CDKN2B-AS1 gene rs4977574 A/G polymorphism and coronary heart disease: A meta-analysis of 40,979 subjects
Yan-yan Li and Hui Wang contribute equally to this work.
Abstract
It has been implied that there is a possible relationship between cyclin-dependent protein kinase inhibitors antisense RNA 1 (CDKN2B-AS1) gene rs4977574 A/G polymorphism and coronary heart disease (CHD) susceptibility. However, as the research results are discrepant, no distinct consensus on this issue has been reached so far. In order to further elaborate the latent association of the CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD, this present meta-analysis was conducted. There were 40,979 subjects of 17 individual studies in the present meta-analysis. The pooled odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were estimated to determine the association strength. Considering the significant heterogeneity among the individual studies, the random-effect models were used. In the current meta-analysis, a significant association between CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD was found under allelic (OR: 1.18, 95% CI: 1.08–1.29, p = 4.83×10−4), recessive (OR: 1.36, 95% CI: 1.11–1.67, p = 0.003), dominant (OR: 0.71, 95% CI: 0.58–0.86, p = 6.26×10−4), heterozygous (OR:1.210, 95% CI: 1.076–1.360, p = 0.001), homozygous (OR: 1.394, 95% CI: 1.163–1.671, p = 3.31×10−4) and additive (OR: 1.180, 95% CI: 1.075–1.295, p = 4.83×10−4) genetic models. A more significant association between them was found in the Asian population than that in the whole population under these genetic models (p < 0.05). However, no significant association between them was found in the Caucasian population (p > 0.05). CDKN2B-AS1 gene rs4977574 A/G polymorphism was associated with CHD susceptibility, especially in the Asian population. G allele of CDKN2B-AS1 gene rs4977574 A/G polymorphism is the risk allele for CHD.
1 INTRODUCTION
Cardiovascular disease (CVD) due to atherosclerosis (AS) is the leading cause of death in humans. In Europe, the United States and other developed countries, deaths caused by coronary heart disease (CHD) account for nearly half of the total number of deaths. Approximately 5.7 million Americans, or 2.5% of the population, have CHD, and one in five American men have a chance of acquiring CHD before the age of 60. In accordance with the CVD report in China in 2019, the prevalence of CVD in China is on the rise. The number of CVD patients has reached 330 million, of which CHD is up to 11 million. In 2017, CVD mortality was the highest, higher than that of cancer and other diseases.1
The main pathological change of CHD is AS, which is marked by the infiltration of inflammatory cells in the artery wall and the abnormal proliferation of vascular smooth muscle cells (VSMCs), leading to vascular endothelial hyperplasia.2 The coronal AS progression is influenced by genetic and environmental factors. Under the coaction of mechanical and chemical injury, vascular endothelial cells are damaged and inflammatory cells stick together, thus promoting the proliferation and migration of vascular endothelial cells and forming AS plaque. Although the aetiology of CHD is incompletely clear, genetic and environmental factors are recognized as the main pathogenesis.3, 4 Environmental factors include gender, age, obesity, high cholesterol, high blood pressure and unhealthy living habits, which can explain 50%-60% of the aetiology of CHD. The remaining 40%-50% of the aetiology of CHD is due to genetic factors.5 As a result, the CHD genetic cause factors become the hot spot in pathogenesis research.
Previous research has confirmed that the cyclin-dependent kinase inhibitor 2A/2B (CDKN2A/2B) gene is associated with the occurrence of coronal AS in the general population.6, 7 Cyclin-dependent kinases (CDKs) could catalyse VSMC proliferation and cause atherosclerotic changes. The protein encoded by the CDKN2A/2B gene could inhibit CDK activities and prevent VSMC cell proliferation and apoptosis. Hence, CDKN2A/2B has the potential protection effect on CHD.
