Phylogeography of the mediterranean green seaweed Halimeda tuna (Ulvophyceae, Chlorophyta)
Abstract
Populations of many Mediterranean marine species show a strong phylogeographic structure, but the knowledge available for native seaweeds is limited. We investigated the genetic diversity of the green alga Halimeda tuna based on two plastid markers (tufA gene and a newly developed amplicon spanning five ribosomal protein genes and intergenic spacers, the rpl2-rpl14 region). The tufA sequences showed that Mediterranean H. tuna represents a single, well-defined species. The rpl2-rpl14 results highlighted a genetic separation between western and eastern Mediterranean populations; specimens collected from widely scattered locations in the Adriatic/Ionian region shared a haplotype unique to this region, and formed a group separated from all western Mediterranean regions. Specimens from Sardinia also formed a unique haplotype. Within the western Mediterranean basin, a gradual shift in the frequency of haplotypes was apparent along a West-East gradient. Our results represent the first clear evidence of an East-West genetic cleavage in a native Mediterranean macroalga and offer an interesting perspective for further research into fine-scale seaweed population structure in the NW Mediterranean Sea.
Abbreviations
-
- MSC
-
- Messinian Salinity Crisis
-
- tufA
-
- elongation factor TU
-
- rpl
-
- ribosomal proteins L
Phylogeography yields insights into the evolutionary history of a species by revealing vicariance events, migration pathways and population expansion or bottleneck (Avise 2000). The effects of these processes on the population structure of benthic marine algae have been increasingly investigated in the last 20 years. Detailed phylogeographic studies are now available for species from many geographical areas, but little information is available for native Mediterranean seaweeds, despite the great macroalgal diversity recorded there (approximately 1,200 species) and the importance of some species as key ecosystem engineers (Pezzolesi et al. 2017, Bermejo et al. 2018).
Most of the phylogeographic work carried out on Mediterranean macroalgae has focused on introduced species, mainly Asparagopsis and Caulerpa (e.g., Famà et al. 2000, Andreakis et al. 2007). Studies on native species were essentially nonexistent until a few years ago (Hu et al. 2016), and while a few studies, usually limited to some parts of the basin, have appeared in recent years (Varela-Álvarez et al. 2015, Buonomo et al. 2017, Pezzolesi et al. 2017, Bermejo et al. 2018, De Jode et al. 2019, Vitales et al. 2019), we still lack clear insight into large-scale population structure and its causes. This lack of knowledge is striking, especially if compared with the detailed phylogeographic insights available for Mediterranean fishes and invertebrates.
The ulvophycean genus Halimeda is globally distributed, occurring from the intertidal to ca. 150 m depth in temperate and tropical seas (Kooistra et al. 2002, Verbruggen et al. 2009). This makes it an ideal model to investigate genus-level biogeographic history associated with paleobiogeographical and macroecological processes, and to characterize evolutionary niche dynamics in the ocean (Hu et al. 2016). For this reason, it has been the subject of some of the most comprehensive evolutionary studies in the field (Kooistra et al. 2002, Verbruggen et al. 2005, 2009). In the Mediterranean, Halimeda is represented by the native Halimeda tuna (Fig. 1A), widespread throughout the basin (besides the recently introduced Halimeda incrassata, Alós et al. 2016). The species is nearly ubiquitous in Mediterranean coastal habitats and important as a producer of carbonate sediments. Therefore, it represents a good model to investigate phylogeographic patterns and processes in Mediterranean seaweeds.
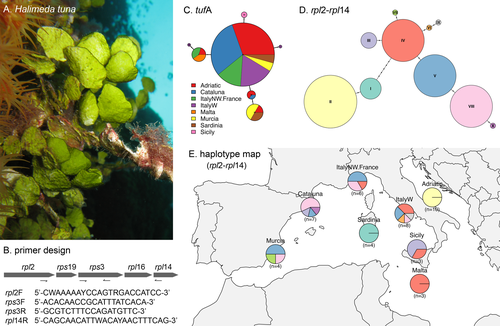
For this study, we obtained 47 samples of Halimeda tuna from 13 locations situated in 8 regions of the western and central Mediterranean Sea (Murcia, Cataluña, northwestern Italy/France, western Italy, Sardinia, Sicily, Malta, Adriatic/Ionian; Table S1 in the Supporting Information). We investigated the genetic diversity of the species in these regions using sequences of two plastid markers, the tufA gene and a newly developed amplicon spanning five ribosomal protein genes and intergenic spacers (rpl2-rpl14 region; Fig. 1B). DNA extraction and amplification of tufA followed Cremen et al. (2016). For the rpl2-rpl14 region, PCR reactions used the primers in Figure 1B and a profile of an initial denaturation (94°C for 3 min), 45 cycles of denaturation (94°C for 1 min), primer annealing (52°C for 1 min), and elongation (72°C for 2 min), and a final elongation (72°C for 4 min). PCR products were purified and sequenced by Macrogen (Seoul, South Korea). The 40 tufA sequences and 44 rpl2-rpl14 sequences generated were manually assembled and aligned in Geneious Prime 2019.1.3 (http://www.geneious.com). The complete data set includes the 84 newly assembled sequences and two sequences previously published by Verbruggen et al. (2005; Table S1). The tufA and rpl2-rpl14 data sets consisted of alignments 822 bp and 1,499 bp long respectively. Haplotype networks were constructed using the haploNet function in pegas v.0.11 (Paradis 2010) in R v.3.6.1 (R Core Team 2019). The rpl2-rpl14 data set contained several simple sequence repeats (SSR) that were re-coded to have each repeat coded as being either present or absent. Sequence alignments and a full knitr log of R analyses are available through FigShare (https://doi.org/10.6084/m9.figshare.11894631.v1).
