Circular RNA hsa_circ_0008003 facilitates tumorigenesis and development of non-small cell lung carcinoma via modulating miR-488/ZNF281 axis
Abstract
As one of the most aggressive malignancies, non-small cell lung carcinoma (NSCLC) has high risks of death. It has been demonstrated that circRNAs accelerate NSCLC progression, but the underlying molecular mechanisms of circRNAs in NSCLC were still obscure. In the first place, the circRNA microarray of NSCLC was investigated in this study, and hsa_circ_0008003 (circ-0008003) was chosen as the research object. Then, it was unveiled that the expression of circ-0008003 examined via qRT-PCR was elevated in tumour tissues relative to the non-tumour tissues, which was associated with TNM stage and lymphatic metastasis in NSCLC. Additionally, the prognosis of NSCLC patients with high circ-0008003 level was poor. Besides, circ-0008003 silencing dampened the invasion and proliferation of NSCLC cells. Next, according to the mechanistic studies, circ-0008003 functioned as a ceRNA of ZNF281 in NSCLC by acting as the endogenous sponge for miR-488, which was proved to be a tumour suppressor in NSCLC. Additionally, ZNF281 overexpression and miR-488 suppression recovered the influences of repressed circ-0008003 on NSCLC cellular processes. It was validated in this research that circ-0008003 triggered tumour formation in NSCLC, which was adjusted via miR-488/ZNF281 axis, casting a novel light on the resultful target for treating NSCLC and predicting the prognosis.
1 INTRODUCTION
Non-small cell lung cancer (NSCLC) is a major cause of death associated with lung cancer, taking up over 85% of all lung cancers.1 The survival of patients can be prolonged by diagnosing and treating the disease in the early stage. Nevertheless, the potential mechanism by which lung cancer evolves remains undocumented. Advances made in RNA research recently have contributed to identification of non-coding RNA (ncRNA) implicated in different biological processes.2, 3 LncRNAs and miRNAs are verified in previous research to function in the evolution of lung cancer, especially in adjustment of apoptosis, proliferation and invasion.4-7 Herein, ncRNAs need to be researched to continuously confirm the mechanism associated with the cancer from the aspect of epigenetics.
As a newfound type of RNAs, circular RNAs (circRNAs) display a wide expression in eukaryotes. As bioinformatics and high-flux sequencing have boomed in recent years, our sight on circRNAs has been notably broadened. Most circRNAs reported recently are non-coding, but some of them encode proteins or polypeptides according to reports.8, 9 Through non-canonical splicing, circRNAs generate with relative resistance to degradation by exonucleases.10, 11
Inextricable associations are found between circRNAs and the development of multiple disorders, especially that of cancer.12, 13 As circRNAs have gradually been extensively analysed, their functions as tumour oncogenes or suppressors are identified through adjusting the apoptosis, migration, proliferation, invasion and metastasis of tumour cells.14-17 For instance, circRNA hsa_circ_0001649 inhibits HCC progression via multiple miRNA sponge18; circRNA 0001785 functions as a competing endogenous RNA (ceRNA) to adjust the mechanism of osteosarcoma by sponging miR-1200 so as to elevate HOXB219; hsa_circ_0091570 acts as a ceRNA to impede the evolution of HCC by sponging hsa-miR-1307.20 However, the effect of circRNAs in NSCLC needs continuous explorations.
In the current research, the expression of circ-0008003 in NSCLC was discussed, and the function and potential regulatory mechanism of circ-0008003 were then investigated. The findings unfolded that circ-0008003 served as a ceRNA to mediate ZNF281 by sponging miR-488 in NSCLC.
2 MATERIALS AND METHODS
2.1 Tissue samples
Thirty NSCLC tissues and 30 matched para-cancerous tissues were collected from NSCLC cases subjected to excision in The Affiliated Huai'an Hospital of Xuzhou Medical University and The Second People's Hospital of Huai'an. Immediately after collection, liquid nitrogen was used to freeze all the samples prior to the extraction of total RNA. All participants signed the informed consent for scientific investigations. This research was conducted in line with the Helsinki Declaration, and it was monitored and approved by the Ethics Committee of The Affiliated Huai'an Hospital of Xuzhou Medical University and The Second People's Hospital of Huai'an.