CDKN2A/2B is an important tumour suppressor gene, belonging to the CDK inhibitor gene family. It is located in the p21.3 band on the short arm of human chromosome 9 and is widely expressed in human cells and tissues as vascular endothelial cells and smooth coronary muscle cells. The CDKN2A/2B gene contains four exons, 1α, 1β, 2 and 3, encoding two different proteins: P16INK4a (P16) and p14ARF (P14). The human CDKN2B-AS1 (also known as ANRIL), located adjacent to the CDKN2A/2B gene cluster, is an underlying gene for CHD that encodes an antisense noncoding RNA. The human CDKN2B-AS1 gene rs4977574 A/G polymorphism is located in the CDKN2A/2B gene intron, where the adenine (A) nucleotide is being replaced with guanine (G) nucleotide. Although it is a nonprotein coding variant, it might influence the CDKN2A/2B gene expression directly. Thus, CDKN2B-AS1 gene rs4977574 A/G polymorphism was reported to be associated with the CHD onset.8
Despite many studies on the association of CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD have been performed, no clear consensus has been reached. In 2013, Sakalar et al. found that the G allele frequency of CDKN2B-AS1 gene rs4977574 polymorphism in myocardial infarction patients was significantly higher than that in controls in a Turkish population.9 Analogously, in 2020, Hua et al. examined the association between CDKN2B-AS1 gene rs4977574 polymorphism and CHD in a Chinese population and found that the G allele is the CHD susceptibility site.10 On the contrary, in 2007, Samani et al.11 reported that the G allele frequency was greatly lower in CHD patients than that in controls in a WTCCC study of a British population.
To verify whether CDKN2B-AS1 gene rs4977574A/G polymorphism is susceptible to CHD development, a meta-analysis of 40,979 subjects from 17 separate studies was conducted (Supplements S1).
2 MATERIALS AND METHODS
2.1 Publication search and inclusion criteria
The terms “CDKN2A/2B,” “CDKN2B-AS1,” “rs4977574,” “coronary heart disease,” “myocardial infarction,” “coronary artery disease” and “polymorphism” were used to perform a search. The electronic databases of Wan Fang, PubMed, VIP, Web of Science, Embase and China National Knowledge Infrastructure were retrieved. The selected papers were released between 2007 and 2020. The retrieval process was updated on May 6, 2021.
The collected studies must meet the next inclusion criteria in the current meta-analysis. The selected studies must evaluate the association of CHD and CDKN2B-AS1gene rs4977574 A/G polymorphism. The CHD diagnosis was defined as that the coronary artery stenosis was over 50% in at least one main coronary artery. The coronary artery stenosis was calculated through dual-source coronary computed tomography or coronary angiography. The selected studies must conform to the Hardy-Weinberg equilibrium (HWE) in genotype distribution in the control group. The included studies were cohort or case-control studies that were publicly released.
2.2 Data extraction
In accordance with a standardized protocol, the information of individual studies was drawn out by three investigators together. Two of them selected the eligible studies. The third one acted as an intercessor to solve any ambiguous issues between them. Studies that deviated from the inclusion criteria were repeatedly published or did not provide valid data were excluded. If analogical data were presented in different papers by an identical author group, the data could be merely used once in the current meta-analysis. The definition of controls included the source of controls, that is either hospital-based or population-based. The quality of studies was assessed by using the Newcastle-Ottawa scale scores.
2.3 Statistical analyses
In this study, six genetic models as allelic (G allele frequency distribution), recessive (GG vs. AA + AG), dominant (AA vs. AG + GG), heterozygous (AG vs. AA), homozygous (GG vs. AA) and additive (G vs. A) were adopted. The ORs and their 95% CIs were calculated to analyse the association of CDKN2B-AS1gene rs4977574 A/G polymorphism and CHD. The heterogeneity among the individual researches was evaluated through the chi-square-based Q-tests.12 If there was obvious heterogeneity, the random-effect model would be applied.13 Otherwise, the fixed-effect model would be utilized.14 The significance was set at p < 0.05 level in all of the above tests.
The potential publication bias was assessed by using the funnel plot and Egger's linear regression test.15 All of the statistical analyses were conducted by utilizing Review Manager 5.3 and Stata 12.0 software.
