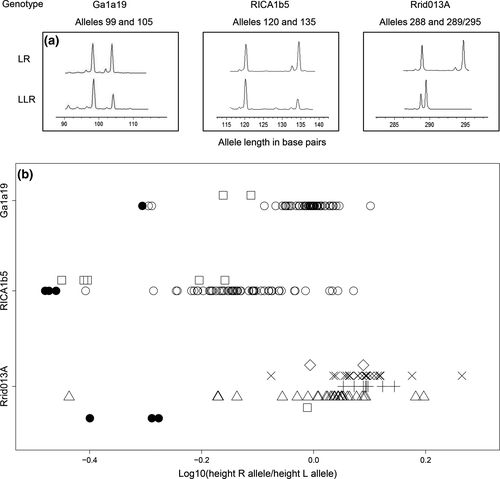
We screened a total of 63 hybrid individuals from all three populations and diagnosed their ploidy level with a set of microsatellite loci (Table 2). The number of different alleles per locus ranged from 1 to 10, with a mean of 5.8. All individuals were indicated to be LR genotype. Three primers (Ga1a19, RICA1b5 and Rrid013A) amplified L and R loci with dosage effect (Fig. 2) demonstrating low levels of polymorphism, often a single L and one R allele were amplified in hybrids. In LLR frogs from a Slovak population, the L peak was clearly higher than the R peak, while in LR frogs, they were of similar height. For each locus, the dosage values clustered into two distinct groups. The three LLR individuals always fell in the left group (Fig. 2) confirming that triploid frogs are detectable by this method. The right group with dosage values around zero are classified as diploid LR frogs. It thus appears that no LRR frogs were present in the sample. The three reference LLR individuals had consistent LLR dosage values at all three loci and were heterozygous for several L-specific alleles at other loci, but the few individuals with LLR dosage values in our study differed for Ga1a19 (N = 2), RlCA1b5 (N = 4) and Rrid013A (N = 1) and did not show heterozygosity for L-specific alleles at other loci. Therefore, those individuals were considered to be diploids.
Null alleles, genetic diversity and clonality in P. esculentus
The highest frequency of null alleles was found in the locus Re1Caga10 (0.313 and 0.217 in the L and R genome, respectively). This locus was subsequently expelled from the analyses of genetic diversity and clonality. Frequency of null alleles in other loci was low, ranging from 0 to 0.116.
Genetic diversity within P. esculentus was slightly higher in L than in R genome. Allelic richness and gene diversity ranged between 1.310–3.680 and 0.051–0.550 in L genome, and 1.900–3.610 and 0.287–0.447 in R genome, respectively (Table 4). Very low values of allelic richness (AR = 1.310) and gene diversity (He = 0.051) were observed in the L genome in the population HFP. Multilocus genotype (MLG) analysis revealed overall four frequent (four or more times encountered) MLGs in R and one in L genome, respectively, although frequency of MLGs varied among the localities (Table 5). In all localities, most hybrids had frequent R MLGs and rare L MLGs, although the frequency of a single L MLGs was high in HFP.
| Locality | L genome | R genome | ||
|---|---|---|---|---|
| AR | He | AR | He | |
| NKC (20) | 3.680 | 0.496 | 2.730 | 0.371 |
| HFP (25) | 1.310 | 0.051 | 3.610 | 0.447 |
| EPMS (18) | 3.330 | 0.550 | 1.900 | 0.287 |
| Mean | 3.140 | 0.469 | 3.010 | 0.389 |
- AR, allelic richness; He, gene diversity
| Multilocus genotypes | NKC | HFP | EPMS |
|---|---|---|---|
| R-A | 3 | 0 | 6 |
| R-B | 1 | 4 | 0 |
| R-C | 4 | 5 | 5 |
| R-D | 4 | 3 | 3 |
| R others | 7 | 12 | 4 |
| L-A | 5 | 20 | 0 |
| L others | 15 | 5 | 18 |
Multilocus disequilibrium (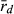 ), a measure of clonality, varied in both genomes (L genome 0.017–1.000; R genome 0.051–0.375) (Table 6). The lowest
), a measure of clonality, varied in both genomes (L genome 0.017–1.000; R genome 0.051–0.375) (Table 6). The lowest 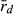 value in the L genome was found at EPMS and co-occurred with the presence of a P. lessonae frog, with exclusively rare L MLGs and with the highest gene diversity (He) in the L genome. Thus, all available information agrees on high levels of recombination in the L genome at EPMS. Similarly, the lowest
value in the L genome was found at EPMS and co-occurred with the presence of a P. lessonae frog, with exclusively rare L MLGs and with the highest gene diversity (He) in the L genome. Thus, all available information agrees on high levels of recombination in the L genome at EPMS. Similarly, the lowest 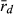 in the R genome was found at HFP, where the highest percentage of P. ridibundus, the highest number of rare R MLGs and the highest genetic diversity in the R genome (both AR and He) were also encountered. Again, the results obtained independently from population composition, genetic diversity and clonality agree with each other indicating high levels of recombination in the R genome at HFP.
