Optimized monoclonal antibody treatment against ELTD1 for GBM in a G55 xenograft mouse model
Abstract
Glioblastoma is an aggressive brain tumour found in adults, and the therapeutic approaches available have not significantly increased patient survival. Recently, we discovered that ELTD1, an angiogenic biomarker, is highly expressed in human gliomas. Polyclonal anti-ELTD1 treatments were effective in glioma pre-clinical models, however, pAb binding is potentially promiscuous. Therefore, the aim of this study was to determine the effects of an optimized monoclonal anti-ELTD1 treatment in G55 xenograft glioma models. MRI was used to assess the effects of the treatments on animal survival, tumour volumes, perfusion rates and binding specificity. Immunohistochemistry and histology were conducted to confirm and characterize microvessel density and Notch1 levels, and to locate the molecular probes. RNA-sequencing was used to analyse the effects of the mAb treatment. Our monoclonal anti-ELTD1 treatment significantly increased animal survival, reduced tumour volumes, normalized the vasculature and showed higher binding specificity within the tumour compared with both control- and polyclonal-treated mice. Notch1 positivity staining and RNA-seq results suggested that ELTD1 has the ability to interact with and interrupt Notch1 signalling. Although little is known about ELTD1, particularly about its ligand and pathways, our data suggest that our monoclonal anti-ELTD1 antibody is a promising anti-angiogenic therapeutic in glioblastomas.
1 INTRODUCTION
Of all malignant gliomas diagnosed in adults, 82% are characterized as glioblastoma (GBM), which has an incidence of 3.19 per 100 000 persons in the United States.1, 2 This high-grade glioma undergoes unregulated vascular angiogenesis and is characterized as being invasive, highly vascular and resistant to apoptosis.3 The current treatment plan is surgical resection followed by a combination of radiation and chemotherapy with temozolomide or bevacizumab.4 However, even with treatments, the median survival for patients is only 12-14 months post-detection, and <5% of patients survive past 5 years post-diagnosis.2, 4 GBMs undergo gene amplification and/or mutation of the epidermal growth factor (EGF) receptor and higher EGFR levels were shown to promote migration, tumour growth and angiogenesis.5 Gliomas rely heavily on angiogenesis for tumour growth, and the new vessels are the key for delivering oxygen and nutrients to the tumour site. Throughout the years, the primary focus among the pro-angiogenic factors was the vascular endothelial growth factor (VEGF) for its role of increasing vascularization in cancer. While the tumour develops, there is an up-regulation of pro-angiogenic cytokines in the region that further increase VEGF-A, along with other microvasculature proliferation factors such as basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF).6 Once up-regulated, VEGF-A binds onto VEGF receptor 2 (VEGFR2) on endothelial cells to initiate a cascade of signalling pathways that promote the formation of new blood vessels.3 Bevacizumab is a monoclonal therapy against VEGF-A approved as a GBM therapeutic agent along with multiple other cancers.4 However, this chemotherapeutic agent has not significantly increased the survival of patients suffering with GBM. Furthermore, bevacizumab has serious adverse side effect such as severe/fatal haemorrhaging that occurs up to fivefold more frequently.7 Due to Bevacizumab's failure to increase patient's survival, it was crucial to shift the focus from VEGF to other angiogenic factors present in GBMs.
