Mitochondrial DNA haplotypes indicate two postglacial re-colonization routes of the spruce bark beetle Ips typographus through northern Europe to Scandinavia
Abstract
enSpecies in northern Europe re-colonized the region after the last glacial maximum via several routes, which could have lingering signatures in current intraspecific trait variation. The spruce bark beetle, Ips typographus, occurs across Europe, and biological differences have been found between southern and northern Scandinavian populations. However, the postglacial history of I. typographus in Scandinavia has not been previously studied at a fine geographical scale. Therefore, we collected specimens across northern Europe and analysed the genetic variation in a quite large mitochondrial fragment (698 bp). A high genetic diversity was found in some of the most northern populations, in the Baltic States, Gotland and central Europe. Detected genetic and phylogeographic structures suggest that I. typographus re-colonized Scandinavia via two pathways, one from the northeast and one from the south. These findings are consistent with the re-colonization history of its host plant, Picea abies. However, we observed low haplotype and nucleotide diversity in southern Scandinavian populations of I. typographus, indicating that (unlike P. abies) it did not disperse across the Baltic Sea in multiple events. Further, the divergence among Scandinavian populations was shallow, conflicting with a scenario where I. typographus expanded concurrently with its host plant from a ‘cryptic refugium’ in the northwest.
Résumé
frLes variations de l'ADN mitochondrial suggèrent l'existence de deux routes postglaciaires du Nord de l'Europe vers la Scandinavie chez le scolyte de l'épicéa Ips typographus.
Après le dernier maximum glaciaire, les espèces européennes ont recolonisé les régions du Nord de l'Europe via différentes routes de migration, ce qui a pu conduire aux variations intra-spécifiques actuellement observées pour un caractère biologique. Le typographe, Ips typographus, est présent dans toute l'Europe et des différences biologiques ont été identifiées entre les populations du Sud et du Nord de la Scandinavie. Cependant, l'histoire postglaciaire d'I. typographus dans le Nord de l'Europe n'a, jusqu'à présent, pas été étudiée à une échelle géographique fine. Dès lors, nous avons récolté des échantillons sur l'étendue de l'Europe du Nord et avons analysé la diversité génétique d'un relativement long fragment de gène mitochondrial (698 pb). Une plus forte diversité génétique a été observée dans certaines des localités situées le plus au Nord ainsi que dans les pays baltes, dans le Gotland et en Europe centrale. La présence d'une structure génétique et phylogéographique suggère qu'I. typographus a recolonisé la Scandinavie via deux routes de migration différentes: à partir du Nord-Est et à partir du Sud-Est. Ces résultats sont en accord avec l'histoire de recolonisation identifiée chez la plante hôte Picea abies. Cependant, nous avons observé une faible diversité haplotypique et une faible diversité nucléotidique chez les populations d'Ips typographus du Sud de la Scandinavie, ce qui indique que (contrairement à P. abies) il n'a pas traversé la Mer Baltique à plusieurs reprises. Enfin, la très faible divergence entre toutes les populations scandinaves ne soutient pas un scénario d'expansion simultanée d'I. typographus et de sa plante hôte depuis ‘un refuge cryptique’ du Nord-Ouest.
Introduction
Climatic oscillations during the Quaternary (the last two million years) have driven repeated extensions and contractions of ice sheets in the Northern Hemisphere. This has been accompanied by migrations of populations to lower latitudes and isolation in refugia (Taberlet et al. 1998; Hewitt 2000), followed by expansion from their refugia and re-colonization of formerly glaciated areas. These contraction and expansion cycles have strongly shaped current spatial distributions of species' lineages (Hewitt 1999; Avise 2000). However, identifying key features of the historical processes, such as refugial sites and migration routes, is challenging due to the diversity of ecological and evolutionary forces acting upon species (Avise 2008).
Here, we address the phylogeography and evolutionary history of the European spruce bark beetle, Ips typographus L. (Coleoptera: Scolytinae), the most serious insect pest of Norway spruce (Picea abies L. Karst). It has killed substantial numbers of spruce trees in Europe (Grégoire and Evans 2004), especially during the last decades of the twentieth century when strong storms triggered outbreaks of the pest across large areas of Scandinavia (Långström et al. 2009; Kärvemo and Schroeder 2010).
The phylogeography of I. typographus has been previously investigated at a large geographic scale by analysing mitochondrial, internal transcribed spacer (ITS) or microsatellite markers in samples collected from locations across Europe (Stauffer et al. 1999; Sallé et al. 2007; Bertheau et al. 2013). These large-scale analyses have provided valuable insights into effects of specific life-history and evolutionary traits on the species' genetic variation. Notably, very weak genetic structure has been detected in microsatellite sequences and was attributed to strong gene flow between populations (Sallé et al. 2007; Gugerli et al. 2008). Accordingly, both field observations (Botterweg 1982; Byers 1995; Franklin and Grégoire 1999) and laboratory experiments (Forsse and Solbreck 1985) indicate that the beetle has high dispersal capacities. In addition, I. typographus appears to have very recent (late Pleistocene) origins in Europe, as all mtDNA haplotypes are poorly differentiated (Bertheau et al. 2013).