2.2 Cell lines and transfection
The Cell Bank of Chinese Academy of Sciences provided five human NSCLC cell lines (CAL-12T, HCC827, A549, H460 and H1299) and one human bronchial epithelioid cell line (16HBE). They were cultured in RPMI 1640 medium (HyClone) with 1% penicillin/streptomycin (Gibco) and 10% foetal bovine serum (FBS; Pan Life Sciences) and maintained under a humid circumstance with 5% CO2 at 37°C.
Subsequently, 1 × 106 recombinant lentivirus-transducing units and 6 μg/mL Polybrene (Sigma) were utilized to infect cells in the case of the fusion of 60%-80%. Treatment with 2 μg/mL puromycin was adopted for the selection of cells with 2 weeks of stable transfection, and these cells were chosen through flow cytometry for later experiments. GenePharma provided lentiviruses and plasmids applied in this research.
2.3 Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
In line with the regimen of the manufacturer, the total RNA extraction from tissues and cells was performed using TRIzol reagent (Invitrogen), followed by qRT-PCR mentioned above.21 Specific primers used in this research were shown below:
GAPDH: forward: 5′-TGACTTCAACAGCGACACCCA-3′,
reverse: 5′-CACCCTGTTGCTGTAGCCAAA-3′;
miR-488: forward: 5′-ACACTCCAGCTGGGTTGAAAGGCTATTTC-3′,
reverse: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAG
TTGAGGACCAAGA -3′
circ-0008003: forward: 5′-TGGGGCTAACAAACTTCACC-3′,
reverse: 5′-GTCCCACCAGAGGTTGTCAC-3′.
2.4 Cell proliferation assay
Based on the regimen of the manufacturer, cell proliferation was examined via Cell Counting Kit-8 (CCK8) (Beyotime) assay. Following 24 hours of culture in a 96-well plate, the cells were subjected to 1 hour of incubation with CCK8 in an incubator for cell culture. A TECAN infinite M200 Multimode microplate reader (TECAN) was employed to test the cell optical density at 450 nm.
For EDU assay, EDU labelled solution (KeyGen Biotech) was used. Nucleus staining was performed by adding DAPI.
2.5 Cell invasion assay
By reference to the regimen of the manufacturer, the invasion capacity of cells was assessed by Transwell assay. Transwell chambers (Millipore Corporation) pre-coated with matrigel were then used. During invasion assessment, the upper chamber was inoculated with 2 × 105 cells in medium with no serum, whereas the lower chamber was added with 600 μL of medium with 10% FBS as a chemoattractant. Following 24 hours of incubation, cells subjected to migration were fixed and dyed with a 5% crystal violet solution (Beyotime), and the images of cells undergoing migration were captured by an optical microscope (Olympus Corporation). Moreover, Image-pro Plus 6.0 (Media Cybernetics) was utilized to count the migrated cells.
2.6 Luciferase reporter assay
Wild-type plasmids circ-0008003-WT and ZNF281-WT containing binding sites for miR-488 and mutant plasmids circ-0008003-MUT and ZNF281-MUT were consolidated into the pGL3 promoter vector (GenePharma). After the seeding of NSCLC cells into a 24-well plate, the cells underwent cotransfection with 80 ng plasmids + 5 ng pRL-SV40 and 50 nmol/L miR-488 mimics or negative control (NC) with the help of Lipofectamine 2000 reagent. After that, the luciferase activity was assessed using the dual-luciferase reporter assay kit (Promega). All experiments were carried out thrice, respectively.
