SCPLPA: An miRNA–disease association prediction model based on spatial consistency projection and label propagation algorithm
Min Chen and Yingwei Deng contributed equally to this work.
Abstract
Identifying the association between miRNA and diseases is helpful for disease prevention, diagnosis and treatment. It is of great significance to use computational methods to predict potential human miRNA disease associations. Considering the shortcomings of existing computational methods, such as low prediction accuracy and weak generalization, we propose a new method called SCPLPA to predict miRNA–disease associations. First, a heterogeneous disease similarity network was constructed using the disease semantic similarity network and the disease Gaussian interaction spectrum kernel similarity network, while a heterogeneous miRNA similarity network was constructed using the miRNA functional similarity network and the miRNA Gaussian interaction spectrum kernel similarity network. Then, the estimated miRNA–disease association scores were evaluated by integrating the outcomes obtained by implementing label propagation algorithms in the heterogeneous disease similarity network and the heterogeneous miRNA similarity network. Finally, the spatial consistency projection algorithm of the network was used to extract miRNA disease association features to predict unverified associations between miRNA and diseases. SCPLPA was compared with four classical methods (MDHGI, NSEMDA, RFMDA and SNMFMDA), and the results of multiple evaluation metrics showed that SCPLPA exhibited the most outstanding predictive performance. Case studies have shown that SCPLPA can effectively identify miRNAs associated with colon neoplasms and kidney neoplasms. In summary, our proposed SCPLPA algorithm is easy to implement and can effectively predict miRNA disease associations, making it a reliable auxiliary tool for biomedical research.
1 INTRODUCTION
MicroRNAs (miRNAs), with a length of approximately 20–25 nucleotides, are a class of non-coding RNAs that do not participate in protein coding,1-3 tissue differentiation,4 cell proliferation2, 3 and cell apoptosis.4-7 However, miR-30a-5p, miR-30d-5p and miR-30c-5p are known to contribute to atherosclerosis and ischemic events, which are related to the development of type 2 diabetes.8 Currently, the understanding of miRNAs is still in its infancy, and the known functions of miRNAs represent only a small fraction. Therefore, identifying miRNAs associated with diseases will help understand the regulatory mechanisms of miRNAs and the mechanisms underlying diseases or tumour development. This work has great significance for human disease prevention and treatment.
In the wake of the discovery of a large number of miRNAs, various databases have been developed to store relevant information about miRNAs. An increasing number of bioinformatics computational methods have been developed to predict associations between miRNAs and diseases and provide assistance for further biological experimental validation. Existing prediction methods can be divided into network, machine learning and matrix factorization-based methods.
Network-based methods mainly aim to construct relationship networks between miRNA and diseases, proteins, environmental factors, etc. Starting from the general hypothesis in biology that ‘functionally similar miRNAs are more likely to be associated with phenotypically similar diseases, and vice versa’, corresponding algorithms are designed based on the topological structure of a relationship network. In 2009, Jiang et al.9 first proposed a computational model based on hypergeometric distribution to predict miRNA–disease associations. They used the relationships between miRNA-regulated target genes to construct an miRNA functional similarity network. Xuan et al.10 and Chen et al.11 predicted unknown miRNA–disease associations by using the K-nearest neighbour algorithm, but the accuracy of these algorithms needs to be improved. Considering that global network similarity can improve prediction accuracy more effectively than local network similarity, Chen et al.12 proposed a method called NetCBI, which uses network consistency to predict associations between miRNAs and diseases. Chen et al. also proposed a series of miRNA–disease association methods13-15 by calculating graph Laplacian scores to obtain network consistency similarity. In 2012, Chen et al.16 proposed a random walk-based association prediction model called RWRMDA, which is simple to implement but cannot predict isolated diseases or new miRNAs without any known associations. Several random walk algorithms, such as MIDP,17 NDBM,18 Mugunga's method,19 GSTRW20 and NPRWR,21 have also been developed and achieved good prediction results. Zhan et al. proposed a model called NDALMA22 based on network distance analysis for predicting lncRNA–miRNA associations, and achieved good predictive performance. However, these algorithms heavily rely on known miRNA–disease (lncRNA–miRNA) associations.
Machine learning-based methods mainly aim to use classification algorithms, such as support vector machines, decision trees, random forests and naive Bayes classifiers, especially popular deep learning methods23 for lncRNA–disease association and miRNA–disease association prediction. For example, Jiang et al.24 and Xu et al.25 achieved good results in using support vector machines for prediction, but the prediction performance of these models is limited by the classifiers used, such as support vector machines and decision trees. Deep learning has also been applied to this field. Zhang et al.26 Ji et al.27 Sujamol et al.28 and Peng et al.29 applied deep autoencoders to predict miRNA–disease associations. Tang et al.30 Dong et al.31 Xuan et al.32 Sun et al.33 and Wang et al.34 respectively applied multi-layer convolutional neural networks for predicting miRNA–disease, metabolite–disease and lncRNA–miRNA associations. Additionally, the graph attention mechanism35, 36 has also been used in the association prediction field. These algorithms have been applied and achieved certain results in this field. However, these models still require positive and negative samples during training and have not solved the problem of selecting negative samples.