Due to the high gene flow and recent origin of I. typographus in Europe, detecting any genetic structure (and thus potentially important indications of its recent population history) is challenging. Use of mitochondrial (mtDNA) markers alone is often insufficient to resolve the complex history of a species, for two main reasons. Firstly, they only provide access to the matrilineal history, due to the uniparental inheritance of mtDNA. Secondly, selection (Dowling et al. 2008), introgression (Ballard and Whitlock 2004) or pseudogenes (Buhay 2009) may bias the inferred history. Nevertheless, as universal mitochondrial primers for specific taxonomic groups are readily available (e.g. the sets compiled for insect analyses by Simon et al. 1994), mtDNA provides convenient material for rapid investigations of matrilineal genealogies, thereby assisting reconstruction of the evolutionary history of populations in specific regions. Furthermore, mtDNA mutates rapidly in animals, and thus, mtDNA markers are valuable for detecting potential structures in species with recent origins (Avise 2000), particularly highly dispersive species, in which intense gene flows can rapidly reduce genetic differentiation among populations.
Recently, mtDNA analysis of intensively sampled populations at a fine geographic scale has provided valuable new insights into the population history of I. typographus in south-eastern Europe. Whereas large-scale approaches identified a single refugium for the whole of southern Europe (Stauffer et al. 1999; Sallé et al. 2007; Bertheau et al. 2013), the fine-scale approach detected at least one additional refugium in the Carpathians during the last glacial maximum (Krascsenitsová et al. 2013). These findings also highlight congruence in the history of local populations of this insect and its host plant, as distinct genetic differentiation between western and south-eastern Carpathian populations of P. abies has been detected (Tollefsrud et al. 2008).
In northern Europe, P. abies apparently survived during the last glaciation in a large refugium in the Russian plains (Schmidt-Vogt 1977; Lagercrantz and Ryman 1990; Tollefsrud et al. 2008) and possibly ‘cryptic’ refugia in the vicinity of the Norwegian coast (Kullman 2002, 2008; Parducci et al. 2012). In contrast, large-scale molecular studies have provided no evidence that the spruce bark beetle expanded concurrently with its host plant from refugia in these northern regions (Stauffer et al. 1999; Sallé et al. 2007; Bertheau et al. 2013). However, phenotypic differences have been reported between southern and northern Scandinavian populations of I. typographus, prompting speculation that they may have different geographic origins. For example, in experiments reported by Komonen et al. (2011), beetles from southern Sweden generally hibernated under spruce bark while those from central Sweden moved to the ground. Similarly, size fluctuation of the beetle's populations in the south and north of mid-Norway is poorly synchronized, and the lack of synchrony is not clearly related to climatic variables (Økland and Bjørnstad 2003).
- If the scenario inferred from large-scale studies of I. typographus is valid, there should be little genetic structure among its populations across Scandinavia, but genetic diversity should decrease from the south to the north, reflecting a single re-colonization route from central Europe across the Baltic Sea into southern Scandinavia (Stauffer et al. 1999; Bertheau et al. 2013).
- Alternatively, if I. typographus followed its host plant during its putative expansion from a Russian refugium, northern and southern Scandinavian populations should be genetically distinct, reflecting two re-colonization routes: one from the south (as above) and one from the north-east via Finland (Lagercrantz and Ryman 1990; Tollefsrud et al. 2008, 2009).
- Finally, if I. typographus expanded concurrently with its host plant from cryptic refugia in mid-Norway (Kullman 2008; Parducci et al. 2012), there should be signs of an ancient phylogeographic divergence, indicative of prolonged isolation of spruce bark beetle populations in this region as it was only reached by the main re-colonization front of P. abies 3000–2000 BP (Tollefsrud et al. 2008).
We tested the three proposed scenarios at fine scale by collecting samples across most parts of northern Europe and then analysing the genetic variation in a 698-bp fragment of a mitochondrial gene both within populations and between regions.
Materials and Methods
Sampling and DNA extraction
We collected 359 adult I. typographus from under the bark of standing or felled P. abies trees or pheromone traps in 44 northern European sites and three central European sites (Fig. 1, Table S1). The central European sites were included as references for this region. Beetles were stored at −20°C or in 96% ethanol at room temperature. Genomic DNA was extracted from the entire body of fresh or up to 1-year-old specimens using a QIAGEN DNeasy Blood & Tissue kit (QIAGEN GmbH, Hilden, Germany) following the manufacturer's protocol. Each extracted DNA sample was eluted and stored in 100 μl of AE buffer (QIAGEN). Reference material is available at the Department of Ecology, Swedish University of Agricultural Sciences and the Laboratory of Biological Control and Spatial Ecology, Université Libre de Bruxelles.