in the R genome was found at HFP, where the highest percentage of P. ridibundus, the highest number of rare R MLGs and the highest genetic diversity in the R genome (both AR and He) were also encountered. Again, the results obtained independently from population composition, genetic diversity and clonality agree with each other indicating high levels of recombination in the R genome at HFP.
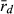 ), a measure of clonality, calculated separately for L and R genomes in hybrid individuals. Sample size at each locality are given in parentheses
), a measure of clonality, calculated separately for L and R genomes in hybrid individuals. Sample size at each locality are given in parentheses| Locality | L genome | R genome |
|---|---|---|
| NKC (20) | 0.107 | 0.221 |
| HFP (25) | 1.000 | 0.051 |
| EPMS (18) | 0.017 | 0.375 |
| Mean | 0.375 | 0.216 |
Discussion
Taxonomic composition of water frogs in Hortobágy National Park
At the three sites investigated in the Hortobágy National Park (HNP), the water frog population comprised all species of the P. esculentus complex. The collected material was dominated by P. ridibundus (61% of all sampled individuals) and P. esculentus (38% of all sampled individuals). In both taxa, males and females were detected. Our results correspond to previous studies reporting P. ridibundus and P. esculentus from the region (Marián 1963; Dely 1967; Murai et al. 1983). At EPMS, we collected a single juvenile P. lessonae individual. Pelophylax lessonae may be more abundant in EPMS or our results were affected by small sample size (N = 21). It is also possible that P. lessonae has suffered population loss and is particularly rare in HNP. Mészáros and Bartos (1978) confirmed the presence of P. lessonae in HFP, but they did not provide sample size data of collected specimens, so we cannot make any conclusions about the past abundance. Given the protected status of the study site, we do not hypothesize that there have been such significant anthropogenic changes inside HNP, implying that regional effects and climate change may play an important role. On the other hand, Mester et al. (2015) reported higher frequencies of P. lessonae at EPMS based on morphological and call monitoring data. This may support our first explanation about low sample size.
At two permanently flooded sites, we found RE populations dominated by P. ridibundus and living sympatrically with P. esculentus, while the site with temporary water level proved to be an LER population, on which both parental species and the hybrid co-occurred. The characteristics of the habitats in HFP and NKC seem to be optimal for P. ridibundus which prefers large and deep water bodies where the water is continuously renewed with oxygen (Morand and Joly 1995; Holenweg and Reyer 2000; Pagano et al. 2001; Mikulíček et al. 2015). This matches our findings, in which 98% (N = 98) of the P. ridibundus individuals were collected from sites with these habitat characteristics. Pelophylax esculentus shows no strict characteristics in habitat preference and it can be found practically in every type of water bodies (Berger 1973; Ildos and Ancona 1994). In the examined populations, hybrids occurred in all types of wetland habitats in HNP, while the only one collected P. lessonae individual was found in the shallow water body with temporary water level in EPMS as previously reported by Mester et al. (2015). After the regulation of the river Tisza in the 19th century at EPMS, the extensive floods have disappeared and erosion intensified, which all led to an accelerated alkalization of the region. Today, the area characterized by extensive Pannonic salt steppes and marshes (Aradi et al. 2003). In the past few decades, the water supply systems of the marshes were reconstructed by the result of a LIFE-Nature programme (Déri et al. 2009). This human-mediated environmental changes may provide possibly new habitats for P. lessonae, which populations show declining trend in Central Europe (Holsbeek and Jooris 2009; Mayer et al. 2013).