Mediterranean Halimeda tuna showed very little variation in the tufA gene, with seven haplotypes recorded and uncorrected pairwise divergences not exceeding 0.36%. Haplotype II was dominant and geographically widespread, occurring in six of the eight regions (Fig. 1C). Three haplotypes (V, VI, and VII), found in two different specimens from western Italy and in two specimens from Sicily, were restricted to a single region (Fig. 1C). The rpl2-rpl14 region was considerably more variable and geographically structured than tufA. Out of ten haplotypes recorded, six were restricted to a single region (Fig. 1, D and E). The Adriatic/Ionian specimens shared a haplotype (II) unique to this region and formed a group separated from all other regions (Fig. 1, D and E). The four specimens from Sardinia also formed a distinct group, characterized by a haplotype (I) exclusive of this region (Fig. 1, D and E). The other haplotypes were shared among several regions but featured a West-East gradient, with some of them (V, VII, VIII) occurring mainly in Murcia, Cataluña and northwestern Italy/France, and others (III, IV, VI, IX) being more frequent in western Italy, Sicily, and Malta (Fig. 1, D and E).
These results provide insights into the genetic diversity of Mediterranean Halimeda tuna at two different levels. The low variation of tufA confirms that in the Mediterranean this alga represents a single, well-defined species; our data do not provide any evidence for the possible existence of cryptic species. It should be noted that beyond our Mediterranean context, the name H. tuna has also been applied in the Caribbean and the Indo-Pacific, but molecular work has shown that these are clearly different species unrelated to the Mediterranean lineage of H. tuna studied here (Verbruggen et al. 2005). The conclusion that within the Mediterranean Sea H. tuna forms a single species should not be considered obvious or banal. Cryptic diversity is a widespread phenomenon in marine organisms including many unrelated seaweeds (Verbruggen 2014), and the complex hydrogeological history of the Mediterranean Sea has promoted cryptic speciation in many taxa. The overall geographical configuration of the Mediterranean Sea came about through the closing of the Tethyan Seaway in the Middle Miocene (13 Mya), but several dramatic changes took place in the last 6 My. The closing of the Strait of Gibraltar caused the extensive drying referred to as Messinian Salinity Crisis (MSC, 5.9-5.3 Mya), which subdivided the Mediterranean into a number of separate hypersaline basins. The hydrogeographic changes that took place in this period and during the Pleistocene glaciations have stimulated speciation, leading to the existence of many (often cryptic) species in several groups of marine animals (e.g., blennies, Carreras-Carbonell et al. 2005; sponges, Blanquer and Uriz 2007; ophiuroids, Boissin et al. 2011; mysid crustaceans, Rastorgueff et al. 2014; gastropod molluscs, Calvo et al. 2015). In seaweeds, two recent studies showed that the coralline alga Lithophyllum stictiforme represents a complex of at least 13 different species (Pezzolesi et al. 2019), eight of which co-exist along French shores (De Jode et al. 2019). The absence of cryptic species in Halimeda suggests that the genus may have gone extinct in the Mediterranean during the MSC and recolonized it from adjacent Atlantic or Indo-Pacific areas (a scenario suggested for another common Mediterranean chlorophyte, Caulerpa prolifera, by Varela-Álvarez et al. (2015)) or, alternatively, might have survived in a single refugium.