3 RESULTS
3.1 Studies and populations
The information was drawn out from 18,430 CHD cases and 22,549 controls (Table 1).9-11, 16-29 Of the 35 obtained literatures after the preliminary search course, 17 met the present meta-analysis including criteria. Among the eighteen eliminated papers, five of them were of review character. Two of them were against the HWE in the control group.30, 31 Nine of them lack the indispensable data.32-40 The rest two excluded papers were not associated with the CDKN2B-AS1 gene rs4977574 A/G polymorphism or CHD (Supplements S2).
| Author | Year | Region | Ethnicity | CHD | Control | Matching criteria | Source of control | NOS score | Genotyping method | sample size (CHD/control) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | |||||||||
| Sakalar C9 | 2013 | Turkey | Caucasian | 8 | 22 | 14 | 13 | 11 | 4 | Age, sex, hypertension, diabetes, hyperlipidaemia family history, ethnicity | HB | 6 | PCR-RFLP | 44/28 |
| Helgadottir A16 | 2007 | Iceland | Caucasian | 554 | 1105 | 556 | 1507 | 2335 | 964 | Ethnicity | PB | 6 | PCR-sequencing | 2215/4806 |
| Helgadottir A16 | 2007 | Philadelphia | Caucasian | 103 | 286 | 180 | 130 | 246 | 119 | Ethnicity | PB | 6 | PCR-sequencing | 569/495 |
| Helgadottir A16 | 2007 | Atlanta | Caucasian | 115 | 274 | 188 | 332 | 597 | 325 | Ethnicity | PB | 6 | PCR-sequencing | 577/1254 |
| Helgadottir A16 | 2007 | Durham | Caucasian | 267 | 549 | 316 | 187 | 383 | 144 | Ethnicity | PB | 6 | PCR-sequencing | 1132/714 |
| KalpanaB17 | 2019 | India | Asian | 23 | 36 | 31 | 106 | 230 | 100 | Hyperlipidaemia, weight, alcohol, ethnicity, smoking, exercise, fruits intake, family history | PB | 6 | Mass Array Technology | 90/436 |
| Hua L10 | 2020 | China | Asian | 149 | 297 | 152 | 87 | 122 | 48 | Drinking, TC, TG, LDL-C, Apo B, Bun, ethnicity | HB | 6 | Mass Array SNP typing | 598/257 |
| SamaniNJ11 | 2007 | UK | WTCCC(Caucasian) | 605 | 937 | 382 | 698 | 1435 | 804 | Ethnicity | PB | 6 | Affymetrix 500K | 1924/2937 |
| SamaniNJ11 | 2007 | UK | German MI study (Caucasian) | 169 | 452 | 239 | 463 | 826 | 354 | Ethnicity | PB | 6 | Affymetrix 500K | 860/1643 |
| Saade S18 | 2011 | Lebanon | Asian | 208 | 685 | 627 | 72 | 195 | 156 | TC, LDL-C, LogTG, ethnicity, BMI | PB | 6 | Illumina chips | 1520/423 |
| Zheng Y19 | 2016 | China | Asian | 422 | 795 | 343 | 540 | 891 | 320 | Age, sex, physical activity, prevalent cholesterolaemia, urban area of residence, individual European admixture proportion | PB | 6 | PCR-ASP | 1560/1751 |