Ploidy level
In this study, we examined the genotypic composition of 63 P. esculentus individuals using microsatellite loci. Nine frogs showed contradictions in their genotype determination. One locus indicated LLR genotype (Fig. 2) either by dosage effect (seven individuals) or by heterozygosity (two individuals) but all the remaining loci in these frogs suggested LR genotype. Thus, we considered all examined hybrids to be LR diploids. In contrast, the three LLR reference frogs from Slovakia were easily recognizable as triploids as they showed dosage effect and/or heterozygosity at 4–5 loci. Although our dataset did not include any LRR individuals, the principle would have been the same. We therefore conclude that any triploid frogs would reliably have been distinguished from LR frogs with the current set of microsatellites as previously demonstrated in several other studies (Christiansen 2005; Hoffmann et al. 2015; Pruvost et al. 2015).
The presence of triploids in Hungarian water frog population was reported by Tunner and Heppich-Tunner (1992) in north-western Hungary. They described a population system comprising P. ridibundus of both sexes, diploid P. esculentus of both sexes and exclusively male triploid (LLR) P. esculentus. The ploidy level of hybrids was confirmed with electrophoresis of the albumin locus and chromosome examination. The observation of Tunner and Heppich-Tunner (1992) was not confirmed recently by the study of Pruvost et al. (2015) and by our field research (J. Vörös, D. Herczeg, L. Choleva, P. Mikulíček; unpublished data) in the same area. Therefore, that was the only report of a triploid population in Hungary; all others (Mészáros and Bartos 1978; Berger et al. 1988; Gubányi and Korsós 1992; this study) encountered only diploid hybrids.
Using population genetics to infer breeding system
The RE-system with syntopic occurence of hybrids with P. ridubundus without P. lessonae tends to be uncommon in Europe; isolated populations observed in eastern Ukraine (Borkin et al. 2004; Biriuk et al. 2016), western and eastern Poland (Rybacki and Berger 2001), Germany (Günther 1990), France (Pagano et al. 1997) and Slovakia (Mikulíček et al. 2015). Whether hybrids in Hungarian RE populations eliminate the R genome and form L gametes, as in some German and Polish populations (Uzzell and Berger 1975; Berger 1977; Uzzell et al. 1980) or the simultaneous production of the L and R gametes, as was observed in populations in Ukraine (Vinogradov et al. 1990, 1991; Biriuk et al. 2016) remains an open question that can only be fully answered by crossing experiments. Alternatively, Hungarian hybridogenetic lineages can produce R gametes (as in the vastly distributed LE system), be reproductively dependent on P. lessonae individuals, which, however, should reach extremely low densities in studied populations. In this case, P. ridibundus would not serve as a host species in hybridogenetic reproduction, but just coexists with hybrids, as was observed in the mid-Rhône drainage in France (Pagano et al. 1997) or in western Slovakia (Mikulíček et al. 2015).
In the absence of crossing experiments, we aimed to settle the question by assessing the level of clonality in the L and the R genome. If all L genomes in hybrids come from sexually reproducing P. lessonae, and the R genomes come partly from clonal gametes of P. esculentus and partly but less frequently from sexually reproducing P. ridibundus, then the L genome should show no clonality (i.e. high number of MLGs and a value of 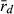 around zero), while the R genome should show some evidence of clonality. Genetic diversity (both AR and He) decreases by clonal reproduction and should therefore be higher in the L than in the R genome.
around zero), while the R genome should show some evidence of clonality. Genetic diversity (both AR and He) decreases by clonal reproduction and should therefore be higher in the L than in the R genome.