The intermittent lowering of sea level and consequent coastline modifications that took place in the course of the subsequent Pleistocene glaciations (Calvo et al. 2015) further contributed to maintain and enhance genetic diversity in many Mediterranean marine species, and it appears that this was the case in Mediterranean Halimeda tuna as well. Our rpl2-rpl14 analyses indicate a genetic separation between western and eastern Mediterranean populations. The specimens from the Adriatic/Ionian region (the eastern Mediterranean samples included in this study) shared the same rpl2-rpl14 haplotype, which was not found in any western Mediterranean specimen; in the haplotype networks this separated the Adriatic/Ionian population from the western Mediterranean populations. The widespread distribution of our sampling locations in the Adriatic/Ionian region implies that this is a robust separation, which agrees with information available for many animal species (e.g., fishes, Bahri-Sfar et al. 2000; limpets, Sá-Pinto et al. 2012; bivalve molluscs, Nikula and Vainola 2003, Cordero et al. 2014; gastropod molluscs, Villamor et al. 2014). The West-East transition represents one of the major biogeographic discontinuities in the Mediterranean Sea (Serra et al. 2010), but evidence for its existence in non-animal species is much more limited. This genetic separation is well-documented in the paleoendemic Mediterranean angiosperm Posidonia oceanica (Arnaud-Haond et al. 2007, Serra et al. 2010), but evidence in macroalgae is weak and based on studies restricted to parts of the basin (e.g., Buonomo et al. 2017). Our results represent the first convincing evidence of an East-West genetic cleavage in a native Mediterranean macroalga. This break represents an imprint of vicariance during the last Pleistocene ice age that is maintained to this day by low gene flow (Arnaud-Haond et al. 2007). During the Last Glacial Maximum (~10,000 ya), the western and eastern Mediterranean were connected only by a narrow channel between Sicily and Tunisia in North Africa. After the sea level started to rise, the Strait of Sicily became larger and deeper, and a new connection opened, the very narrow Strait of Messina. Although resulting from past vicariance events, the genetic separation between western and eastern Mediterranean populations is believed to be presently maintained by low levels of gene flow, which are typical of species with low dispersal (Serra et al. 2010). Notably for Halimeda, this agrees with the general notion that species of this genus have limited dispersal, a scenario drawn by previous biogeographic studies (e.g., Verbruggen et al. 2009). Our results also suggest that in H. tuna the biogeographical boundary between western and eastern Mediterranean is concordant with that recognized for P. oceanica. The precise placement of the West-East boundary has been debated in the literature, with the eastern coast of Sicily and Calabria belonging either to the western or to the eastern basin (Serra et al. 2010 and references therein). The specimens from Malta sequenced in this study shared a haplotype (IV) that occurred also in three western Mediterranean regions (northwestern Italy/France, western Italy, Sicily) but not in the Adriatic/Ionian samples, suggesting that Malta populations genetically belong to the western Mediterranean. For P. oceanica, the results of Serra et al. (2010) established the southeastern tip of Calabria (the southernmost region of continental Italy) as boundary of the transition, thus including the whole of Sicily and Malta in the western Mediterranean. Our data suggest that a similar situation probably applies to H. tuna, although finer-scale sampling in this area would be needed for confirmation.
The genetic individuality of the specimens from Sardinia, all sharing the rpl2-rpl14 haplotype I not found in any other region, is a surprising finding that does not match phylogeographic patterns for other marine species studied to date, as to our knowledge none have shown evident genetic discontinuity between Sardinian populations and populations from nearby western Mediterranean regions. We did only sequence four specimens from Tavolara and Molara, two small islands located on the northeast coast of Sardinia, and expect that extending the sampling to other portions of the Sardinian shoreline would reveal additional haplotypes present in other western Mediterranean regions.
The apparent frequency shift in the distribution of rpl2-rpl14 haplotypes in the western Mediterranean is an intriguing finding. Some biogeographical sectors are recognized along the coasts of the northwestern Mediterranean (Alboran Sea, Balearic Sea, Gulf of Lion and Ligurian Sea). These discontinuities are generally considered to be less important from a phylogeographic point of view, although small-scale genetic differences among localities within these sectors have been highlighted (e.g., in Lithophyllum stictiforme; De Jode et al. 2019). Our data for Halimeda appear to support this trend with a gradient in haplotype frequencies along the East-West axis, offering an interesting perspective for further research into fine-scale population structure in the NW Mediterranean Sea.
Part of the samples were collected in the course of a P.R.I.N. 2010–2011 project (Coastal bioconstructions: structure, function and management), for which FR acknowledges financial support from the Italian Ministry for Education, Universities and Research. We are very grateful to Fabio Badalamenti, Claudio Battelli, Fabio Bulleri, Carlo Cerrano, Olivier De Clerck, Edward Drew, Laura Pezzolesi and Massimo Ponti for collecting and providing specimens of Halimeda tuna.