2.7 RNA reduction assay
RNAs were labelled with SP6/T7 RNA polymerase (Roche) and the Biotin RNA labelling mix (Roche Diagnostics) in vitro and treated with RNase-free DNase I (Roche). Then, RNeasy Mini Kit (Qiagen) was utilized for the purification of RNAs labelled with biotin (Bio-miR-488-Mut, Bio-miR-488-WT and Bio-NC). Thereafter, the mixed solution (50 pmol of biotinylated RNA and 1 mg NSCLC cell) was added with washed beads (6 mL) containing streptavidin agarose (Life Technologies), and 1 hour of incubation was carried out at room temperature. After the rinsing of beads, the amplification of eluted RNAs was conducted, followed by measurement of them via qRT-PCR.
2.8 RNA immunoprecipitation (RIP) assay
By reference to the guidance of the manufacturer, RIP assay was carried out with the use of the Magna RIP kit (Millipore). Specifically, lysis buffer (Gibco) containing RNase inhibitor was used to lyse H460 and H1299 cells. After that, protein A/G agarose beads with AGO2 antibody were added to incubate cell lysates, with normal rabbit IgG as NC. Then, the RNAs that were co-precipitated underwent washing, purification and examination using qRT-PCR.
2.9 RNA-Fluorescence In Situ Hybridization (RNA-FISH)
RNA-FISH was conducted using circ-0008003 probes conjugated with fluorescence by reference to the former research.22 Then, DNA probe sets (Biosearch Technologies) were employed for hybridization based on the regimen of Biosearch Technologies. Ultimately, a FV1000 confocal laser microscope (Olympus) was utilized for probing cells.
2.10 In vivo assays
Shanghai SLAC Laboratory Animal Co. Ltd. provided female nude mice aged 4 weeks, which were randomly allocated into two groups: 1 × 107 circ-0008003 shRNA H460 cells in 100 μL of medium with no serum were injected at the left flank in one group (n = 4), while control cells treated with NC were injected in the other group (n = 4). Afterwards, a calliper was applied to test and calculate the tumour size every other day on the basis of the formula: size = length×width2/2. Prior to tumour excision and photography, the mice were killed after 3 weeks. Then immunohistochemistry (IHC) staining was carried out with the fixed tumours. Based on the Care and Use of Laboratory Animals of the National Institutes of Health, assays were conducted.
2.11 Immunohistochemistry staining
Ki-67 antibody and PCNA antibody (Abcam) were used for IHC staining by reference to the regimen mentioned above.23
2.12 Statistical analysis
Experimental data (obtained from experiments conducted thrice) were expressed as the mean ± standard deviation (SD). Intergroup comparison was carried out using the Student's t test, and Kaplan-Meier (K-M) curves were plotted for the comparison of overall survival via log-rank test. To figure out the correlation between clinicopathological characteristics and circ-0008003 expression, Fisher's exact test or chi-square test was employed. Besides, miR-488's association with circ-0008003/ZNF281 expression was determined by Pearson's correlation analysis. P < .05 meant a statistical significance.
3 RESULTS
3.1 Abnormal circ-0008003 expression was observed in NSCLC tissues and cells
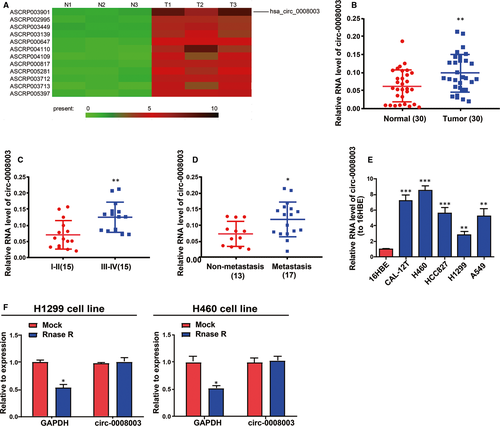
GSE112214 microarray analysis demonstrated that circ-0008003 displayed a high expression in NSCLC tissues. On the basis of the criteria of P < .05 and fold change >4, the different expression of circRNAs including circ-0008003 was confirmed in NSCLC tissues (Figure 1A). To verify the role of circ-0008003 in NSCLC, qRT-PCR was adopted to test its expression in NSCLC tissues and matched non-tumour tissues. According to the findings, NSCLC tissues had remarkably raised circ-0008003 expression relative to non-tumour tissues (Figure 1B). And circ-0008003 level in TNM stage Ⅰ-Ⅱ was evidently lower than that in TNM stage Ⅲ-Ⅳ (Figure 1C). Besides, tissues undergoing lymphatic metastasis had higher circ-0008003 expression than those undergoing no lymphatic metastasis (Figure 1D). Additionally, circ-0008003 relative expression in NSCLC cells was evidently raised relative to human bronchial epithelioid cell line 16HBE (Figure 1E). Because of the highest or lowest circ-0008003 expressions in H460 and H1299 cells, they were picked for later assays.24, 25 Circ-0008003 has resistance to RNase R digestion, so it was considered to possess circRNA features (Figure 1F). These results suggest the elevated aberrant expression of circ-0008003 and its role in NSCLC as an oncogene.