The three localities analysed produced different results. In both, EPMS and NKC, as expected, the L genome was indeed less clonal (lower score of 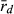 , higher number of MLGs) and had the higher level of genetic diversity than the R genome. This suggests that at EPMS and NKC, populations belong to the LE system typical in central Europe, in which hybrids make R gametes and only can produce new hybrids by mating with P. lessonae. This effect was especially apparent in EPMS. This makes sense, as EPMS was the locality with typical P. lessonae-specific habitat, where P. ridibundus was rare, where the only P. lessonae in the present study was found and where Mester et al. (2015) suggested a higher proportion of P. lessonae. In contrast, at NKC, the habitat was more similar to P. ridibundus preferences, P. ridibundus dominated in numbers and no P. lessonae was found there. If only few individuals of P. lessonae are present and have parented all the hybrids, genetic diversity in the L genome of the hybrids should indeed be low, as was observed at NKC. An even lower number of P. lessonae parents should produce a genetic signal in the hybrid offspring that is indistinguishable from clonal reproduction (low number of multilocus genotypes and high value of
, higher number of MLGs) and had the higher level of genetic diversity than the R genome. This suggests that at EPMS and NKC, populations belong to the LE system typical in central Europe, in which hybrids make R gametes and only can produce new hybrids by mating with P. lessonae. This effect was especially apparent in EPMS. This makes sense, as EPMS was the locality with typical P. lessonae-specific habitat, where P. ridibundus was rare, where the only P. lessonae in the present study was found and where Mester et al. (2015) suggested a higher proportion of P. lessonae. In contrast, at NKC, the habitat was more similar to P. ridibundus preferences, P. ridibundus dominated in numbers and no P. lessonae was found there. If only few individuals of P. lessonae are present and have parented all the hybrids, genetic diversity in the L genome of the hybrids should indeed be low, as was observed at NKC. An even lower number of P. lessonae parents should produce a genetic signal in the hybrid offspring that is indistinguishable from clonal reproduction (low number of multilocus genotypes and high value of 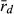 ). This could explain why HFP could also belong to the LE system, although the L genome had a lower number of multilocus genotypes, a higher score of
). This could explain why HFP could also belong to the LE system, although the L genome had a lower number of multilocus genotypes, a higher score of 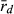 , and a lower level of genetic diversity than the R genome.
, and a lower level of genetic diversity than the R genome.
The application of 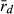 to infer breeding system of water frogs in the present study is to our knowledge novel. Previously,
to infer breeding system of water frogs in the present study is to our knowledge novel. Previously, 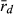 was used to demonstrate genetic recombination in triploid hybrids in all-hybrid populations, but not to infer breeding system (Christiansen and Reyer 2009). We emphasize that the most accurate way to assess the type of gametes produced by hybrids and to determine the breeding system of water frogs is by crossing experiments; however, this is very time consuming. Therefore, our novel way of applying
was used to demonstrate genetic recombination in triploid hybrids in all-hybrid populations, but not to infer breeding system (Christiansen and Reyer 2009). We emphasize that the most accurate way to assess the type of gametes produced by hybrids and to determine the breeding system of water frogs is by crossing experiments; however, this is very time consuming. Therefore, our novel way of applying 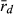 in relation to breeding system seems to have great potential for use in future genetic water frog studies and might thus help extend our understanding of breeding system and their distribution. This method clearly has its limitations related to the number and polymorphism of selected genetic markers which might explain that it did not provide the expected result for all populations tested, but these first results represented a valuable first contribution to gathering experience with the method for the benefit of future studies.
in relation to breeding system seems to have great potential for use in future genetic water frog studies and might thus help extend our understanding of breeding system and their distribution. This method clearly has its limitations related to the number and polymorphism of selected genetic markers which might explain that it did not provide the expected result for all populations tested, but these first results represented a valuable first contribution to gathering experience with the method for the benefit of future studies.
In conclusion, additional efforts need to be carried out (e.g. sampling during mating activity) to clarify the distribution of P. lessonae at HNP and this may strengthen our assumption about the breeding system of water frogs inhabited in this nature reserve.
Acknowledgements
This research was supported by the European Union and cofinanced by the European Social Fund through the Social Renewal Operational Programme under the projects TÁMOP-4.2.2/B-10/1-2010-0024 and SROP-4.2.2.B-15/1/KONV-2015-0001. Additional financial support was provided by the Campus Hungary Short Term Study Program (B2/1R/14652 and B2/1R/17869). During the project, JV was financed by the Hungarian Scientific Research Fund (OTKA K77841) and the Bolyai János Research Scholarship of the Hungarian Academy of Sciences. PM was supported by the Slovak Research and Development Agency under the contract no. APVV-0147-15. The authors are indebted to Virág Krízsik, Orsolya Márton and Mária Tuschek in the Molecular Taxonomy Laboratory of the Hungarian Natural History Museum for their technical assistance and Zsolt Végvári and Béla Mester who helped us collecting frogs during fieldwork. We also acknowledge Edvárd Mizsei for implementing the map with QGIS. Furthermore, we are thankful to Attila Fülöp and Csongor I. Vágási in MTA-DE Lendület Behavioural Ecology Research Group and who, with their useful insights, helped us progress with the data processing, and Daniel R. Brooks who kindly helped to improve the language of the manuscript. Research permit was issued by the Tisza Region Inspectorate of Environment, Nature Conservation and Water Management (4633/OH/2012).