Matrix factorization-based methods have also attracted researchers' attention. In 2017, Li et al.37 used matrix completion algorithms to construct an MCMDA model for prediction of miRNA–disease associations. Chen et al. improved MCMDA and developed models such as IMCMDA38 and NCMCMDA.39 Many researchers have combined matrix factorization algorithms with other methods for prediction; in particular, the NIMCGCN40 model combines matrix completion algorithms with graph convolutional networks, the NIMGSA41 model combines graph autoencoders with self-attention mechanisms and the MDA-AENMF42 model combines a five-layer autoencoder. These models can solve the sparsity problem of heterogeneous biological data networks, but they have not effectively addressed the parameter selection problem. Additionally, many scholars have conducted extensive research in related fields,43-48 which is also of reference value.
In summary, existing prediction models can be used to predict miRNA–disease associations but still have shortcomings, such as complex algorithm design, high computational complexity and difficulty in parameter selection. Further research is thus needed in predicting miRNA–disease associations. In the present work, a novel method, namely, SCPLPA, is proposed for prediction of miRNA–disease associations and was developed starting from the perspective of the structure of heterogeneous graphs and the heterogeneity of content.
This study constructs a heterogeneous disease similarity network composed of a disease semantic similarity network and a disease Gaussian interaction spectrum kernel similarity network as well as a heterogeneous miRNA similarity network composed of an miRNA functional similarity network and an miRNA Gaussian interaction spectrum kernel similarity network. The label propagation algorithm is then implemented in both heterogeneous networks, and their results are integrated as the initial prediction scores for miRNA–disease associations. The matrices of the heterogeneous disease similarity network and the heterogeneous miRNA similarity network are projected into the initial prediction score matrix. The two spatial projection scores are integrated as the final prediction score. As a result, multiple evaluation metrics, including AUC, AUPR, ACC, MCC and F1, indicate that SCPLPA outperforms other state-of-the-art methods in terms of predictive performance. In addition, SCPLPA can predict the relationships between isolated diseases and new miRNAs. The AUC values for predicting isolated diseases and new miRNAs are 0.8412 and 0.8289, respectively. Two case results further validate the ability of SCPLPA to predict unknown miRNA associations related to diseases.
2 MATERIALS AND METHODS
2.1 Human miRNA–Disease association data
The experimentally validated miRNA–disease association data are from HMDD v2.049 . If there is a known association between a miRNA and a disease node , it is set to 1; otherwise, it is set to 0. The variables nm and nd represent the number of diseases and miRNAs, respectively.
2.2 Disease semantic similarity
Many scholars have proposed methods to measure the semantic similarity of diseases based on disease classification information described in MeSH (Medical Subject Headings).50 In this method, each disease is represented as a directed acyclic graph (DAG) , where represents the ancestor node set of disease (including the disease itself) and represents the set of related connections. The similarity between diseases is calculated as follows:
The data are downloaded from the literature51 and named as .
2.3 miRNA functional similarity
The functional similarity between diseases is calculated based on the semantic similarity of diseases. The specific process is described as follows.52
represents the semantic similarity value between disease and disease .
In the above equation, m and n refer to the number of diseases associated with miRNA and miRNA , respectively.
The matrix is used to represent the miRNA functional similarity matrix.
2.4 Gaussian interaction spectral kernel similarity
2.5 Integration of disease similarity and miRNA similarity
This heterogeneous disease similarity network is represented by the matrix .
This heterogeneous miRNA similarity network is represented by the matrix .
2.6 SCPLPA
The algorithm consists of three steps. The first step involves constructing accurate disease similarity networks and miRNA similarity networks by using heterogeneous data sources (Equations 6-11). The second step involves using the label propagation algorithm to obtain estimated scores for miRNA–disease associations. The third step involves using the spatial consistency projection algorithm to obtain precise scores for miRNA–disease associations. The flowchart is shown in Figure 1.
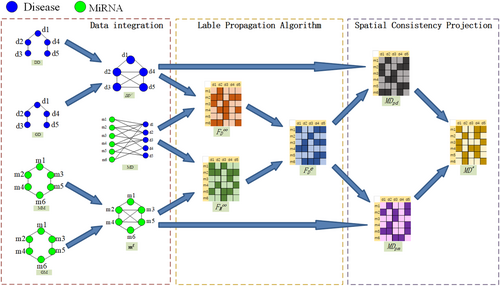
2.6.1 Estimated scores for miRNA–Disease associations
The label propagation algorithm is applied separately to the heterogeneous disease similarity network and the heterogeneous miRNA similarity network to obtain initial scores for miRNA–disease associations. These initial scores are combined to obtain the estimated scores.
is iterated until , and the iteration is then stopped. The predicted result is the initial score for miRNA–disease associations based on the heterogeneous disease similarity network, represented by the matrix .
is iterated until , and the iteration is then terminated. The probability space reaches a stable state and is denoted as . This value is the initial score for miRNA–disease associations based on the heterogeneous miRNA similarity network.