| Qiao L20 | 2017 | China | Asian | 43 | 114 | 69 | 26 | 35 | 18 | Age, BMI,TC,LDL-C, HDL-C, ethnicity | HB | 6 | PCR-sequencing | 226/79 |
| Wang YQ21 | 2014 | China | Asian | 583 | 1139 | 595 | 777 | 1325 | 482 | Sex, WHR, alcohol intake, ethnicity | HB | 6 | PCR-ASP | 2317/2584 |
| Cao XL22 | 2016 | China | Asian | 117 | 272 | 176 | 152 | 255 | 134 | Age, sex, smoking, LDL-C, ApoB, ethnicity | HB | 6 | PCR-RFLP, direct sequencing | 565/541 |
| Matsuoka R23 | 2015 | Japan | Asian | 448 | 898 | 476 | 651 | 1132 | 501 | SBP, ethnicity | PB | 6 | Luminex bead-based multiplex assay | 1822/2284 |
| Beigi SSH24 | 2015 | Iran | Asian | 22 | 44 | 34 | 17 | 44 | 32 | Age, sex, Height, Weight, BMI, TC,TG,FBS, HDL,LDL,SBP, DBP, ethnicity | HB | 6 | TaqMan SNP genotyping assay | 100/93 |
| Tang OS25 | 2017 | China | Asian | 37 | 136 | 116 | 38 | 134 | 166 | Age, sex, DBP, ethnicity | HB | 6 | real-time PCR | 289/338 |
| Lee IT26 | 2014 | China | Asian | 198 | 479 | 248 | 181 | 318 | 135 | BMI, HDL,SBP, DBP, ethnicity | HB | 6 | StepOnePlus Real-Time PCR | 925/634 |
| Qi L27 | 2012 | China | Asian | 35 | 64 | 43 | 46 | 94 | 52 | Ethnicity | HB | 6 | PCR-RFLP | 142/192 |
| Xu JJ28 | 2018 | China | Asian | 202 | 253 | 429 | 287 | 182 | 438 | Age, Bleeding, TC, RBC, ethnicity | HB | 6 | iMLDR | 884/907 |
| Temel SG29 | 2019 | Turkey | Caucasian | 24 | 33 | 14 | 39 | 76 | 38 | Age, sex, Glucose, TC, ethnicity | HB | 6 | PCR-RFLP | 71/153 |
- Abbreviations: CDKN2A/2B: Cyclin-dependent kinase inhibitor 2A/2B; NOS score: Newcastle-Ottawa scale scores; PCR-RFLP: Polymerase chain reaction-restriction fragment length polymorphism; PCR-ASP: Polymerase chain reaction-allelic-specific primer; iMLDR: improved multiplex ligase detection reaction; TC: total cholesterol; TG: triglyceride; LDL-C: low-density lipoprotein cholesterol; Apo B: apolipoproteins B; Bun: blood urea nitrogen; BMI: body mass index; WHR: waist hip rate; SBP: systolic blood pressure; DBP: diastolic blood pressure; RBC: red blood cell.
3.2 Pooled analyses
In the current meta-analysis, a significant association between CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD was found under allelic (OR: 1.18, 95% CI: 1.08–1.29, p = 4.83×10−4), recessive (OR: 1.36, 95% CI: 1.11–1.67, p = 0.003), dominant (OR: 0.71, 95% CI: 0.58–0.86, p = 6.26×10−4), heterozygous (OR:1.210, 95% CI: 1.076–1.360, p = 0.001), homozygous (OR: 1.394, 95% CI: 1.163–1.671, p = 3.31×10−4), and additive (OR: 1.180, 95% CI: 1.075–1.295, p = 4.83×10−4)genetic models. (Table 2, Figures 1-6).