DNA amplification and sequencing
We sequenced a 789-bp fragment of the mitochondrial cytochrome c oxidase subunit I (COI) gene corresponding to the region spanning nucleotides 2201 to 2990 of the Drosophila yakuba sequence in each sample (Clary and Wolstenholme 1985). Both strands of the fragment were sequenced to verify its identity, using the primer pair Jerry (C1-J-2183)/Pat (L2-N-3014) published by Simon et al. (1994) as follows. The fragments were amplified in 20 μl reaction mixtures consisting of 4.0 mM MgCl2, 300 μM dNTP, 0.5 μM of each primer, 0.5 units of HotstarTaq DNA polymerase and approximately 10–40 ng DNA in a thermocycler (Eppendorf AG, Hamburg, Germany). The amplification protocol consisted of 15 min denaturation at 95°C followed by 36 cycles of 30 s denaturation at 94°C, 30 s annealing at 47°C, 1 min 30 s extension at 72°C and a final 10 min extension step at 72°C. The amplified fragments were electrophoretically separated on a 1% agarose gel and stained with ethidium bromide to verify their quality. The target PCR product was purified, using a QIAquick PCR Purification kit (QIAGEN GmbH) or ExoSAP-IT (USB, Cleveland) following the manufacturers' instructions, then sequenced at Uppsala Genome Center (http://www.genpat.uu.se/facilities/genome_center/) or by Macrogen (Seoul, Korea; http://www.macrogen.com).
After removing low-quality sections at the end of each sequenced fragment, we aligned the acquired sequences using the ClustalW algorithm (Thompson et al. 1994) as implemented in BioEdit (Hall 2007) and then manually edited the alignment. To ensure that the data set included no sequences of nuclear copies of the mtDNA COI gene (so-called NUMTs; Lopez et al. 1994), we translated the sequences into amino acids and checked that the resulting electropherograms indicated no unexpected stop codons or double peaks (Song et al. 2008). Three sequences that yielded ambiguous electropherograms (e.g. double peaks) were removed from the dataset prior to analysis. In addition, we removed 14 sequences with identical overlaps to two identified NUMT sequences, JN133882 and JN133883, published by Bertheau et al. (2011).
Genealogical relations between haplotypes
To reconstruct the genealogical relations between haplotypes, we constructed a haplotype network using the median-Joining algorithm implemented in Network 4.611 with epsilon set to 10 (Bandelt et al. 1999, http://www.fluxus-engineering.com) and a phylogenetic tree using the Bayesian method implemented in beast v1.7.5 (Drummond and Rambaut 2007) with the HKY model (Hasegawa et al. 1985) defined by Bertheau et al. (2013). We fixed a Yule speciation process as prior of tree, and default options were used for all other prior and operator settings. Two independent MCMC analyses were run for 50 million iterations of posterior sampling with logging to file every 10 000 iterations. Equilibrium was confirmed by effective sample size (ESS) values larger than 200 as calculated in Tracer v1.5 (Rambaut and Drummond 2009). When two independent runs converged to the same posterior distribution and same estimates, we combined tree files and discarded 10% of the sampled trees as burn-in in LogCombiner v1.7.5 (http://beast.bio.ed.ac.uk/LogCombiner). Remaining trees were summarized in the form of a maximum clade credibility tree using TreeAnnotator implemented in beast, and the resulting tree was visualized in FigTree v1.4.0. (http://tree.bio.ed.ac.uk/software/figtree).
Genetic structure
To assess the genetic structure among populations, we applied spatial analysis of molecular variance as implemented in samova 1.0 (Dupanloup et al. 2002) to identify geographically homogeneous clusters that are maximally differentiated from others. We applied a simulated spatial annealing procedure (calculated with geographical distances) for K = 2–8 to identify the optimal number of population groups. We then selected the optimal K-value, defined as the value that yielded the maximum ΦCT value (plateau) while excluding configurations with single-population groups (which indicate disappearance of group structure; Magri et al. 2006).
In addition to samova, we applied discriminant analysis of principal component (DAPC) (Jombart 2008). DAPC discriminates individuals associated with pre-defined groups according to a model that maximizes the variance between groups while minimizing the variance within groups. The groups used for the DAPC were determined a priori with the K-mean clustering algorithm. The analyses were run through the ape (Paradis et al. 2004) and adegenet (Jombart 2008) packages implemented in r software (v 2.14; R Development Core Team 2011) on a standardized allele frequency table, obtained by scaleGen, with the binomial method. We selected the optimal number of groups using the minimum Bayesian Information Criterion (BIC) and a configuration without single-population groups as criteria. As DAPC assigns individuals to groups and samova sites to groups, we obtained comparable results from averaging individual posterior membership probabilities of DAPC at the site level. Results of the optimal configuration retained for clustering analyses were plotted on a map (Fig. 3).
Phylogenetic reconstructions suggested the presence of three clades, and the genetic structure analyses identified 2–3 clusters. Thus, we tested for the presence of phylogeographic structure by comparing two estimates of genetic variation: Gst (based solely on differences in haplotype frequencies; Pons and Petit 1995) and Nst (based on differences in haplotype frequencies and the genetic distances between haplotypes; Pons and Petit 1996). According to Pons and Petit (1996), a significantly higher value of Nst indicates that genealogically closely related haplotypes co-occur within populations more often than random expectations. samova group assignments were treated as populations for these analyses. The computed indices and their significance were assessed with 1000 permutations in PermutCpSSR 2.0 (http://www.pierroton.inra.fr/genetics/labo/Software/Permut/).