3.2 The promotive effect of circ-0008003 on NSCLC cellular processes
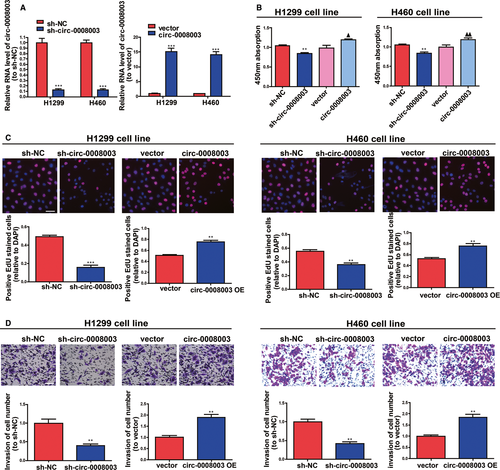
Because of the confirmed increase in circ-0008003 expression in NSCLC, its influences in NSCLC cell biological processes were tested by silencing or raising circ-0008003 expressions in H460 and H1299 cells. QRT-PCR findings illustrated that circ-0008003 was remarkably pulled down or raised in NSCLC cells after the addition of circ-0008003 shRNA or overexpressing vector (Figure 2A). According to CCK8 and 5-Ethynyl-2'-deoxyuridine (EDU) assays, the proliferation ability of NSCLC cells was decreased after silencing circ-0008003 and increased after up-regulating circ-0008003 (Figure 2B-C). Besides, the invasion abilities of the two cell lines became weak after silencing circ-0008003 and strong after up-regulating circ-0008003 unfolded by Transwell assay (Figure 2D). These data reveal that circ-0008003 promotes NSCLC cell invasion and proliferation in vitro.
3.3 Tumour growth in vivo was slowed down by reduced circ-0008003 expression
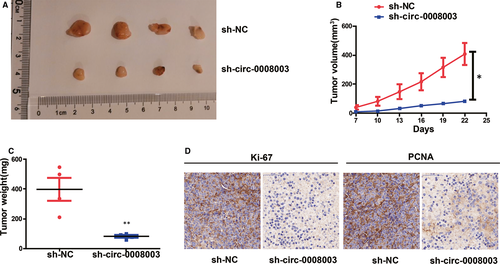
H460 cells treated with circ-0008003 shRNA or NC were used for in vivo experiments to continuously prove the effect of circ-0008003 in tumour formation of NSCLC. In comparison with those in NC group, repressed circ-0008003 resulted in relatively small xenografts, identical to the results in vitro (Figure 3A). Moreover, relative to those in control group, tumours derived from H460 cells with repressed circ-0008003 had limited weight and size (Figure 3B,C). And IHC staining was adopted to examine the expressions of Ki67 and PCNA, the markers of cell proliferation.26, 27 Consequently, there were substantially less stained PCNA and Ki67 in tumours in circ-0008003-repressed cell group relative to those in NC-treated H460 cell group (Figure 3D). It can be deduced that circ-0008003 repression inhibits tumour formation in vivo.