The epidermal growth factor, latrophilin, and seven transmembrane domain-containing protein on chromosome 1 (ELTD1), alternatively known as the adhesion G protein-coupled receptor L4 (ADGRL4), was first discovered in developing cardiomyocytes.8 ELTD1, a novel regulator of brain angiogenesis, was found to promote tumour growth and metastasis.9 We previously reported that ELTD1 was highly expressed in high-grade gliomas and was expressed on both endothelial and tumour cells.8 Furthermore, ELTD1 expression was shown to be regulated by the two main angiogenic pathways, where VEGF increased ELTD1 expression, and DLL4-Notch signalling decreased ELTD1 expression in normal vasculature.9 Further investigation into ELTD1 demonstrated that increased signalling from VEGF-A resulted in an increase of ELTD1 expression in endothelial cells, and that targeting ELTD1 had decreased VEGFR2 expression in a glioma model.10, 11
There are approximately 17 000 new GBM diagnoses every year, increasing the need for new and more effective cancer therapeutics.1 Our group found that polyclonal antibody (pAb) treatments against ELTD1 in orthotropic GL261 and human G55 xenograft glioma pre-clinical models were successful in decreasing tumour volumes (TV), increasing survival and decreasing microvessel density levels (MVD) when compared to untreated (UT) control.12 However, batch-to-batch variabilities as well as potential promiscuity of the pAb posed concerns about specificity as long-term treatment for patients. Monoclonal antibodies (mAb) are produced from a single B cell clone that allows for homogeneous antibodies and are established as a successful class of targeted treatments for various cancers and chronic inflammatory diseases.13 Emerging mAb treatments bind onto growth factors overexpressed on the tumour to disrupt downstream signalling effects to decrease tumour cell growth, proliferation and migration.14
Previous research has demonstrated that pAb treatment against ELTD1 was an effective treatment in GBM pre-clinical models. This study used an optimized monoclonal antibody (mAb) against ELTD1 that has a higher specificity by only binding to the external region of the receptor (430 AA) overcoming the limitations set by the pAb treatments in hopes of obtaining a more specific and profound effect on a G55 glioma pre-clinical model.
2 MATERIALS AND METHODS
2.1 Preparation of recombinant extracellular domain of ELTD1 human Ckappa fusion protein
To construct extracellular domain of human ELTD1 and mouse ELTD1 expression vectors, genes encoding the human ELTD1 (Glu20-Leu406) and mouse ELTD1 (Glu20-Leu455) were chemically synthesized (Genscript, Picataway, NJ, USA). The genes were subcloned into the modified pCEP4 vector encoding Cκ domain (human immunoglobulin κ light chain constant domain) at the 5′ region as reported previously.15
The expression vectors encoding extracellular domain of human ELTD1 and mouse ELTD1 were transfected into HEK293F cells (Invitrogen, Carlsbad, CA, USA) using 25-kD linear polyethyleneimine (Polyscience), as reported previously.16 Human and mouse ELTD1 Cκ fusion proteins were purified from the culture supernatants by affinity chromatography using KappaSelect resin (GE Healthcare) according to the manufacturer's instructions.
2.2 Generation of anti-ELDT1 antibody
White leghorn chickens were immunized with human ELTD1 Cκ fusion proteins. A phage-displayed chicken single-chain variable fragment (scFv) library was constructed using total RNA isolated from the bone marrow, spleen and bursa of Fabricius of immunized chickens, as described previously.17 Positive clones were enriched by biopanning and screened in a phage enzyme immunoassay, as described previously.18 Phage clones showing cross reactivity against human and mouse ELTD1 were selected, and their nucleotide sequences were determined by Sanger sequencing. The gene of selected scFv clone was subcloned into a modified mammalian expression vector encoding the hinge region of human IgG1 and the CH2-CH3 domains of rabbit IgG at the 3′ region as reported previously.19 The expression vectors encoding anti-ELTD1 scFv-rFc fusion were transfected into HEK293F cells (Invitrogen) as described above. The scFv-rFc fusion protein was purified from the culture supernatants of transiently transfected HEK293F cells using protein A Sepharose column (Repligen) according to the manufacturer's instructions.
2.3 Enzyme immunoassay
96-well microtiter plate wells (Corning Inc, Corning, NY, USA) were coated with human ELTD1 or mouse ELTD1 Cκ fusion protein in coating buffer (0.1M NaHCO3, pH 8.6) and then blocked with 3% (w/v) BSA in phosphate-buffered saline (PBS). After incubation with serially, 10-fold diluted anti-ELTD1 scFv-rFc fusion protein (0.01-100 nM) and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Fc specific) (Jackson Immuno Research, Inc) were added to each well. After washing with 0.05% (v/v) Tween 20 in PBS (PBST), ABTS HRP substrate solution (Thermo-Scientific Pierce) was added and the absorbance was measured at 405 nm with a Multiscan Ascent microplate reader (LabSystems).