Genetic differentiation among populations was quantified by comparing pairwise fixation indices (FST), and proportions of genetic variation within and between the identified geographical groups of samples were estimated by analysis of molecular variance (amova) using Arlequin 3.5 software (Excoffier and Lischer 2010).
Summary statistics
For populations at each locality, we calculated the following diversity indices using DnaSP 5 (Librado and Rozas 2009): number of haplotypes, haplotype diversity (Hd), nucleotide diversity (Pi) and the mean number of pairwise differences (MNPD). We also calculated allelic richness (r) using the rarefaction method proposed by El Mousadik and Petit (1996), as implemented in Contrib 1.02 (http://www.pierroton.inra.fr/genetics/labo/Software/Contrib).
To explore regional diversity, the sampling sites were classified into seven geographical groups: north-western Scandinavia (NWS), north-eastern Scandinavia (NES), south-western Scandinavia (SWS), south-eastern Scandinavia (SES), the Baltic States and Russia (BSR), Denmark (DEN) and central Europe (CE) (Table 1). We then tested for possible differences in summary statistics calculated for the geographical groups using nonparametric Kruskal–Wallis analysis of variance (as we could not be certain that the data were normally distributed).
| Code | N | Voucher specimen | Region | Nb (HT) | Haplotype ID | Hd (SD) | MNPD (SD) | Pi (SD) | Ar [3] | FS | D | SAM | DAPC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NLS | 4 | INLS1-4 | NWS | 4 | 8, 16, 21, 28 | 1.00 (0.18) | 2.50 (1.69) | 0.0036 (0.0007) | 2.00 | −1.51 | −0.80 | 1 | 1 (3); 2 (1) |
| NLV | 8 | INLV1-8 | NWS | 3 | 13, 17, 20 | 0.46 (0.20) | 1.11 (0.80) | 0.0016 (0.0007) | 0.75 | 0.39 | −0.18 | 1 | 1 (6); 2 (2) |
| OVE | 7 | IOVE1-7 | NWS | 4 | 11, 13, 20, 24 | 0.86 (0.10) | 1.33 (0.94) | 0.0019 (0.0004) | 1.40 | −0.91 | 0.40 | 1 | 1 (3); 2 (4) |
| VER | 9 | IVER1-9 | NWS | 4 | 9, 11, 20, 25 | 0.78 (0.11) | 1.22 (0.85) | 0.0017 (0.0005) | 1.50 | −0.63 | −0.69 | 1 | 1 (6): 2 (3) |
| ORK | 7 | IORK1-7 | NWS | 3 | 8, 20, 23 | 0.52 (0.21) | 0.57 (0.52) | 0.0008 (0.0004) | 0.86 | −0.92 | −1.24 | 1 | 1 (6); 2 (1) |
| ROV | 7 | IROV1-7 | NES | 4 | 1, 11, 20, 30 | 0.81 (0.13) | 1.33 (0.94) | 0.0019 (0.0005) | 1.46 | −0.91 | 0.40 | 2 | 1 (2); 2 (5) |
| KAL | 10 | IKAL1-10 | NES | 6 | 8, 11, 20, 22, 29, 36 | 0.78 (0.14) | 1.00 (0.73) | 0.0014 (0.0004) | 1.42 | −3.88** | −1.74* | 1 | 1 (8); 2 (2) |
| VIN | 10 | IVIN1-10 | NES | 7 | 11, 13, 20, 26, 27, 31, 34 | 0.87 (0.11) | 1.76 (1.11) | 0.0025 (0.0006) | 1.63 | −3.71** | −1.64* | 1 | 1 (8); 2 (2) |
| SOL | 9 | ISOL1-9 | NES | 4 | 1, 11, 19, 20 | 0.69 (0.15) | 1.00 (0.74) | 0.0014 (0.0004) | 1.20 | −1.04 | −0.36 | 1 | 1 (5); 2 (4) |
| LIK | 10 | ILIK1-10 | SES | 7 | 1, 11, 16, 20, 23, 25, 35 | 0.91 (0.08) | 1.49 (0.98) | 0.0021 (0.0004) | 1.74 | −4.29** | −0.63 | 1 | 1 (7); 2 (3) |
| UKK | 7 | IUKK1-7 | SES | 5 | 1, 11, 20, 23, 32 | 0.91 (0.10) | 1.43 (0.98) | 0.0020 (0.0020) | 1.71 | −2.31* | −0.60 | 1 | 1 (4); 2 (3) |
| POR | 5 | IPOR1-5 | SES | 2 | 1, 27 | 0.40 (0.24) | 1.20 (0.91) | 0.0017 (0.0010) | 0.60 | 1.69 | −1.05 | 2 | 1 (1); 2 (4) |
| LAP | 5 | ILAP1-5 | SES | 3 | 1, 3, 20 | 0.70 (0.22) | 1.20 (0.91) | 0.0017 (0.0007) | 1.20 | −0.19 | −1.05 | 2 | 1 (1); 2 (4) |