3.4 Circ-0008003 was considered to be an endogenous sponge for miR-488 in NSCLC
CircRNAs expressed in the cytoplasm have been demonstrated to substantially affect pathological and physiological processes by sponging miRNAs to some extent.28, 29 To ascertain the cellular localization of circ-0008003, cells were isolated into the nucleus and cytoplasm fractions, in which the controls, U6 and GAPDH, could be seen. QRT-PCR validated that circ-0008003 was monitored mainly in the cytoplasm fractions in NSCLC cells (Figure S1A). Identically, a substantial staining of circ-0008003 in the cytoplasm of H1299 cells was also determined in RNA-FISH (Figure S1B). It can be seen that circ-0008003 probably functions in NSCLC at the post-transcriptional level. To forecast circ-0008003 miRNAs with underlying interaction, the online bioinformatics tool Circinteractome was employed. As a result, circ-0008003 was unveiled to bind to miR-488 with the highest potential, and the sequences of wild-type circ-0008003 (with forecasted binding sites), mutant circ-0008003 (with mutant sites) and miR-488 were displayed in Figure 4A. Subsequently, circ-0008003's interaction with miR-488 primarily appeared in the cytoplasm rather than the nucleus of H460 and H1299 cells (Figure S1C,D), thus validating the above forecasting.
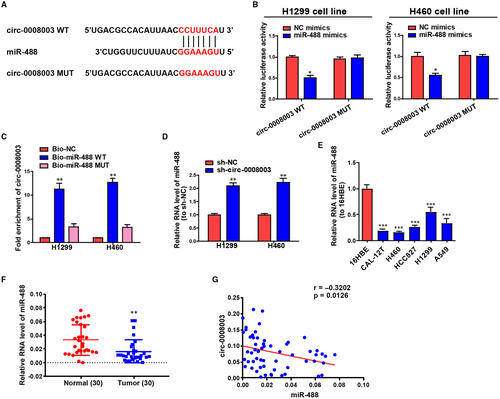
Then, luciferase reporter assay was conducted to ascertain the above-mentioned interaction. It was illustrated that the luciferase activity of circ-0008003-WT could be repressed by miR-488 mimics, but that of circ-0008003-MUT remained unchanged (Figure 4B). According to the RNA reduction experiment, circ-0008003 only gathered in complexes repressed not by Bio-miR-488-Mut or Bio-NC but by Bio-miR-488-WT (Figure 4C). And repressing circ-0008003 expression led to the elevated level of miR-488 in H460 and H1299 cells (Figure 4D). Further, miR-488 expression in NSCLC cells and tissues was investigated through qRT-PCR, the results of which ascertained that miR-488 was markedly pulled down in NSCLC cells and tissues relative to the controls (Figure 4E,F). Eventually, the inverse relationship between the level of miR-488 and circ-0008003 expression was uncovered by Pearson's correlation analysis in NSCLC patients (Figure 4G). These findings uncover that circ-0008003 may promote NSCLC progression through the inverse adjustment of miR-488 expression.
3.5 Aberrant miR-488 expression slowed down NSCLC progression
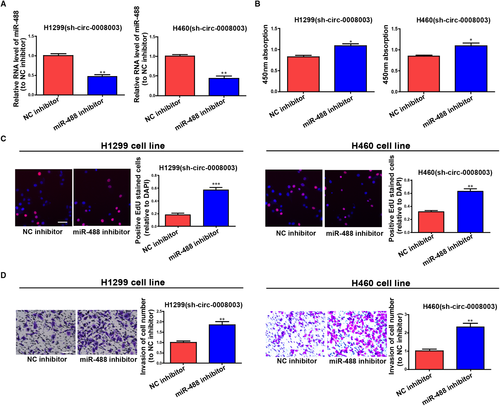
Figure S2A illustrated that the expression of miR-488 was greatly inhibited or enhanced in NSCLC cells treated with miR-488 inhibitor or mimics relative to that in cells treated with NC. Besides, miR-488 impeded cell invasion and proliferation (Figure S2B-D). Collectively, the results indicate that miR-488 influences NSCLC evolution as a tumour suppressor.