2.4 G55 Xenograft model and treatment
All animal studies were conducted with the approval (protocol 17-48) of the Oklahoma Medical Research Foundation Institutional Animal Care Use Committee policies, which follow NIH guidelines. Human G55 xenograft cells were implanted intracerebrally in 2-month-old male mice (Hsd:Athymic Nude-Foxn1nu mice; Harlan Inc), as previously described.10, 12 The animals were divided into three groups: UT, pAb and mAb anti-ELTD1 treated. Once tumours reached 6-7 mm3 (determined via MRI), mice were either left UT or were treated with 2 mg/kg of either polyclonal anti-ELTD1 (Bioss, ETL/ELTD1 Polyclonal Antibody, bs-13111R) or an optimized mAb against ELTD1 every 3-4 days (treated M/Th, T/F, W/Sat). All mice were euthanized when tumours reached ≥150 mm3.
2.5 In vivo magnetic resonance (MR) techniques
2.5.1 Morphological imaging
Mice were anesthetized and positioned in a cradle. A 30-cm horizontal bore Bruker Biospin magnet operating at 7 T (Bruker BioSpin GmbH) was used. A BA6 gradient set and mouse head coil were used to perform all MRI experiments as previously described.12 All animals were imaged every 2-3 days until the end of the study starting at 10 days post-G55 implantation surgery.
2.5.2 Perfusion imaging
The perfusion imaging method, arterial spin labelling, was used as previously described.20 Perfusion maps were obtained on a single axial slice of the brain located on the point of the rostro-caudal axis where the tumour had the largest cross section. Five regions of interest (ROIs) were manually outlined around the tumour, and appropriate ROIs were also taken from the contralateral side of the brain for comparison purposes. To calculate the differences in (rCBF) values, tumour rCBF values were obtained at late (prior to termination) and early (at tumour detection) tumour stages and normalized to rCBF values in the contralateral brain region of corresponding animals.
2.5.3 Molecular-targeted MR imaging (mt-MRI)
The contrast agent, biotin-BSA (bovine serum albumin)-Gd (gadolinium)-DTPA, was prepared as previously described by our group,12 based on the modification of the method developed by Dafni et al21, 22 pAb anti-ELTD1 (Bioss) or mAb anti-ELTD, were conjugated to the albumin moiety through a sulfo-NHS-EDC link according to the protocol of Hermanson.23 mt-MRI was performed when tumour volumes were around 130-180 mm3. Molecular probes with a biotin-albumin-Gd-DTPA construct bound to anti-ELTD1 antibodies were injected via a tail vein catheter in mice. A non-specific mouse immunoglobulin IgG Ab (Alpha Diagnostics) was used with the biotin-albumin-Gd-DTPA construct as a negative control. MRI was done as previously described.10, 21 Relative probe concentrations were calculated to assess the levels of ELTD1and the non-specific IgG contrast agent in each animal. Contrast difference images were created from the pre- and (90 minutes) post-contrast datasets for the slice of interest, by computing the difference in T1 relaxation times between the post-contrast and the pre-contrast image on a pixel basis. From difference images, ten ROIs of equal size (0.05 cm2) were drawn within areas with the highest T1 relaxation at the TR 800 ms, in the tumour parenchyma and contralateral side of the brains of each animal, after anti-ETLD1 probe injections. T1 values obtained from the ROIs in the tumour regions were normalized to the corresponding contralateral sides. The T1 relaxation values of the specified ROIs were computed from all pixels in the ROIs, by the following equation taken (processed by ParaVision 5.0, Bruker): S (TR) = S0 (1 − e−TR/T1), where TR is the repetition time, S0 is the signal intensity (integer machine units) at TR, T1 and TE = 0 and T1 is the constant of the longitudinal relaxation time.24 Overlays of contrast difference images and T1-weighted images were generated using Photoshop software (version C.S 6).
2.6 Immunohistochemistry and standard staining
All mice were euthanized after the last MRI examination. The brain of each animal was removed, preserved in 10% neutral buffered formalin, and processed routinely. Haematoxylin-eosin staining: tissues were fixed in 10% neutral buffered formalin, dehydrated and embedded in paraffin. Sections were deparaffinized, rehydrated and stained according to standard protocols. Several reagents were produced by Vector Labs Inc (VLI).