| HAM | 8 | IHAM1-8 | SES | 2 | 1, 27 | 0.54 (0.02) | 1.61 (1.06) | 0.0023 (0.0005) | 0.80 | 2.99 | 1.60 | 2 | 1 (3); 2 (5) |
| TUU | 7 | ITUU1-7 | SES | 2 | 1, 15 | 0.29 (0.20) | 0.57 (0.52) | 0.0008 (0.0006) | 0.43 | 0.86 | −1.24 | 2 | 1 (0); 2 (7) |
| ALA | 10 | IALA1-10 | SWS | 4 | 1, 4, 20, 39 | 0.53 (0.18) | 1.11 (0.79) | 0.0016 (0.0006) | 0.89 | −0.65 | −0.82 | 2 | 1 (2); 2 (8) |
| LIL | 11 | ILIL1-11 | SWS | 3 | 8, 11, 20 | 0.64 (0.09) | 0.73 (0.58) | 0.0010 (0.0002) | 1.03 | −0.02 | 0.20 | 2 | 1 (5); 2 (6) |
| TOK | 5 | ITO1-5 | SWS | 4 | 1, 7, 11, 20 | 0.90 (0.16) | 1.40 (1.02) | 0.0023 (0.0006) | 1.70 | −1.65 | −0.18 | 2 | 1 (2); 2 (3) |
| AAS | 7 | IAAS1-7 | SWS | 2 | 1, 6 | 0.48 (0.17) | 0.48 (0.46) | 0.0007 (0.0002) | 0.71 | 0.59 | 0.56 | 2 | 1 (0); 2 (7) |
| ARS | 7 | IARS1-7 | SWS | 2 | 1, 21 | 0.29 (0.20) | 0.57 (0.52) | 0.0012 (0.0008) | 0.43 | 0.86 | −1.24 | 2 | 1 (1); 2 (6) |
| DEL | 7 | IDEL1-7 | SWS | 4 | 1, 11, 20, 23 | 0.71 (0.18) | 1.33 (0.94) | 0.0019 (0.0006) | 1.26 | −0.91 | 0.40 | 1 | 1 (2); 2 (5) |
| SIL | 11 | ISIL1-11 | SWS | 3 | 1, 11, 20 | 0.56 (0.13) | 0.95 (0.70) | 0.0014 (0.0003) | 0.91 | 0.48 | 1.18 | 2 | 1 (3); 2 (8) |
| NAS | 10 | INAS1-10 | SWS | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (10) |
| TIE | 5 | ITIE1-5 | SWS | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (5) |
| GRA | 5 | IGRA1-5 | SWS | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (5) |
| CHA | 8 | ICHA1-8 | SWS | 3 | 1, 11, 20 | 0.61 (0.16) | 0.96 (0.73) | 0.0014 (0.0004) | 1.00 | 0.14 | 0.93 | 2 | 1 (2); 2 (6) |
| SKI | 7 | ISKI1-7 | SWS | 2 | 1, 38 | 0.29 (0.20) | 0.57 (0.52) | 0.0008 (0.0006) | 0.43 | 0.86 | −1.24 | 2 | 1 (0); 2 (7) |
| VAT | 10 | IVAT1-10 | SWS | 2 | 1, 13 | 0.20 (0.15) | 0.40 (0.40) | 0.0006 (0.0004) | 0.30 | 0.59 | −1.40 | 2 | 1 (0); 2 (10) |
| ORS | 9 | IORS1-9 | SWS | 3 | 1, 13, 20 | 0.42 (0.19) | 0.83 (0.65) | 0.0012 (0.0006) | 0.60 | 0.02 | −0.94 | 2 | 1 (1); 2 (8) |
| TIB | 9 | ITIB1-9 | SWS | 2 | 1, 20 | 0.22 (0.17) | 0.44 (0.43) | 0.0006 (0.0005) | 0.33 | 0.67 | −1.36 | 2 | 1 (1); 2 (8) |
| OMB | 7 | IOMB1-7 | SWS | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (7) |
| GOT | 4 | IGOT1-4 | SWS | 4 | 1, 5, 11, 27 | 1.00 (0.18) | 2.17 (1.50) | 0.0031 (0.0009) | 2.00 | −1.74 | −0.07 | 2 | 1 (1); 2 (3) |
| SAV | 4 | ISAV1-4 | SWS | 2 | 1, 11 | 0.50 (0.26) | 0.50 (0.52) | 0.0007 (0.0004) | 0.75 | 0.17 | −0.61 | 2 | 1 (0); 2 (4) |
| MAR | 5 | IMAR1-5 | SWS | 2 | 1, 2 | 0.60 (0.17) | 0.60 (0.56) | 0.0009 (0.0002) | 0.90 | 0.63 | 1.23 | 2 | 1 (0); 2 (5) |
| ASA | 7 | IASA1-7 | SWS | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (7) |
| BNS | 9 | IBNS1-9 | SWS | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (9) |
| TON | 5 | ITON1-5 | SWS | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (5) |
| NOR | 8 | INOR1-8 | BSR | 5 | 1, 5, 11, 20, 36 | 0.86 (0.11) | 1.46 (0.99) | 0.0021 (0.0005) | 1.59 | −1.86* | −0.22 | 2 | 1 (0); 2 (8) |
| RUG | 7 | IRUG1-7 | BSR | 2 | 1, 11 | 0.57 (0.12) | 0.57 (0.52) | 0.0008 (0.0002) | 0.86 | 0.86 | 1.34 | 2 | 1 (0); 2 (7) |
| E2B | 3 | IE2B1-3 | BSR | 1 | 1 | 0.00 (0.00) | 0.00 (0.00) | 0.0000 (0.0000) | 0.00 | 0.00 | 0.00 | 2 | 1 (0); 2 (3) |
| LL | 10 | ILL1-10 | BSR | 6 | 1, 11, 20, 27, 33, 36 | 0.84 (0.10) | 1.84 (1.15) | 0.0026 (0.0004) | 1.58 | −2.05* | 0.18 | 2 | 1 (5); 2 (5) |