3.6 Circ-0008003 accelerated NSCLC development by repressing miR-488
To ascertain that the role of circ-0008003 required miR-488 reduction in a direct manner, miR-488 inhibitor was transfected into circ-0008003-repressed H460 and H1299 cells, and the observably reduced miR-488 was discovered by qRT-PCR (Figure 5A). A foregone conclusion was obtained from CCK8 and EDU assays that the proliferation capacities of circ-0008003-repressed H460 and H1299 cells were promoted by miR-488 suppression (Figure 5B,C). Lastly, silencing miR-488 was proved by Transwell assay to evidently boost the invasion capacities of circ-0008003-repressed H460 and H1299 cells (Figure 5D). The above discovery implies that miR-488 participates in the carcinogenic process regulated by circ-0008003 in NSCLC cells.
3.7 ZNF281 was a direct target of miR-488
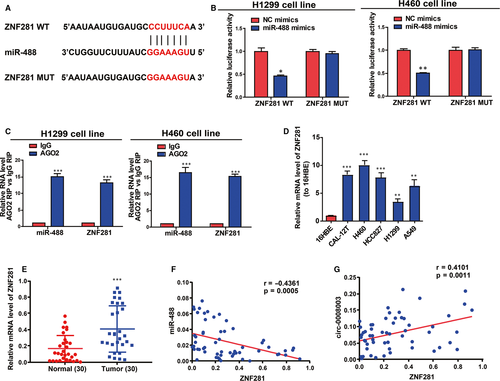
Next, the specific potential mechanism of circ-0008003/miR-488 axis in stimulating NSCLC evolution was discussed. With bioinformatics prediction tools (TargetScan, miRDB and Starbase), ZNF281, a carcinogene in cancers, was found to be a target of miR-488 (Figure 6A). Besides, the direct binding of miR-488 to ZNF281 3'UTR at the forecasting sites was ascertained in NSCLC cells (Figure 6B). From immunoprecipitate conjugated with Ago2 in H460 and H1299 cells, miR-488 and ZNF281 mRNA were obtained, validating the existence of the interaction between miR-488 and ZNF281 mRNA in RNA-induced silencing complex (RISC) adjusted by miR-488 (Figure 6C). Next, the expression of ZNF281 was disclosed to notably rise in NSCLC cells and tissues relative to controls (Figure 6D-E). What's interesting was that miR-488 level was shown as an inverse correlator while circ-0008003 expression as a positive correlator of ZNF281 expression in NSCLC tissues (Figure 6F,G). As a result, ZNF281 may affect circ-0008003/miR-488 axis in NSCLC evolution.
3.8 ZNF281 positively adjusted circ-0008003 in NSCLC evolution
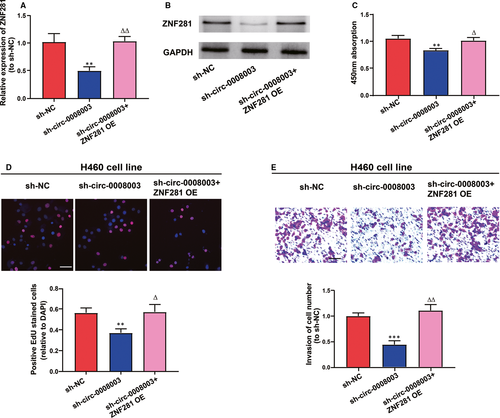
To figure out the adjustment of ZNF281 on the role of circ-0008003 in NSCLC evolution, the function of ZNF281 overexpression in the biological processes of circ-0008003-repressed H460 cells was analysed. In the first place, it was revealed that mRNA and protein expression levels of ZNF281 were remarkably impeded after the repression of circ-0008003 and became normal after cotransfection with ZNF281 overexpression vector (Figure 7A,B). Afterwards, circ-0008003-repressed H460 cells predominantly regained their repressed proliferation abilities following cotransfection with ZNF281 overexpression vector (Figure 7C,D). Additionally, ZNF281 overexpression alleviated the hampering of circ-0008003-repressed invasion processes in H460 cells (Figure 7E). Basically, circ-0008003 facilitates NSCLC evolution via circ-0008003/miR-488/ZNF281 axis.