2.6.2 Accurate scores for miRNA–Disease Associations
In the above formula, is the 2-norm of .
3 RESULTS
3.1 Evaluation metrics
We evaluated the performance of SCPLPA using LOOCV (leave-one-out cross-validation), where each miRNA–disease association was selected as a test sample object once, with all other miRNA–disease associations used as the training set until all miRNA–disease associations were tested once. By setting different thresholds and plotting the ROC (receiver operating characteristic) curve with TPR (true positive rate or sensitivity) as the y-axis and FPR (false positive rate or 1—Specificity) as the x-axis, the AUC (area under the ROC curve) was calculated. The curve plotted with recall rate on the x-axis and precision on the y-axis is known as the PR (precision-recall) curve. The area under the PR curve is referred to as the AUPR (area under the PR curve) value.
The TP (true positive) in the above formulas refers to the number of correctly predicted positive samples, that is the number of positive samples predicted as positive. FP (false positive) refers to the number of incorrectly predicted positive samples, that is the number of negative samples predicted as positive. TN (true negative) refers to the number of correctly predicted negative samples, that is the number of negative samples predicted as negative. FN (false negative) refers to the number of incorrectly predicted negative samples, that is the number of positive samples predicted as negative.
3.2 Effect of parameter selection
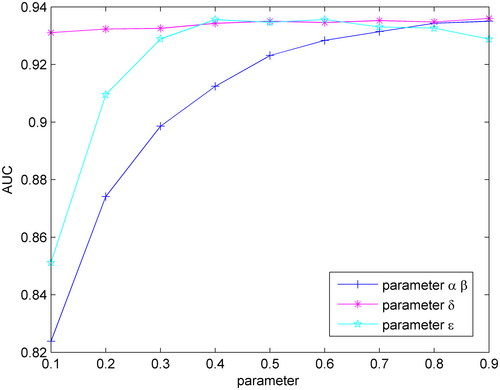
In the Equations 12 and 14, and represent the probabilities of receiving initial label information in the label propagation algorithm for miRNA–disease associations, while and control the rate at which information from neighbours is retained. For simplicity, and are set to be the same size. The estimated score for miRNA–disease associations is calculated by weighting the prediction results and from the heterogeneous miRNA network and the heterogeneous disease network by using the label propagation algorithm, with representing the proportion of the two prediction results. The precision score for miRNA–disease associations is calculated by weighting the prediction scores based on miRNA spatial consistency projection and disease network spatial consistency projection, with representing the proportion of the two prediction results. This section mainly discusses the effect of these parameters on the predictive performance of SCPLPA.
In the first step, the optimal values for and are determined. Here, parameters and are initially set to 0.5, with a step size of 0.1. Parameters (or ) are increased from 0.1 to 0.9 with a step size of 0.1, and leave-one-out cross-validation is performed to calculate AUC (Figure 2). When is set to 0.9, the AUC value is maximized at 0.9335. Therefore, parameters and are set to 0.9. The optimal value for is then determined. Based on the obtained values of = = 0.9, the parameter is set to 0.5 and then the parameter is increased to 0.9 with a step size of 0.1. The cross-validation is performed again to calculate the AUC values. When is 0.6, the AUC is maximized at 0.9346 (Figure 2). Therefore, let = 0.9. Finally, in the case of = = 0.9 and = 0.9, the parameter is increased from 0.1 to 0.9 with a step size of 0.1. When is 0.6, the AUC value is maximized at 0.9356. Thus, the following optimal parameter values are obtained: = = 0.9, = 0.9, = 0.6.