Histological sections (5 µm) embedded in paraffin and mounted on HistoBond®Plus slides (Statlab Medical Products) were rehydrated and washed in phosphate-buffered saline (PBS). The sections were processed using the ImmPRESS™ VR Reagent Anti-Rabbit IgG Peroxidase (VLI cat #MP-6401). Antigen retrieval (pH6 citrate antigen unmasking solution; VLI cat#H-3300) was accomplished via 20 minutes in a steamer followed by 30 minutes cooling at room temperature. Sections were treated with a peroxidase blocking reagent (Bloxall, VLI cat#SP-6000), followed by 2.5% normal horse serum to inhibit non-specific binding. Rabbit Anti-CD34 antibody (abcam81289; 5.28 µg/mL) or Rabbit Anti-NOTCH 1 (abcam52627; 11 µg/mL) was applied to each section and following incubation overnight (4°C) in a humidified chamber, sections were washed in PBS, the ImmPRESS VR reagent was applied according to the manufacturer's directions.
To characterize MVD and Notch expression levels, five ROIs, captured digitally (20×), were identified in each case. Only areas containing tumour tissue were analysed, excluding areas with necrosis and/or significant artifacts. The number of positive pixels was divided by the total number of pixels (negative and positive) in the analysed area. ROIs were analysed and imaged using Aperio ImageScope (Leica Biosystems).
Sections for streptavidin horse radish peroxidase (SA-HRP) were processed as above, except they were incubated overnight with ready to use (RTU) Strp-HRP (VLI cat#SA-5704). Appropriate washes were in PBS. Slides were incubated with NovaRed® (VLI cat#SK-4805) chromogen for visualization. Counterstaining was carried out with Hematoxylin QS Nuclear Counterstain (VLI). Appropriate positive and negative tissue controls were used.
2.7 RNA isolation and preparation
Mice were euthanized after the last MRI examination. Brains were removed, snap frozen and stored at −80°C. Total RNA from tumour tissues from all groups was purified with a RNeasy Mini Kit (Qiagen) and quantified by spectrophotometry (Nanodrop).
Concentration of RNA was ascertained, and overall quality of RNA was verified. Sequencing libraries were generated (Lexogen Quantseq FWD library prep kit) according to the manufacturer's protocol. Briefly, the first strand of cDNA was generated using 5′-tagged poly-T oligomer primers, and following RNase digestion, the second strand was generated using 5′-tagged random primers. A subsequent PCR step with additional primers added the complete adapter sequence to the initial 5′ tags, added unique indices for demultiplexing of samples and amplified the library. Final libraries for each sample were assayed (Agilent Tapestation) for appropriate size and quantity. These libraries were pooled in equimolar amounts (fluorometric analyses). Final pools were absolutely quantified using qPCR (Roche LightCycler 480 instrument with Kapa Biosystems Illumina Library Quantification reagents). Sequencing was performed (Illumina Nextseq 500 instrument) with High Output chemistry and 75-bp single-ended reads.
2.8 Bioinformatics analysis
Paired-end fastq files were checked for quality using multiQC,25 for which the following mean (standard deviation) descriptive values were 54 million reads (12), 68.8% (2.8%) duplicate reads and a GC content of 51.3% (0.7%). The indexing and alignment were run with kallisto26 against build 38 of the human reference genome from the Genome Reference Consortium (GRCh38). Assignment of counts to exon features and normalization were performed along with the alignment via the biojupies pipeline to provide a counts matrix. Significant differential genes were determined by DESeq227 for genes having both a Benjamini-Hochberg adjusted P-value < .05 and an absolute log fold change of >1.3. Gene set enrichment analysis was performed on the identified differential genes via enrichR28 (gseapy API).
2.9 Statistical analysis
Survival curves were analysed using Kaplan-Meier curves. Tumour volumes, perfusion changes, and immunohistochemistry protein levels, and molecular-targeted MRI data were analysed and compared by one- or two-way ANOVA with multiple comparisons (Tukey's or Sidak's, respectively). Data were represented as mean ± SD, and P-values of either *<.05, **<.01, ***<.001, ****<.0001 were considered statistically significant.
3 RESULTS
Prior studies demonstrated that non-specific IgG antibody treatments as a control group did not differ from untreated; therefore, in this study, we only utilized untreated animals as the control group.12 G55 glioma-bearing mice treated against ELTD1, both pAb (P = .0207) and mAb (P = .0024), significantly increased the survival compared with untreated (average survival ~ 9 days post-tumour detection) as shown in Figure 1A. TVs 9 days post-tumour detection, monitored via MRI, were significantly lower with the monoclonal (P = .0067) and polyclonal (P = .0384) anti-ELTD1 treatment compared with controls (Figure 1B). Representative images of tumour-bearing mice from all treatment groups are shown in Figure 1C.