4 DISCUSSION
CircRNAs have been demonstrated to substantially influence different biological processes in cells.30, 31 According to reports, aberrant adjustment of circRNAs exerts effects on cancer evolution and is associated with patient's prognosis.32, 33 CircRNAs have been recently proved to pivotally affect NSCLC.34 For example, circRNA hsa_circ_0008305 (circPTK2) impedes EMT and metastasis triggered by TGF-β through controlling TIF1γ in NSCLC35; circRNA F-circEA-2a originated from EML4-ALK fusion gene boosts invasion and migration of NSCLC cells.36 In this research, elevation of circ-0008003 in NSCLC tissues and cells was discovered, and high level of circ-0008003 had a positive relationship to poor prognosis, TNM stage and lymphatic metastasis. Moreover, repression of circ-0008003 hampered tumour growth in vivo as well as the cell invasion and proliferation in vitro. The findings validate that circ-0008003 affects NSCLC development and progression like an oncogene.
Verified by multiple reports, circRNAs adjust different cancers through targeting mRNAs or miRNAs.37, 38 CircRNA hsa_circ_0091570 blocks HCC progression by sponging hsa-miR-1307 like a ceRNA.20 CircRNA hsa_circRNA_100290 adjusts cell growth in oral squamous cell carcinoma by GLUT1 and glycolysis like a ceRNA for miR-378a.39 CircRNA CircCACTIN accelerates the evolution of gastric cancer by sponging miR-331-3p and mediating the expression of TGFBR1.40 Also, circRNAs serve as an endogenous sponge for miRNAs in NSCLC. For instance, circRNA 100146 exerts an oncogenic effect by binding to miR-615-5p and miR-361-3p in NSCLC in a direct manner.41 Currently, it was uncovered that circ-0008003 with expression primarily in the cytoplasm accelerates NSCLC evolution by sponging miR-488 that represses NSCLC,42, 43 which was identified as a suppressor gene in NSCLC in this research.
ZNF281 is located at chromosome 1q32.1 and is a critical regulator of embryonic stem cell differentiation and tissue development.44 Multiple studies have demonstrated the diverse roles of ZNF281 in stem cell pluripotency and development processes.45, 46 Therefore, ZNF281 is regarded as a relevant gene for cellular stemness. Notably, recent investigations have revealed that ZNF281 is a novel oncoprotein that is frequently overexpressed in human malignancies, including colorectal and pancreatic cancers.47, 48 More importantly, high ZNF281 expression in cancer tissues enhances metastatic potential and reduces patient survival.46, 49, 50 However, its actual role and underlying mechanism in NSCLC have not yet been elucidated. This research showed that ZNF281 was a target of miR-488 and circ-0008003 affected NSCLC via adjusting miR-488/ZNF281 signalling like an oncogene.
To sum up, circ-0008003 plays a carcinogenic role in NSCLC development via sponging miR-488, thus releasing and increasing ZNF281. This research firstly evidences circ-0008003's identities as an underlying treatment marker and a prognostic target in NSCLC, but its clinical significance still needs in-depth investigations in the future.
ACKNOWLEDGEMENTS
This work was supported by the grants from the Foundation of the Top-Notch Scientific Research of the High-Level Health Talents Project (Grant No. LGY2018042).
CONFLICT OF INTEREST
The authors state that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
Runhuan Gu: Investigation (equal); Writing-original draft (equal). Koufeng Shao: Methodology (equal); Validation (equal). Qiaoxia Xu: Data curation (equal); Software (equal); Visualization (equal). Xue Zhao: Formal analysis (equal); Software (equal); Visualization (equal). Haibing Qiu: Project administration (equal); Resources (equal); Writing-review & editing (equal). Haibo Hu: Conceptualization (lead); Resources (lead).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