| JUR | 8 | IJUR1-8 | BSR | 5 | 1, 11, 20, 36, 37 | 0.86 (0.11) | 1.29 (0.90) | 0.0018 (0.0004) | 1.59 | −2.17* | −0.73 | 1 | 1 (5); 2 (3) |
| FEL | 7 | IFEL1-7 | DEN | 2 | 1, 10 | 0.29 (0.20) | 0.29 (0.34) | 0.0004 (0.0003) | 0.43 | −0.10 | −1.01 | 2 | 1 (0); 2 (7) |
| RUD | 7 | IRUD1-7 | DEN | 4 | 1, 10, 14, 20 | 0.71 (0.18) | 1.33 (0.94) | 0.0019 (0.0006) | 1.26 | −0.91 | −0.88 | 2 | 1 (1); 2 (6) |
| SLO | 3 | ISLO1-3 | CE | 3 | 1, 11, 13 | 1.00 (0.27) | 1.33 (1.10) | 0.0019 (0.0006) | 2.00 | −1.22 | 0.00 | 2 | 1 (0); 2 (3) |
| WIE | 5 | IWIE1-5 | CE | 3 | 11, 12, 18 | 0.80 (0.16) | 1.00 (0.80) | 0.0014 (0.0004) | 1.40 | −1.69 | 0.99 | 3 | 1 (0); 2 (5) |
| SOP | 9 | ISOP1-9 | CE | 5 | 1, 8, 11, 18, 20 | 0.86 (0.09) | 1.39 (0.94) | 0.0020 (0.0003) | 1.59 | −0.48 | 0.24 | 3 | 1 (2); 2 (7) |
| MEAN | 7 | 39 | 0.71 (0.02) | 1.75 (1.11) | 0.0019 (0.0001) | −0.51 | −0.26 |
- Codes indicate localities (see Fig. 1 and Table S1). Number of individuals per site (N); number of haplotypes [Nb (HT)]; haplotype diversity (Hd); mean number of pairwise differences (MNPD); nucleotide diversity (Pi); allelic richness with rarefaction to 3 (Ar); Fu's statistic (Fs); Tajima's statistic (D); samova cluster (SAM.); DAPC clusters (DAPC, with the number of individuals assigned to one DAPC cluster with p > 0.8). Asterisks indicate significance: *p < 0.05 and **p < 0.01.
Neutrality departure tests
Finally, to detect signatures of possible demographic expansion events in the sampled populations' evolutionary history, we calculated two statistics: Fu's FS (Fu 1997) and Tajima's D (Tajima 1989) using simulations in Arlequin and DnaSP 5.0, respectively. These statistics indicate whether populations are in mutation-drift equilibrium (Wright–Fisher model) or whether there are signs of processes that distort the pattern (Ramos-Onsins and Rozas 2002). In the absence of selection, negative values of these statistics indicate that the number of alleles exceeds random expectations, potentially due to population expansion. Fu's statistic reportedly detects population expansion more sensitively than Tajima's (Fu 1997).
Results
Genetic diversity
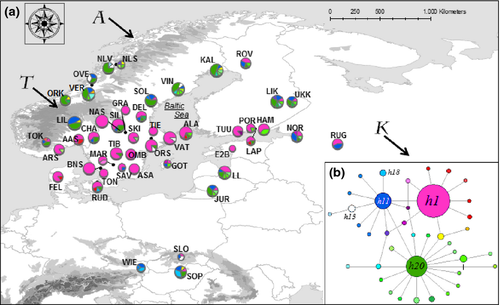
The final alignment consisted of sequences of the targeted 698-bp fragment of the mtDNA COI gene obtained from 342 individuals. In total, 33 polymorphic nucleotides and 39 haplotypes were identified (Figs 1 and 2). The identified haplotypes are available in GenBank database (Accession numbers JX845179- JX845217).
Three major haplotypes were found, designated h1, h20 and h11. These were detected in 160 (47%), 63 (18%) and 39 (11%) of the 342 analysed individuals, respectively. The other haplotypes mostly consisted of single mutation variants of these three major haplotypes (Fig. 1). As only 448 bp overlapped with sequences published by Bertheau et al. (2011), we could not fully recover all of the haplotypes. However, the three most abundant haplotypes (h1, h11 and h20) overlapped with the most common haplotype detected by Bertheau et al. (2011), HTI, and two others (h13 and h18) overlapped with the other two major haplotypes they reported (It1 and HTII). Not surprisingly, as haplotype HTII was only found in southern European populations in large-scale studies (Stauffer et al. 1999; Bertheau et al. 2013), we detected it in Central European populations but not in any of the Scandinavian populations (Fig. 1).