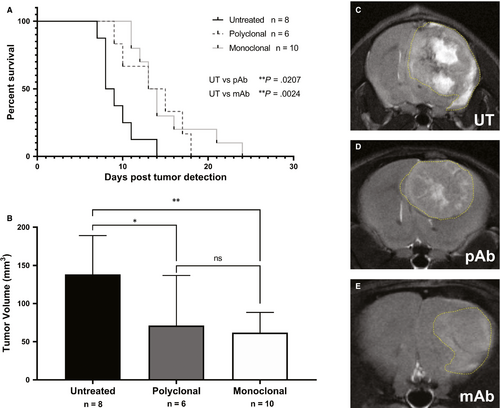
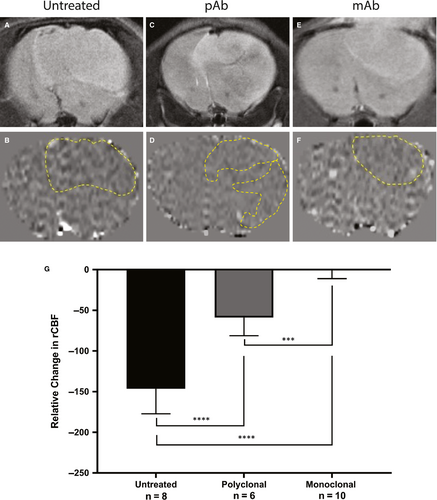
MRI perfusion measures the relative cerebral blood flow (rCBF) and can be used to assess the microvasculature alterations associated with tumour angiogenesis. Healthy normal tissue has a set rCBF; however, as the tumour grows, it disrupts the vasculature, and therefore decreases the perfusion rate. Differences in rCBF demonstrated that the untreated mice had a decrease in rCBF in the tumour regions depicting increased angiogenesis while the anti-ELTD1-treated animals had a normalization of perfusion values. Representative morphological MRIs along with the corresponding perfusion maps of the brain are depicted in Figure 2A-F. The decrease in perfusion (depicted as decreased normalized rCBF) as a result of the tumour, outlined by the yellow-dashed line in 2B, 2D, 2F, is demonstrated by the dark areas. Our monoclonal and polyclonal treatments were successful in decreasing angiogenesis and therefore increasing perfusion within the tumour region. The polyclonal anti-ELTD1 treatment was able to minimize the decrease in rCBF (P < .0001) compared with UT mice. The mAb treatment was significantly more effective in decreasing the rCBF compared with both pAb treatment (P = .0001) and UT (P < .0001) (Figure 2G) and normalized the rCBF within the tumour region to contralateral levels.
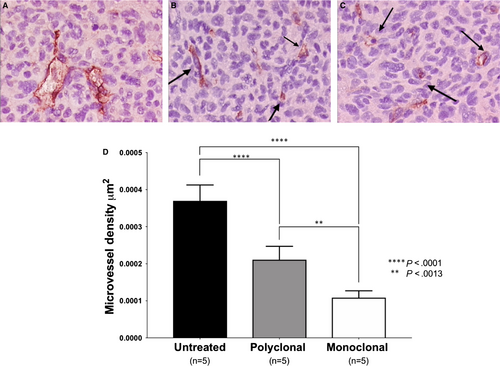
ELTD1 has been linked with pathological angiogenesis. Therefore, we analysed MVD to determine whether the anti-ELTD1 treatments would alter the tumour vasculature. Representative CD34 IHC images for each treatment groups are shown in Figure 3A-C. CD34 analysis demonstrated that the anti-ELTD1 treatments significantly decreased the MVD levels (P < .0001) compared with control (Figure 3D). The monoclonal anti-ELTD1 treatment further decreased the MVD levels compared with the polyclonal anti-ELTD1 treatment (P = .0013) and was able to return the MVD to near normal levels.