| Genetic model | Pooled OR (95% CI) | Z value | p Value | Literature number | CHD size | control size | Pheterogeneity(I 2%) |
|---|---|---|---|---|---|---|---|
| Allelic genetic model | 1.18 (1.08–1.29) | 3.49 | 4.83×10–4* | 17 | 18430 | 22549 | <0.00001 (88%)* |
| Caucasian subgroup | 1.18 (0.96–1.46) | 1.55 | 0.12 | 4 | 7392 | 12030 | <0.00001 (95%)* |
| Asian subgroup | 1.20 (1.13–1.27) | 5.78 | 7.47×10−9* | 13 | 11038 | 10519 | 0.03(47%)* |
| Recessive genetic model | 1.36 (1.11–1.67) | 2.99 | 0.003* | 17 | 18430 | 22549 | <0.00001 (92%)* |
| Caucasian subgroup | 1.40 (0.93–2.11) | 1.60 | 0.11 | 4 | 7392 | 12030 | <0.00001(96%)* |
| Asian subgroup | 1.40 (1.16–1.70) | 3.46 | 5.40×10−4* | 13 | 11038 | 10519 | <0.00001(81%)* |
| Dominant genetic model | 0.71 (0.58–0.86) | 3.42 | 6.26×10−4* | 17 | 18430 | 22549 | <0.00001 (93%)* |
| Caucasian subgroup | 0.70 (0.45–1.11) | 1.51 | 0.13 | 4 | 7392 | 12030 | <0.00001 (97%)* |
| Asian subgroup | 0.67 (0.59–0.77) | 5.62 | 1.91×10−8* | 13 | 11038 | 10519 | 0.0002(68%)* |
| Heterozygous genetic model | 1.210 (1.076–1.360) | 3.19 | 0.001* | 17 | 18430 | 22549 | <0.00001 (76.6%)* |
| Caucasian subgroup | 1.175 (0.928–1.489) | 1.34 | 0.181 | 4 | 7392 | 12030 | <0.00001 (87.4%)* |
| Asian subgroup | 1.247 (1.109–1.401) | 3.70 | 2.16×10−4* | 13 | 11038 | 10519 | 0.011(53.9%)* |
| Homozygous genetic model | 1.394 (1.163–1.671) | 3.59 | 3.31×10−4* | 17 | 18430 | 22549 | <0.00001 (87.9%)* |
| Caucasian subgroup | 1.393 (0.913–2.124) | 1.54 | 0.124 | 4 | 7392 | 12030 | <0.00001 (94.9%)* |
| Asian subgroup | 1.451 (1.304–1.613) | 6.86 | 6.89×10−12* | 13 | 11038 | 10519 | 0.112(33.7%) |
| Additive genetic model | 1.180 (1.075–1.295) | 3.49 | 4.83×10−4* | 17 | 18430 | 22549 | <0.00001 (88.4%)* |
| Caucasian subgroup | 1.182 (0.956–1.461) | 1.55 | 0.122 | 4 | 7392 | 12030 | <0.00001 (95.0%)* |
| Asian subgroup | 1.199 (1.127–1.274) | 5.78 | 7.47×10−9* | 13 | 11038 | 10519 | 0.03 (47.3%)* |
- Abbreviations: CHD: coronary heart disease; CI: confidence interval; OR: odds ratio; CHD size: the total number of CHD cases; control size: the total number of control group; Allelic genetic model: G allele distribution frequency; Dominant genetic model: AAvs.AG + GG; Recessive:GGvs.AA + AG; Heterozygous genetic model: AG vs. AA; Homozygous genetic model: GG vs. AA; Additive genetic model: total G allele vs. total A.
- *p ≤ 0.05.
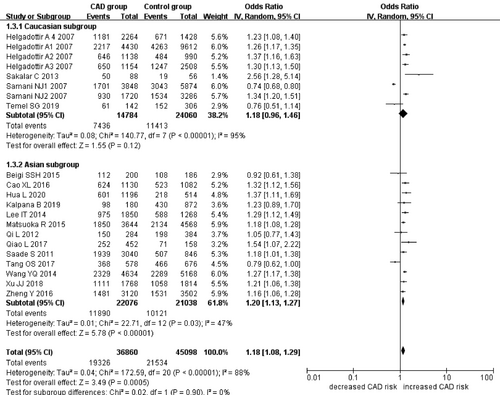
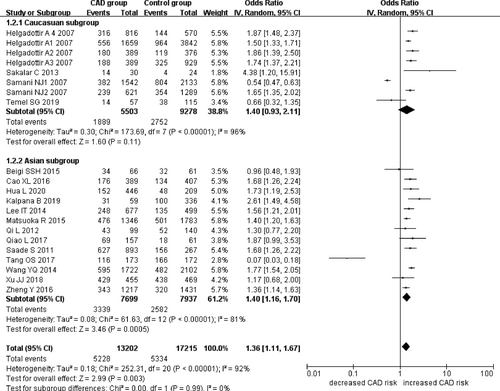
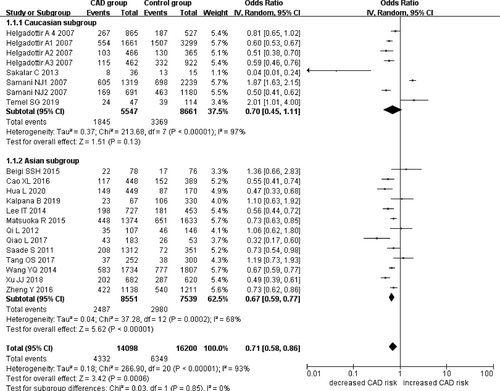