Analysis of all aligned sequences yielded haplotype diversity (Hd) and nucleotide diversity (Pi) values of 0.71 (SD = 0.02) and 0.002 (SD = 0.000), respectively (Table 1). Both of these variables differed among geographical regions according to a Kruskal–Wallis anova by ranks test (H = 14.48, p = 0.05 and H = 13.24, p = 0.04, respectively). However, no significant differences were detected in either of them in pairwise comparisons of the regions' populations (data not shown).
As shown in Table 1, allelic richness (Ar) was particularly low in populations of the South Swedish mainland (ASA, BNS, TON, OMB, GRA, NAS and TIE) and highest in three populations of the Baltic States (LL, 1.58; JUR, 1.59 and NOR, 1.59), two populations of southern Finland (LIK, 1.74 and UKK, 1.71), two populations of the Norwegian part of the Scandes Mountains (NLS, 2.00 and TOK, 1.70), the Gotland island population (GOT, 2.00), one population of Central Sweden (VIN, 1.63) and the three populations of central Europe (SLO, 2.00; WIE, 1.40; SOP, 1.59).
Genealogical relations
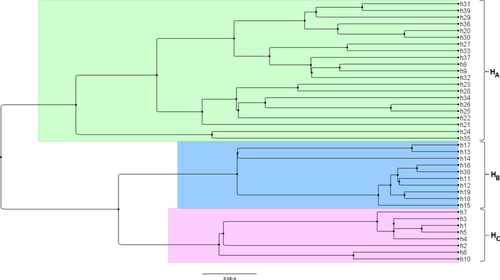
The phylogenetic tree reconstructed by beast revealed three clades (designated HA, HB and HC) corresponding to the three main haplogroups (Fig. 2). When plotted on a map, the haplotype frequencies revealed a partition between northern and southern Scandinavia, with HC and HA haplogroups mainly present in the southern and northern parts, respectively (Figs 1 and 2).
Genetic and geographic structures
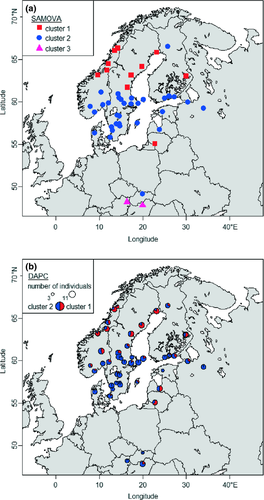
The spatial analysis of molecular variation (samova) provided highest support for three genetic clusters, designated 1, 2 and 3 (Table 1). Cluster 1 included all northern samples, except those from the Finnish (ROV) and Lithuanian (JUR) locations. Cluster 2 included all samples from Denmark, southern Norway, Sweden, Finland, the Baltic States, Slovakia and Russia. Cluster 3 included the remaining two populations from central Europe (WIE and SOP). It also indicated that 42% of the total variation was distributed among clusters, just 3% among populations within clusters and most (55%) within populations (Table 2). For a K-value of 3, FCT was 0.42 (p < 0.001), FST was 0.45 (p < 0.001) and FSC was 0.05 (p < 0.001). The global FST (with no grouping of samples) was 0.35 (p < 0.001), and FIS was 0.10 (p < 0.001).
| Source of variation | df | Sum of squares | Variance components | % of variation | F-stat |
|---|---|---|---|---|---|
| Among clusters | 2 | 58.77 | 0.36 Va | 42.23 | FCT = 0.42 |
| Among populations within clusters | 44 | 28.78 | 0.03 Vb | 3.09 | FSC = 0.05 |
| Within populations | 295 | 136.90 | 0.46 Vc | 54.67 | FST = 0.45 |
| Total | 341 | 224.45 | 0.85 |
The DAPC analysis favoured a structure with two genetic clusters. The BIC curve decreased continuously between K = 2 and K = 7, but values over K = 2 led to additional clusters with a single individual, and thus, there was no support for a structure with more than two clusters. DAPC with K = 2 gave very similar partitioning to the samova with K = 3 (Fig. 3), except that the latter grouped two central European samples into an additional cluster.
When considering groups identified by the samova analysis as populations, phylogeographic structure was detected with a (slightly) significant difference (p = 0.02) between Gst (0.215) and Nst (0.290) statistics.
Pairwise fixation indices between populations are shown in Figure S1. Within geographical regions, differentiations were mostly low to moderate (70% of pairwise comparisons within a region gave FST values <0.15). Among geographical regions, those from northern Europe were generally well differentiated from those of Central Europe (CE) (70% of pairs >0.15), but few pairwise FST values exceeded 0.5 (indicating <15% differentiation). However, populations from south-western Scandinavia were clearly differentiated from those of neighbouring northern regions, as 90% of the pairwise calculated FST values between SWS and NWS or NES populations exceeded 0.5.
Demographic expansion
Fu's test of analysis of departure from neutrality detected signs of expansion in seven populations, all from geographical groups in the eastern part (Baltic States and Russia, BSR; south-eastern and north-eastern Scandinavia, SES and NES) of the study region (Table 1). However, Tajima's D statistics were negative for populations in only two of these locations (KAL and VIN, in northern Sweden).