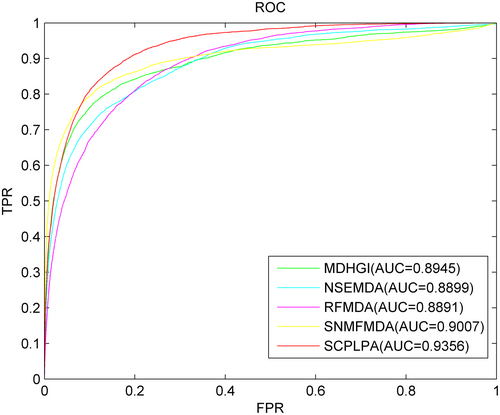
3.3 Comparison with state-of-the-art methods
To the best of our knowledge, MDHGI,54 NSEMDA,55 RFMDA56 and SNMFMDA57 are excellent computational methods used to predict miRNA–disease associations. These methods utilize information similar to SCPLPA and can be used for predicting associations between isolated diseases and new miRNAs. Here, SCPLPA is compared with these methods through the parameter selection described in their respective papers. The AUC value is used as the performance metric to evaluate the prediction performance. LOOCV is performed to compare the prediction results (Figure 3). The AUC values for SCPLPA, MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.9356, 0.8945, 0.8899, 0.8891 and 0.9007, respectively. To enhance the persuasiveness of our experiments, we compared SCPLPA with several other models based on AUPR, ACC, MCC and F1 values. As shown in Table 1, the AUPR value of SCPLPA is 0.4596, while MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.3367, 0.3198, 0.3345 and 0.3489, respectively. SCPLPA is, respectively, higher than the other control methods by 26.74%, 30.42%, 27.22% and 24.09%. The ACC values of SCPLPA, MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.5503, 0.5607, 0.5321, 0.5215 and 0.5317, respectively. SCPLPA is 1.89% lower than that of MDHGI, but respectively higher than NSEMDA, RFMDA and SNMFMDA by 3.31%, 5.23% and 3.38%.The MCC values of SCPLPA, MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.1762, 0.1507, 0.1472, 0.1356 and 0.1681, respectively. SCPLPA is higher than the other comparison methods by 14.47%, 16.46%, 23.04% and 4.60%, respectively. The F1 values of SCPLPA, MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.1102, 0.1023, 0.1054, 0.1012 and 0.1045, respectively. SCPLPA is higher than the other comparison methods by 7.17%, 4.36%, 8.17% and 5.17%, respectively. From these indicators, we can see that the performance of SCPLPA is significantly better than the other four methods. Overall, SCPLPA outperforms the other prediction models in terms of predictive performance.
| Method | AUC | AUPR | ACC | MCC | F1 |
|---|---|---|---|---|---|
| SCPLPA | 0.9356 | 0.4596 | 0.5503 | 0.1762 | 0.1102 |
| MDHGI | 0.8945 | 0.3367 | 0.5607 | 0.1507 | 0.1023 |
| NSEMDA | 0.8899 | 0.3198 | 0.5321 | 0.1472 | 0.1054 |
| RFMDA | 0.8891 | 0.3345 | 0.5215 | 0.1356 | 0.1012 |
| SNMFMDA | 0.9007 | 0.3489 | 0.5317 | 0.1681 | 0.1045 |
3.4 Prediction of new miRNAs and isolated diseases
New miRNAs have not been widely associated with specific diseases or biological functions in existing literature or databases. These miRNAs may be newly discovered, or their functions and mechanisms may not be fully understood. Rapid and accurate identification of the relationship between new miRNAs and diseases would greatly enhance our understanding of the molecular mechanisms of diseases. However, predicting the association between new miRNAs and diseases poses a significant challenge because of unknown association information. Therefore, the model cannot be directly used for prediction. The following procedure is performed once for each miRNA to further evaluate the performance of the SCPLPA model in predicting new miRNA–disease associations: first, the known associations between miRNAs to be queried and all diseases are removed, and it is simulated as a new miRNA; SCPLPA is then used for prediction. This process is repeated until each new miRNA is used as a test sample. The prediction results are evaluated using the ROC curve and AUC value. Figure 4 shows that SCPLPA achieves an AUC value of 0.8412, indicating good performance in predicting new miRNA–disease associations.
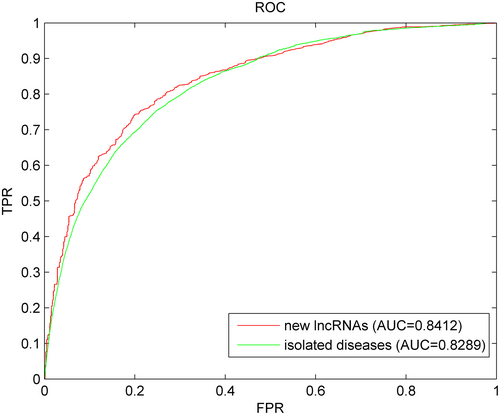
Diseases with completely unknown association information with miRNAs are named as isolated diseases. The prediction of the association between isolated diseases and miRNA is a challenging but promising research area. The association data between the disease to be predicted and all miRNAs are removed, and SCPLPA is used for prediction until each miRNA is tested once. From Figure 4, it can be seen that the AUC value is 0.8289, indicating that SCPLPA can effectively address the problem on the prediction of associations between isolated diseases and miRNAs.
3.5 Case analysis
Colon and kidney neoplasms were selected as case studies to demonstrate the predictive ability of the proposed SCPLPA model for disease–miRNA associations. All of the prediction results were validated in two independent databases, namely, HMDD v3.258 and dbDEMC 2.0.59
Colon neoplasm is a tumour that poses a threat to human health and presents a complex pathological and physiological landscape.60 Identifying miRNAs associated with colon neoplasms plays a crucial role in understanding the pathogenesis, treatment and prognosis of these tissues. The HMDD v2.0 database contains 78 known miRNA–colon neoplasm associations, which were used as training samples to predict potential miRNAs associated with colon neoplasms. Table 2 lists the top 50 predicted miRNAs related to colon neoplasms and their supporting evidence obtained using the SCPLPA model. Among these miRNAs, 49 candidate genes were confirmed in the HMDD v3.2 and dbDEMC 2.0 databases, and only hsa-mir-367 was not validated. We believe that in the near future, biologists will further reveal the relationship of these miRNAs to colon neoplasms through experiments.