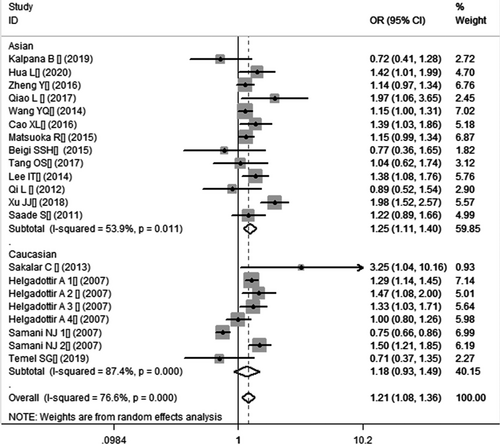

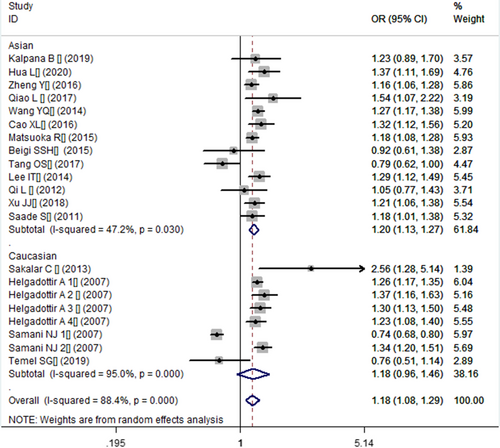
In the subsequent subgroups analysis stratified by the ethnicity, the relationship between the CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD was found more prominent in the Asian subgroup than that in the whole population. In the Asian subgroup, there was a significant relationship under allelic (OR: 1.20, 95% CI: 1.13–1.27, p = 7.47×10−9), recessive (OR: 1.40, 95% CI: 1.16–1.70, p = 5.40×10−4), dominant (OR: 0.67, 95% CI: 0.59–0.77, p = 1.91×10−8), heterozygous (OR: 1.247, 95% CI: 1.109–1.401, p = 2.16×10−4), homozygous (OR: 1.451, 95% CI: 1.304–1.613, p = 6.89×10−12) and additive (OR: 1.199, 95% CI: 1.127–1.274, p = 7.92×10−9) genetic models. In the Caucasian subgroup, no significant association was detected between CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD under allelic (OR: 1.18, 95% CI: 0.96–1.46, p = 0.12), recessive (OR: 1.40, 95% CI: 0.93–2.11, p = 0.11), dominant (OR: 0.70, 95% CI: 0.45–1.11, p = 0.13), heterozygous (OR: 1.175, 95% CI: 0.928–1.489, p = 0.181), homozygous (OR: 1.393, 95% CI: 0.913–2.124, p = 0.124) and additive (OR: 1.182, 95% CI: 0.956–1.461, p = 0.122) genetic models.
In the overall population, the heterogeneity among the individual studies was detected significant under all of the genetic models (p < 0.05). However, in the subgroup analysis stratified by the ethnicity, the heterogeneity was decreased under these genetic models in the Asian subgroup. Among them, the heterogeneity even did not exist any longer under the homozygous genetic models (p > 0.05). However, in the Caucasian subgroup, the heterogeneity remained significant and the I2(%) was still more than 87.4%. Hence, it was shown that the heterogeneity might be originated from the ethnicity.
3.3 Bias diagnostics
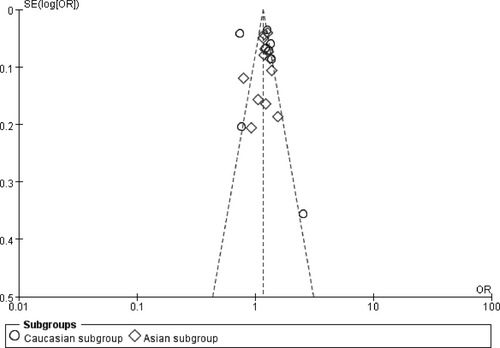
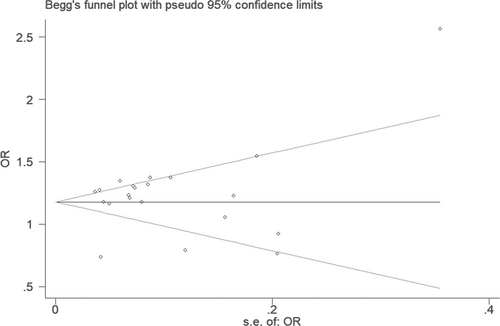
In the funnel plot under the allelic genetic model, no publication bias was visualized (Figure 7). Moreover, no significant difference was found by utilizing Egger's test under the additive genetic model. It was suggested that there was no publication bias in the current meta-analysis under the additive genetic model (T =0.79, p = 0.448) (Figure 8).
3.4 Sensitivity analysis
Sensitivity analysis was performed under the additive genetic model. After any individual study was removed from the current meta-analysis, the primary analysis result was unaffected. This result was comparatively steady (Figure 9).