Discussion
Our fine-scale survey of mitochondrial haplotypes of northern European Ips typographus populations provides new insights into the postglacial history of this forest pest. We identified not only genetic structure but also phylogeographic structure between the northern and southern parts of Scandinavia. We also detected high genetic diversity in northern and eastern populations, but low genetic diversity in southern populations of Scandinavia.
These phylogeographic patterns suggest that the current maternal lineages of Scandinavia have at least two different geographic origins. Neither the phylogeographic structure nor the genetic diversity distributions are consistent with northward expansion via a single re-colonization route from an entry point in southern Scandinavia. Thus, we rejected scenario 1, postulating a single postglacial re-colonization route to Scandinavia from the south (Stauffer et al. 1999; Bertheau et al. 2013).
The observed patterns are also inconsistent with scenario 3, which incorporates prolonged isolation of I. typographus populations in one of the cryptic north-western refugia of its host plant (e.g. Andøya and Trøndelag; Fig. 1) (Kullman 2008; Parducci et al. 2012). This is because the divergence observed between the northern and southern population pools is quite shallow, and all haplotypes differed only by one or two mutational steps. Furthermore, we detected no signs of population expansion in populations of the north-western coast as none of the Tajima and Fu's statistics were significantly negative for this area.
Therefore, the observed phylogeographic patterns are more consistent with scenario 2, which postulates postglacial re-colonization of inland Scandinavia via a southern and a northern route. One of these routes is similar to the southern re-colonization route suggested by Stauffer et al. (1999) for I. typographus, which was also apparently used by its host plant P. abies (Giesecke and Bennett 2004; Tollefsrud et al. 2009) and other species, for example, the bush cricket (Kaňuch et al. 2013). However, in contrast to its host plant, the genetic diversity of I. typographus observed in southern Scandinavia is particularly low, suggesting that few dispersal events across the Baltic Sea were successful, at least for matrilineal lineages. Considering the geographic distribution of the HA haplotypes and the cluster assignments (both of which identified a second genetic pool along the Gulf of Finland and in the Scandes Mountains), the second re-colonization route could correspond to a postulated expansion of its host plant from a north-eastern refugium, probably located in the Russian plains (Kostroma, Fig. 1) (Lagercrantz and Ryman 1990; Tollefsrud et al. 2008). Furthermore, samples from sites near the border between southern Finland and southern Russian Karelia, which are among the closest to Kostroma, yielded some of the highest diversity indices and significantly negative Tajima and Fu's statistics, indicative of an expansion event.
Particularly, high diversity was also observed at some locations in central Sweden and southern mid-Norway (e.g. TOK and DEL sites), probably because they were in a zone of secondary contact between the southern and northern gene pools. Further, we detected relatively weak differentiation between neighbouring populations but relatively high mean numbers of pairwise differences in this region, which may be indicative of a zone where populations from different isolated refugia mixed (Petit et al. 2003).
The putative northern re-colonization route has not been detected by large-scale molecular surveys (Stauffer et al. 1999; Sallé et al. 2007; Bertheau et al. 2013), probably because they did not include high latitude sampling sites in northern Scandinavia. Our use of a relatively large gene fragment, 140 bp longer than mitochondrial fragments sequenced in the large-scale surveys (Stauffer et al. 1999; Bertheau et al. 2013), may also have contributed to the fine resolution of genetic variation we obtained.
In conclusion, the findings of this study are consistent with the previous conclusion that fine-scale analysis of mitochondrial markers is valuable for high-resolution exploration of the population history of highly dispersive species such as I. typographus (Krascsenitsová et al. 2013). Our application of this approach to Scandinavian populations provided the first indications that the region was re-colonized after the last glaciation via two pathways: from the north-east to the north-west through Finland and across the Baltic Sea to southern Scandinavia. The two routes explain the current partitioning between southern and northern populations of I. typographus, and may be related to observed differences in behaviour between them. They also provide potentially useful information for improving pest management. For instance, removing attacked trees should be more efficient for managing populations dominated by the southern gene pool as they seem to hibernate under spruce bark more often than their northern counterparts (Komonen et al. 2011). In addition, future models for predicting effects of climate change on the species' population dynamics may benefit by applying specific scenarios to the populations from the two gene pools. However, it has been suggested that the weaker dispersal of females than males of the species could have caused discrepancies between large-scale mitochondrial and nuclear patterns (Sallé et al. 2007). Therefore, further investigations should utilize already-identified microsatellite markers (Sallé et al. 2007; Stoeckle and Kuehn 2011) to assess, at fine-scale, if the phylogeographic patterns observed in this study are representative of the whole species' population history or are limited to matrilineal lineages.
Acknowledgements
This study was funded by a special grant from SLU for bark beetle research and would not have been possible without assistance of the persons listed in Supplementary Table 1, who contributed samples. We are grateful to Celia K. Boone and three anonymous reviewers for valuable comments on this study. F. Mayer was supported by a doctoral grant from the Belgian Fonds pour la Formation a la Recherche dans l'Industrie et l'Agriculture (FRIA) and a Van Buuren award from the ULB.