To determine where our Ab was localizing in vivo, we synthesized a biotin-BSA (bovine serum albumin)-Gd-DTPA probe attached to either non-specific IgG, polyclonal anti-ELTD1 or monoclonal anti-ELTD1 antibodies (Figure 4A). The molecular probes were injected via tail vein catheter into untreated G55-glioma-bearing mice, T1 relaxation times and signal intensity were calculated via MRI. T1 relaxation is an MRI contrast parameter that is reduced in the presence of our molecular probe. The results shown in Figure 4B demonstrate the presence of our molecular probe as the per cent relative expression, due to its effect on T1 relaxation. Both T1 (P = .0002) and signal intensity (P = .008) were significantly increased by the monoclonal anti-ELTD1-attached probe compared with the non-specific IgG-attached probe. Our polyclonal anti-ELTD1-attached probe significantly increased the T1 relaxation (P = .0307) but did not significantly affect the signal intensity (P = .0602).
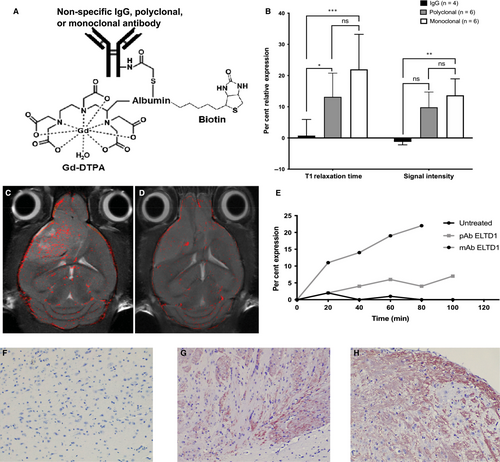
Molecular-targeted MRIs of representative monoclonal anti-ELTD1 and non-specific IgG probe data were overlaid onto morphological images of untreated G55 tumour-bearing animals. Figure 4C demonstrates that the monoclonal anti-ELTD1-attached probe had an increased binding specificity against the tumour region. However, the non-specific IgG probe mainly clustered around the blood vessels (Figure 4D). After monitoring the expression of the molecular probes, we saw that the mAb-attached probe had a more profound and sustained effect when compared to both the non-specific IgG and polyclonal anti-ELTD1-attached probe (Figure 4E).
Our molecular probe had an attached biotin tag to further localize it in the tissue. Once molecular targeting was concluded, the animal was terminated and their tissue was taken for histology. By staining the tumour tissue with SA-HRP, we confirmed our molecular targeting results. Our polyclonal- and monoclonal-attached probes were localized in the tumour tissue post-termination while there were no traces of our non-specific IgG-attached probes in the tissue. (Figure 4F-H). These data demonstrate that the mAb probe has significantly higher binding specificity against the tumour region than both the polyclonal-attached probe and IgG probe.
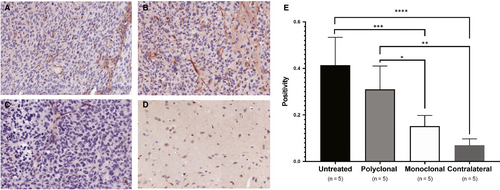
Notch signalling is important for cell differentiation, proliferation, as well as tumour angiogenesis, and normal vasculature has been shown to decrease ELTD1 expression.29 Therefore, we examined whether Notch levels changed with anti-ELTD1 treatment. Positivity analysis of the stained samples demonstrated that the untreated glioma tumour samples had the highest amount of Notch1. Our monoclonal anti-ELTD1 treatment significantly decreased Notch1 expression levels compared with both pAb treatments (P = .0357) and untreated control (P = .0006) and brought down the expression to contralateral levels (Figure 5E).
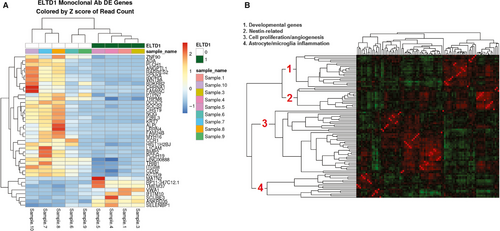
Our in vivo data demonstrated that the mAb treatment against ELTD1 was more effective in the G55 xenograft model; therefore, we only examined the effect that our mAb treatment (compared to untreated) had on the genes in the tumour region. From all of the genes found in Figure 6A, ADA, SCN5A, L1CAM, BMP2, ALPL, TRPM8, SELENBP1 have been directly associated with gliomas. While other genes were associated with various other cancers such as hepatocellular carcinoma (VWA130), lung cancer (SCUBE3,31 PLCH1,32 CHRNA1,33 CDH234) and breast cancer (IFITM10,35 DCDC2,36 CHST9,37 CDH238). To see whether some of the genes down-regulated upon anti-ELTD1 Ab treatment had been similarly co-regulated in other experiments, we first calculated gene-gene Pearson's correlations using experiments from the microarray platform GPL570, which are publicly available as part of NCBI's GEO database. Figure 6B shows the clustered gene-gene correlations of our down-regulated genes using the GPL570 data. Roughly, 4 clusters (developmental genes, nestin-related, cell proliferation/angiogenesis, astrocyte microglia inflammation) are apparent, indicating that groups of genes seen as differentially expressed in our experiment have also been observed in other experiments.