4 DISCUSSION
In this meta-analysis, a remarkable association between CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD was observed in the entire population under the allelic (OR: 1.18), recessive (OR: 1.36), dominant (OR: 0.71), heterozygous (OR: 1.210), homozygous (OR: 1.394) and additive (OR: 1.180) genetic models. In addition, the G allele of CDKN2A/2B gene rs4977574 A/G polymorphism was susceptible to CHD development. Given the evident heterogeneity among individual studies (p < 0.00001), a subgroup analysis by ethnicity was performed. In the subgroup analysis, the relationship between CDKN2B-AS1 gene rs4977574 A/G polymorphism and CHD was more prominent in the Asian subgroup than in the Caucasian subgroup, while the heterogeneity in the Asian subgroup was lower than that in the Caucasian subgroup. The heterogeneity source might be associated with the different ethnicities.
Recent studies have found that the CDKN2A/2B gene is only approximately 100 kb apart from the chromosome 9p21 risk gene, which is generally expressed in many tissue and cell types. The two kinds of proteins encoded by the CDKN2A/2B gene are the INK4 family member p16 (p16INK4a) and p14arf, which act as tumour suppressors by regulating the cell cycle. p16 inhibits CDK4 and CDK6, thus activates the retinoblastoma family protein, and then stops the G1 phase of cell transition to S phase. The function of p14ARF (known asp19ARF in mice) is activating p53 tumour suppressor 2. Somatic mutations in CDKN2A are common in human cancers, and CDKN2A is estimated to be the second most frequently inactivated gene in cancer after p53. One of the homologous genes in CDKN2A is CDKN2B, and they have the same biological function.41
CDKN2A/2B mainly inhibits VSMC proliferation in the cell cycle regulation, which is closely related to the pathogenesis of AS of CHD.42 The possible mechanisms of CDKN2B-AS1 gene rs4977574 A/G polymorphism affecting the AS formation remain unclear. It might be closely related to the direct influence of intron variation on CDKN2A/2B gene expression.8 The product of this CDKN2B-AS1 gene is a functional RNA molecule that interacts with polycomb repressive complexes 1 and 2 resulting in CDKN2A/2B’epigenetic silencing.43 CDKN2B-AS1 gene rs4977574 A/G mutation may induce overexpression of CDKN2B-AS1 transcript, thereby inhibiting the expression of CDKN2A/2B. Related studies have shown that the CDKN2B-AS1 gene expression level is increased in peripheral blood mononuclear macrophages containing the G allele of rs4977574 polymorphism, which can lead to biological behaviours, such as increased cell proliferation and adhesion, and AS.44 Ding et al.45 reported that CHD patients had lower expression of CDKN2A and CDKN2B than healthy people, which suggested that the decreased CDKN2A and CDKN2B level was associated with the CHD pathogenesis. Previous studies have shown that the transcription level of CDKN2B-AS1 is strongly correlated with the severity of AS. CDKN2B-AS1 knockdown in vascular smooth muscle changes the gene expression, which is involved in extracellular matrix remodelling. CDKN2B-AS1 plays a pivotal role in the reconstitution of vascular structure and function alterations.46
Qiao et al. performed a correlation analysis of rs4977574 polymorphism and biomarker. They found that the risk of high HbA1c in GG + GA genotype subjects was significantly increased (OR = 2.08, 95%CI: 1.11–3.89, p = 0.022).20 They speculated that rs4977574 polymorphism might be involved in the regulation of glucose metabolism. The G allele of rs4977574 polymorphism might be associated with impaired pancreatic beta cell function and decreased insulin secretion. CDK4 is an effective beta cell proliferative regulator of the pancreas. Abnormal CDKN2A/2B gene expression could lead to pancreatic hypoplasia and diabetes.47 Type 2 diabetes mellitus is a disease with an equal risk to that of CHD. Abnormal blood glucose metabolism increases the risk of early CHD. Some studies have found that, under the influence of risky SNP loci, certain transcription products of the CDKN2B-AS1 gene can regulate the expression of ADIPOR1, VAMP3 and C11ORF10; the above-mentioned genes play an important role in the regulation of metabolic activities, such as blood glucose and lipid.48 Therefore, rs4977574 might be involved in the occurrence and development of early-onset CHD by affecting blood glucose and lipid metabolism through related signalling pathways.20