| Rank | miRNA name | Evidences | Rank | miRNA name | Evidences |
|---|---|---|---|---|---|
| 1 | hsa-mir-135a | dbDEMC | 26 | hsa-mir-34b | dbDEMC |
| 2 | hsa-mir-135b | HMDD, dbDEMC | 27 | hsa-mir-193a | dbDEMC |
| 3 | hsa-mir-18b | HMDD, dbDEMC | 28 | hsa-mir-425 | dbDEMC |
| 4 | hsa-mir-625 | dbDEMC | 29 | hsa-mir-129 | dbDEMC |
| 5 | hsa-mir-139 | dbDEMC | 30 | hsa-mir-99a | dbDEMC |
| 6 | hsa-mir-185 | dbDEMC | 31 | hsa-mir-149 | dbDEMC |
| 7 | hsa-mir-375 | HMDD, dbDEMC | 32 | hsa-mir-34c | dbDEMC |
| 8 | hsa-mir-497 | dbDEMC | 33 | hsa-mir-409 | dbDEMC |
| 9 | hsa-mir-215 | HMDD, dbDEMC | 34 | hsa-mir-373 | dbDEMC |
| 10 | hsa-mir-25 | HMDD, dbDEMC | 35 | hsa-mir-103a | dbDEMC |
| 11 | hsa-mir-27a | HMDD, dbDEMC | 36 | hsa-mir-429 | HMDD, dbDEMC |
| 12 | hsa-mir-224 | HMDD, dbDEMC | 37 | hsa-mir-124 | dbDEMC |
| 13 | hsa-mir-302c | dbDEMC | 38 | hsa-mir-96 | HMDD, dbDEMC |
| 14 | hsa-mir-186 | dbDEMC | 39 | hsa-mir-148a | HMDD, dbDEMC |
| 15 | hsa-mir-338 | dbDEMC | 40 | hsa-mir-339 | HMDD, dbDEMC |
| 16 | hsa-mir-151a | dbDEMC | 41 | hsa-mir-93 | HMDD, dbDEMC |
| 17 | hsa-mir-183 | dbDEMC | 42 | hsa-mir-182 | dbDEMC |
| 18 | hsa-mir-542 | dbDEMC | 43 | hsa-mir-335 | HMDD, dbDEMC |
| 19 | hsa-mir-345 | dbDEMC | 44 | hsa-mir-320a | dbDEMC |
| 20 | hsa-mir-708 | dbDEMC | 45 | hsa-mir-203 | HMDD, dbDEMC |
| 21 | hsa-mir-194 | HMDD, dbDEMC | 46 | hsa-mir-100 | dbDEMC |
| 22 | hsa-mir-130a | HMDD, dbDEMC | 47 | hsa-mir-153 | dbDEMC |
| 23 | hsa-mir-199b | dbDEMC | 48 | hsa-mir-526a | dbDEMC |
| 24 | hsa-mir-200a | HMDD, dbDEMC | 49 | hsa-mir-302d | dbDEMC |
| 25 | hsa-mir-367 | Unconfirmed | 50 | hsa-mir-95 | dbDEMC |
Kidney neoplasm is a common tumour that has an increasing incidence rate. It has multiple histological subtypes, each has its own unique molecular characteristics. The most common subtype is clear cell renal cell carcinoma, which accounts for 75% of all cases. The 5-year survival rate of clear cell renal cell carcinoma is less than 10%.61 Hence, predicting miRNAs associated with kidney neoplasms is of great practical significance.
The HMDD v2.0 database contains only seven known miRNA–kidney neoplasm-associated pairs. These pairs were used as known information to implement SCPLPA and predict potential miRNAs associated with kidney neoplasms for the discovery of new molecular associations as prognostic or therapeutic markers. As shown in Table 3, all the top 50 predicted kidney neoplasm-related miRNAs have been confirmed in HMDD v3.2 and dbDEMC 2.0. The two cases demonstrate that the SCPLPA model exhibits satisfactory performance in predicting new potential miRNA–disease associations.