4 DISCUSSION
Through a global microarray meta-analysis (GAMMA),39 we identified ELTD1, an angiogenic marker, to be highly expressed in high-grade gliomas and other groups have suggested that high ELTD1 expression levels may correlate with the aggressiveness of the glioma.29, 40 Previous studies have demonstrated that anti-ELTD1 treatments with pAb were effective in mouse GL261 and human G55 xenograft glioma models.12 Other groups have also discovered that microRNA-139-5p directly binds onto and targets ELTD1 to inhibit cell proliferation in gliomas.41
This study focuses on an optimized mAb therapy against ELTD1 in a human G55 xenograft glioma mouse model. G55 is a stable xenograft cell line that was initially taken from a human GBM and passaged through nude mice.42, 43 Historically, this cell line has many characteristics of primary human GBM such as hypervascularity and necrosis and has been used by numerous studies focusing on invasive intracranial tumours.42-45 Our data have shown that repetitive IV treatments with both pAb and mAb against ELTD1 led to a significant decrease in tumour volumes and increase in survival. Prior published work from our laboratory showed a survival increase of 7-10 days with the pAb ELTD1 treatment; however, this current study only showed an average increase of 5 days.12 The discrepancy between studies is due to the different doubling times between our G55 cells. The 2017 study used high-passaged G55 cells in which the untreated mice had a doubling time of 2.5 days with an average survival of 18 days; however, this current study used low-passaged G55 cells that appeared more aggressive due to their faster doubling period of 2 days and an average survival of 10 days. The mAb treatment against ELTD1, however, was able to increase the doubling time to approximately 2.7 days even in this more aggressive glioma model.
Our optimized mAb treatment was shown to not only return the perfusion levels, depicting a normalization of the vasculature within the tumour region, but was also effective in significantly decreasing the MVD levels. Together, these data demonstrate that our mAb therapy against ELTD1 has a more profound effect on the tumour-related microvasculature when compared to both untreated and pAb-treated animals. Prior studies using human G55 cells have shown increased haemorrhaging using anti-VEGF therapy in mice, and patients undergoing bevacizumab treatment have shown an increased risk of intracranial haemorrhaging.7, 12 We did not see any haemorrhaging in our pAb- nor mAb-treated tumour regions on our MR images nor when stained with Prussian blue. Therefore, this suggests that anti-ELTD1 treatment could potentially be safer for use in the clinic.
Together, our data demonstrated that ELTD1 is an important angiogenic marker in high-grade gliomas. By using an optimized mAb treatment against ELTD1, we were able to significantly increase survival, decrease TV and normalize the tumour-associated vasculature. This study demonstrated that our optimized mAb against ELTD1 had higher binding specificity when compared to a commercially used pAb confirmed through molecular targeting and histology.
Prior studies have stated that VEGF increases ELTD1 expression and DLL4-Notch decreases the expression in normal vasculature; however, our study has demonstrated that this relationship may be more complex in the tumour environment.9 We were also able to significantly reduce Notch1 levels within the tumour and return them to near contralateral levels with our mAb treatment. Furthermore, our RNA-sequencing data demonstrated that 3 of the genes (SCN5A, L1CAM, BMP2) affected with the anti-ELTD1 treatment that were directly associated with gliomas influenced and interacted with Notch signalling. Therefore, suggesting that ELTD1 has a more complex relationship with Notch1 than previously understood. However, the RNA-sequence analysis did not show specific gene changes with Notch.