Similar meta-analyses on CDKN2B-AS1 gene rs4977574 polymorphism and CHD have been conducted in the past. In 2014, Huang et al.30 performed a case-control study and a meta-analysis on the association between them. They found a strong association between rs4977574 and CHD risk. However, their case-control study was against HWE. Hence, their conclusion was not as reliable as that deduced from the current meta-analysis. In 2018, Xu et al.49 performed a meta-analysis and found that rs4977574 polymorphism was associated with the CHD risk in Asian population. The G allele might contribute to a high risk of CHD. Although they obtained a similar conclusion to that of the current meta-analysis, they limited the studies in the Asian population. They also included studies against HWE. In the current meta-analysis, the research scope was expanded, which included Asian and Caucasian studies. Several new studies published after 2018 were also included. In 2020, Zhang et al. performed a meta-analysis on this subject and obtained a similar conclusion.50 Nevertheless, they also included studies deviating from HWE. In addition, they mixed Asian and Caucasian studies and did not divide them into two subgroups. The individual studies included in their meta-analysis were not as complete as those in the current meta-analysis. In the current meta-analysis, a subgroup analysis stratified by ethnicity was performed. Yuan et al. also performed a meta-analysis on rs4977574 polymorphism and CHD in 2020 and obtained similar results.51 However, in their meta-analysis, individual studies deviating from HWE were included. Although they divided the participating subjects into three subgroups, namely West Asian, East Asian, and Caucasian, the included individual studies were not complete yet. Therefore, the current meta-analysis is much more comprehensive and convincing than the previous ones.
Nonetheless, this research still has some shortcomings. Multiple large-scale studies remain insufficient to determine the exact association of CDKN2B-AS1 gene rs4977574 polymorphism and CHD. Environmental factors, such as smoking, lack of exercise, excessive eating and obesity, also have a significant effect on the CHD process. The microeffects of other genes involved in CHD have yet to be adequately clarified (eg PCSK9 gene E670G polymorphism and TGF-β1 gene-509C/T polymorphism).52, 53 The CDKN2B-AS1 gene rs1333040C>T, rs1333042 A>G and rs10757274 A>G variants might also affect the pathogenesis of CHD patients.50
In sum, CDKN2B-AS1 gene rs4977574 polymorphism was evidently relevant to CHD risk, particularly in the Asian population. The G allele of CDKN2B-AS1 gene rs4977574 polymorphism might confer the CHD risk to the population. Additional large-scale studies on the association of CDKN2B-AS1 gene rs4977574 polymorphism and CHD should be conducted to verify this viewpoint in the near future.
ACKNOWLEDGEMENTS
This work was funded by the National Natural Science Foundation of China (NSFC 81100073 to Dr Yan-yan Li), Excellent Young and Middle-Aged Teachers Assistance Program of Nanjing Medical University for Dr Yan-yan Li (2013-2015, JX2161015034), Jiangsu Overseas Research & Training Program for University Prominent Young & Middle-aged Teachers and Presidents, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). This work was also funded by the Natural Science Foundation of Jiangsu Province (BK 2012648 to Dr Hui Wang), ‘six talent peaks’ project in Jiangsu Province (2015-WSN-033). Thank all our colleagues working in the Department of geriatrics, the First Affiliated Hospital of Nanjing Medical University.
CONFLICT OF INTEREST
The authors have no other conflicts of interest to disclose.
AUTHOR CONTRIBUTIONS
Yan-yan Li: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Funding acquisition (lead); Investigation (lead); Methodology (lead); Project administration (lead); Resources (lead); Software (lead); Supervision (lead); Validation (lead); Visualization (lead); Writing-original draft (lead); Writing-review & editing (lead). Hui Wang: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Funding acquisition (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (supporting); Software (supporting); Supervision (supporting); Validation (supporting); Visualization (supporting); Writing-original draft (supporting); Writing-review & editing (supporting). Yang-yang Zhang: Conceptualization (equal); Data curation (equal); Formal analysis (supporting); Funding acquisition (supporting); Investigation (supporting); Methodology (equal); Project administration (equal); Resources (equal); Software (supporting); Supervision (supporting); Validation (supporting); Visualization (supporting); Writing-original draft (supporting); Writing-review & editing (supporting).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