| Rank | miRNA name | Evidences | Rank | miRNA name | Evidences |
|---|---|---|---|---|---|
| 1 | hsa-mir-155 | HMDD, dbDEMC | 26 | hsa-mir-134 | dbDEMC |
| 2 | hsa-mir-146a | dbDEMC | 27 | hsa-mir-7 | dbDEMC |
| 3 | hsa-mir-122 | HMDD, dbDEMC | 28 | hsa-mir-17 | HMDD, dbDEMC |
| 4 | hsa-mir-34a | HMDD, dbDEMC | 29 | hsa-mir-142 | dbDEMC |
| 5 | hsa-mir-221 | dbDEMC | 30 | hsa-mir-708 | HMDD |
| 6 | hsa-mir-125b | dbDEMC | 31 | hsa-mir-9 | dbDEMC |
| 7 | hsa-mir-16 | dbDEMC | 32 | hsa-mir-184 | dbDEMC |
| 8 | hsa-mir-29a | dbDEMC | 33 | hsa-mir-106b | dbDEMC |
| 9 | hsa-mir-210 | HMDD, dbDEMC | 34 | hsa-mir-148a | dbDEMC |
| 10 | hsa-mir-31 | dbDEMC | 35 | hsa-mir-19a | dbDEMC |
| 11 | hsa-mir-29b | dbDEMC | 36 | hsa-mir-27a | HMDD, dbDEMC |
| 12 | hsa-mir-199a | HMDD, dbDEMC | 37 | hsa-mir-1207 | dbDEMC |
| 13 | hsa-mir-26a | dbDEMC | 38 | hsa-mir-19b | dbDEMC |
| 14 | hsa-mir-145 | dbDEMC | 39 | hsa-mir-373 | dbDEMC |
| 15 | hsa-mir-133a | dbDEMC | 40 | hsa-let-7b | dbDEMC |
| 16 | hsa-mir-222 | dbDEMC | 41 | hsa-mir-200a | HMDD, dbDEMC |
| 17 | hsa-mir-196a | dbDEMC | 42 | hsa-mir-126 | HMDD, dbDEMC |
| 18 | hsa-mir-206 | dbDEMC | 43 | hsa-mir-137 | dbDEMC |
| 19 | hsa-mir-20a | dbDEMC | 44 | hsa-mir-30b | dbDEMC |
| 20 | hsa-mir-1 | dbDEMC | 45 | hsa-mir-34c | dbDEMC |
| 21 | hsa-mir-200b | dbDEMC | 46 | hsa-mir-212 | dbDEMC |
| 22 | hsa-mir-15b | dbDEMC | 47 | hsa-let-7a | dbDEMC |
| 23 | hsa-mir-218 | dbDEMC | 48 | hsa-mir-92a | dbDEMC |
| 24 | hsa-mir-29c | dbDEMC | 49 | hsa-mir-124 | dbDEMC |
| 25 | hsa-mir-223 | dbDEMC | 50 | hsa-mir-204 | dbDEMC |
All miRNA associations related to the disease to be validated were removed before implementing SCPLPA to test its predictive performance for isolated diseases. For colon neoplasms, 78 known colon neoplasm–miRNA associations were deleted and SCPLPA was used to predict potential miRNA–lung neoplasm associations. All the top 50 predicted miRNAs were supported by evidence in HDMM3.2 and dbDEMC databases (Table 4). Similarly, seven known kidney neoplasm–miRNA associations were deleted, and the SCPLPA model was used to predict kidney neoplasm-related miRNAs. The top 50 predicted associations were supported by evidence in HDMM3.2 and dbDEMC (Table 5).
| Rank | miRNA name | Evidences | Rank | miRNA name | Evidences |
|---|---|---|---|---|---|
| 1 | hsa-mir-145 | HMDD, dbDEMC | 26 | hsa-let-7b | HMDD, dbDEMC |
| 2 | hsa-mir-218 | HMDD, dbDEMC | 27 | hsa-mir-101 | HMDD, dbDEMC |
| 3 | hsa-mir-200c | HMDD, dbDEMC | 28 | hsa-mir-19a | HMDD, dbDEMC |
| 4 | hsa-mir-126 | HMDD, dbDEMC | 29 | hsa-mir-221 | HMDD, dbDEMC |
| 5 | hsa-mir-125b | HMDD, dbDEMC | 30 | hsa-mir-210 | HMDD, dbDEMC |
| 6 | hsa-let-7a | HMDD, dbDEMC | 31 | hsa-mir-124 | dbDEMC |
| 7 | hsa-mir-34a | HMDD, dbDEMC | 32 | hsa-mir-222 | HMDD, dbDEMC |
| 8 | hsa-mir-200b | HMDD, dbDEMC | 33 | hsa-mir-148a | HMDD, dbDEMC |
| 9 | hsa-mir-21 | HMDD, dbDEMC | 34 | hsa-mir-203 | HMDD, dbDEMC |
| 10 | hsa-mir-16 | HMDD, dbDEMC | 35 | hsa-let-7c | HMDD, dbDEMC |
| 11 | hsa-mir-143 | HMDD, dbDEMC | 36 | hsa-let-7d | HMDD, dbDEMC |
| 12 | hsa-mir-31 | HMDD, dbDEMC | 37 | hsa-mir-25 | HMDD, dbDEMC |
| 13 | hsa-mir-34c | dbDEMC | 38 | hsa-mir-214 | dbDEMC |
| 14 | hsa-mir-27a | HMDD, dbDEMC | 39 | hsa-mir-199a | dbDEMC |
| 15 | hsa-mir-155 | HMDD, dbDEMC | 40 | hsa-mir-135a | dbDEMC |
| 16 | hsa-mir-183 | dbDEMC | 41 | hsa-mir-181a | HMDD, dbDEMC |