Aside from the possible relationship with Notch, the RNA-seq data gave further insight as to what pathways the anti-ELTD1 treatment is targeting. ADA and BMP2 expression are correlated with poor prognosis in glioma patients,46, 47 which were both down-regulated with the anti-ETLD1 treatments. Furthermore, the anti-ELTD1 treatment worked to down-regulate SCN5A, TRPM8 and BMP2, which were all shown to increase glioma cell proliferation, migration and invasion.48-50 Alkaline phosphatase (ALPL) is a stem cell marker, that is highly expressed around necrotic areas within the tumour, and high expression of ALPL and CD133 (another stem cell marker) has been associated with poor prognosis for patients.51 The mAb therapy against ELTD1 was successful in down-regulating ALPL. CD133+ glioma cells show neurosphere-like growth, drive tumour formation and are resistant to standard therapies.52 L1CAM, a gene down-regulated by anti-ELTD1 treatment, is overexpressed in GBMs and CD133+ glioma cells and regulates neural cell growth, migration and survival during development.53, 54 Furthermore, targeting and inhibiting L1CAM in CD133+ glioma cells suppressed tumour growth and increased the survival in a glioma xenograft model.54 Glioma stem cells (GSC) are the main cause of GBM recurrence after therapy and are characterized by CD133.55 However, recent reports have shown that CD133 may not be a robust marker for GSC and CD133 cells that possess GSC properties may give rise to aggressive tumours.55, 56 Nestin was first characterized as a neuronal stem cell marker and found on the surface of both CD133-positive and -negative cells and may serve as a more efficent GSC marker in GBMs.55, 56 Furthermore, nestin has been shown to be a key player in proliferation, migration and survival of GBMs and other cancers.57-59 Interestingly, cluster gene-gene analysis from the genes down-regulated by the anti-ELTD1 treatment suggests that there is a down-regulation response for nestin-related pathways.
Our data demonstrate that an optimized mAb therapy against ELTD1 is a potential anti-angiogenic therapy against GBMs. Although discovered in 2001, there are various unknowns about ELTD1, including its mechanism of action and its ligand. However, we hope that through our newly obtained RNA-sequence data, possible ELTD1 pathways of interaction can be examined. Future studies will further focus on ELTD1's relationship with Notch as well as possible ligands of ELTD1.
ACKNOWLEDGEMENTS
This work was supported in part by the Oklahoma Medical Research Foundation (RT) and the Seoul National University Foundation (JC); National Institutes of Health (1S10OD023508-01 to RT, 2P20GM103636 to JW). Whole slide scanning and image analysis are kindly provided by Peggy and Charles Stephenson Cancer Center at the University of Oklahoma Health Sciences Center, Oklahoma City, OK, an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (P20 GM103639) and a Cancer Center Supporting Grant Award from the National Cancer Institute (P30 CA225520). MZ performed the research and wrote the manuscript. The manuscript was edited by RT, NS, DS, JW, JC and RG. JZ, DS and SR assisted during the animal surgery. MR imaging and treatment were conducted by MZ, JZ, NS, DS, SR, LT and RG. Molecular-targeted MR imaging and RNA isolation were conducted by NS and MZ. JC, KH and JJ generated the anti-ELTD1 monoclonal antibody. MZ, ML, KMF and JC performed and assisted with the IHC and analysis. JW and CB performed RNA-seq and bioinformatics analyzes. MZ and SR analysed the MRI data. RT was the principle investigator.
CONFLICT OF INTEREST
None of the authors have any conflict of interest.
AUTHOR CONTRIBUTIONS
MZ performed the research and wrote the manuscript. The manuscript was edited by RT, NS, DS, JW, JC and RG. JZ, DS, NM and SR assisted during the animal surgery. MR imaging and treatment were conducted by MZ, JZ, NS, NM, DS, SR, LT and RG. Molecular-targeted MR imaging and RNA isolation were conducted by NS and MZ. JC, KH and JJ generated the anti-ELTD1 monoclonal antibody. MZ, ML, KMF and JC performed and assisted with the IHC and analysis. JW and CB performed RNA-seq and bioinformatics analyzes. MZ and SR analysed the MRI data. RT was the principle investigator that conceived the study design, oversaw the entire study and performed the data interpretation.
Open Research
DATA AVAILABILITY STATEMENT
Data will be made available upon reasonable request.