| 17 | hsa-mir-20a | HMDD, dbDEMC | 42 | hsa-mir-196a | HMDD, dbDEMC |
| 18 | hsa-mir-200a | HMDD, dbDEMC | 43 | hsa-mir-18b | HMDD, dbDEMC |
| 19 | hsa-mir-17 | HMDD, dbDEMC | 44 | hsa-mir-125a | HMDD, dbDEMC |
| 20 | hsa-mir-92a | HMDD, dbDEMC | 45 | hsa-mir-146b | dbDEMC |
| 21 | hsa-mir-34b | dbDEMC | 46 | hsa-mir-205 | HMDD, dbDEMC |
| 22 | hsa-mir-375 | HMDD, dbDEMC | 47 | hsa-mir-107 | HMDD, dbDEMC |
| 23 | hsa-mir-182 | dbDEMC | 48 | hsa-mir-142 | HMDD, dbDEMC |
| 24 | hsa-mir-18a | HMDD, dbDEMC | 49 | hsa-mir-127 | HMDD, dbDEMC |
| 25 | hsa-mir-10b | HMDD, dbDEMC | 50 | hsa-mir-9 | dbDEMC |
| Rank | miRNA name | evidences | Rank | miRNA name | evidences |
|---|---|---|---|---|---|
| 1 | hsa-mir-145 | dbDEMC | 26 | hsa-mir-18a | dbDEMC |
| 2 | hsa-mir-218 | dbDEMC | 27 | hsa-let-7b | dbDEMC |
| 3 | hsa-mir-200c | HMDD, dbDEMC | 28 | hsa-mir-10b | dbDEMC |
| 4 | hsa-mir-126 | HMDD, dbDEMC | 29 | hsa-mir-182 | dbDEMC |
| 5 | hsa-mir-125b | dbDEMC | 30 | hsa-mir-221 | dbDEMC |
| 6 | hsa-mir-200b | dbDEMC | 31 | hsa-mir-210 | HMDD, dbDEMC |
| 7 | hsa-mir-34a | HMDD, dbDEMC | 32 | hsa-let-7c | dbDEMC |
| 8 | hsa-let-7a | dbDEMC | 33 | hsa-mir-203 | HMDD, dbDEMC |
| 9 | hsa-mir-21 | HMDD, dbDEMC | 34 | hsa-mir-375 | dbDEMC |
| 10 | hsa-mir-34c | dbDEMC | 35 | hsa-mir-127 | dbDEMC |
| 11 | hsa-mir-200a | HMDD, dbDEMC | 36 | hsa-mir-9 | dbDEMC |
| 12 | hsa-mir-143 | dbDEMC | 37 | hsa-mir-124 | dbDEMC |
| 13 | hsa-mir-20a | dbDEMC | 38 | hsa-let-7f | dbDEMC |
| 14 | hsa-mir-27a | HMDD, dbDEMC | 39 | hsa-mir-199a | HMDD, dbDEMC |
| 15 | hsa-mir-92a | dbDEMC | 40 | hsa-let-7i | dbDEMC |
| 16 | hsa-mir-155 | HMDD, dbDEMC | 41 | hsa-mir-222 | dbDEMC |
| 17 | hsa-mir-16 | dbDEMC | 42 | hsa-mir-19b | dbDEMC |
| 18 | hsa-mir-101 | dbDEMC | 43 | hsa-mir-100 | dbDEMC |
| 19 | hsa-mir-17 | HMDD, dbDEMC | 44 | hsa-mir-142 | dbDEMC |
| 20 | hsa-mir-31 | dbDEMC | 45 | hsa-mir-214 | HMDD, dbDEMC |
| 21 | hsa-let-7d | dbDEMC | 46 | hsa-mir-146b | dbDEMC |
| 22 | hsa-mir-183 | HMDD, dbDEMC | 47 | hsa-mir-223 | dbDEMC |
| 23 | hsa-mir-34b | dbDEMC | 48 | hsa-mir-125a | dbDEMC |
| 24 | hsa-mir-19a | dbDEMC | 49 | hsa-mir-148a | dbDEMC |
| 25 | hsa-mir-205 | dbDEMC | 50 | hsa-mir-146a | dbDEMC |
The above experimental results further demonstrate the reliability of SCPLPA in predicting miRNAs related to isolated diseases. The model also addresses the limitation of many current miRNA–disease association prediction models in predicting miRNAs related to isolated diseases.
4 DISCUSSION
The association between miRNAs and diseases has attracted research attention. Variations and dysregulation of miRNAs can lead to various diseases. As such, identifying and predicting the association between miRNAs and diseases is beneficial for understanding the function and pathogenesis of miRNAs. Existing biological experimental methods for identifying miRNA–disease associations are time consuming and labour intensive. Computational prediction methods can serve as effective supplementary tools for experimental validation. Predicting potential miRNA–disease associations through computational methods has become a hot topic in bioinformatics, resulting in the development of related prediction models. However, future works should address few issues, such as low prediction accuracy, difficulty in obtaining negative samples and challenges in predicting associations for isolated diseases and new miRNAs.
This paper proposes an SCPLPA model based on network consistency projection and a label propagation algorithm to predict potential miRNA–disease associations. SCPLPA not only performs well in predicting unknown miRNA–disease interactions but also effectively predicts isolated diseases and new miRNAs.SCPLPA was compared with four state-of-the-art models, namely, MDHGI, NSEMDA, RFMDA and SNMFMDA, to evaluate its performance. The ACC value of SCPLPA is 0.5503, while MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.5607, 0.5321, 0.5215 and 0.5317, respectively. The AUC values of the five models obtained through LOOCV are 0.9356, 0.8945, 0.8899, 0.8891 and 0.9007, respectively. Furthermore, the AUPR value of SCPLPA is 0.4596, while MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.3367, 0.3198, 0.3345 and 0.3489, respectively. SCPLPA's AUPR outperforms various state-of-the-art models by at least 24.09%. This indicates that in the given datasets with imbalanced positive and negative samples, SCPLPA's predictive performance has a clear advantage over other state-of-the-art models, demonstrating better robustness in handling imbalanced datasets. Additionally, the MCC values of SCPLPA, MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.1762, 0.1507, 0.1472, 0.1356 and 0.1681, respectively. The F1 values of SCPLPA, MDHGI, NSEMDA, RFMDA and SNMFMDA are 0.1102, 0.1023, 0.1054, 0.1012 and 0.1045, respectively. SCPLPA also has a slight lead in F1 and MCC values. In conclusion, compared to the other four state-of-the-art models, SCPLPA can improve robustness in imbalanced datasets while maintaining high prediction accuracy, showing superior performance in miRNA–disease association tasks. Each disease (miRNA) was simulated as an isolated disease (new miRNA) to evaluate the prediction performance of SCPLPA for new miRNAs and isolated diseases. Cross-validation was then performed for each disease (miRNA). The AUC values of SCPLPA are 0.8289 (0.8412). Colon and kidney neoplasms were selected for case analysis to further validate the reliability of the SCPLPA model in predicting the relationship between potential miRNAs and diseases. In the top 50 rankings and the corresponding disease-related miRNA predictions, the accuracy levels verified by the HDMM3.2 and dbDEMC databases are 98% and 100%, respectively. In the prediction of the isolated disease cases, all the top 50 rankings were confirmed by the two databases. The reliable predictions of SCPLPA provide insights for the identification of potential miRNA biomarkers and contribute to future research on the involvement of miRNAs in human disease mechanisms.
The outstanding predictive performance of SCPLPA is mainly due to two reasons. First, it integrates disease semantic similarity data and disease Gaussian interaction profile kernel similarity data to construct a heterogeneous disease similarity network. It also integrates miRNA functional similarity data and miRNA Gaussian interaction profile kernel similarity data to construct a heterogeneous miRNA similarity network, which can more accurately characterize the similarity between diseases and miRNAs. Second, the SCPLPA method combines the label propagation algorithm and network consistency projection sub-models. The label propagation algorithm estimates lncRNA–disease associations, alleviates the sparsity of known miRNA–disease association data and addresses the positive and unlabelled learning problem. Consistency information between different networks is obtained, thereby solving the problems on predicting isolated diseases and new miRNAs and improving the accuracy of predicting potential miRNA–disease associations. Although SCPLPA can effectively predict miRNA–disease associations but has certain limitations. First, integrating more omics data can construct more accurate disease similarity networks and miRNA similarity networks. Second, our algorithm is based on the prediction of known miRNA–disease associations, which may lead to biased results towards diseases with known associated miRNAs. Inspired by various association prediction methods such as drug–target interaction prediction62 and ligand–receptor interactions,63-66 we plan to explore boosting-based or deep learning-based models to enhance microRNA–disease prediction in future research.
AUTHOR CONTRIBUTIONS
Min Chen: Conceptualization (equal); formal analysis (equal); methodology (equal); resources (equal); software (equal); supervision (equal); writing – original draft (equal); writing – review and editing (equal). Yingwei Deng: Conceptualization (equal); formal analysis (equal); investigation (equal); methodology (equal); resources (equal); software (equal); supervision (equal); writing – original draft (equal); writing – review and editing (equal). Zejun Li: Funding acquisition (equal); resources (equal). Yifan Ye: Investigation (equal); validation (equal); visualization (equal). Lijun Zeng: Project administration (equal); visualization (equal). Ziyi He: Investigation (equal); validation (equal); visualization (equal). Guofang Peng: Visualization (equal).
ACKNOWLEDGEMENTS
The work was supported by the Nature Science Foundation of Hunan Province, China (Grant No. 2024JJ7115) and the National Natural Science Foundation of China (Grant No. 62172158).
CONFLICT OF INTEREST STATEMENT
The authors confirm that there are no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
All datasets generated for this study are included in the article/supplementary material.




