Past and future climate effects on population structure and diversity of North Pacific surfgrasses
Abstract
Aim
Understanding the impacts of past and future climate change on genetic diversity and structure is a current major research gap. We ask whether past range shifts explain the observed genetic diversity of surfgrass species and if future climate change projections anticipate genetic diversity losses. Our study aims to identify regions of long-term climate suitability with higher and unique seagrass genetic diversity and predict future impacts of climate change on them.
Location
Northeast Pacific.
Time Period
Analyses considered a timeframe from the Last Glacial Maximum (LGM; 20 kybp) until one Representative Concentration Pathway (RCP) scenario of future climate changes (RCP 8.5; 2100).
Major Taxa Studied
Two seagrass species belonging to the genus Phyllospadix.
Methods
We estimated population genetic diversity and structure using 11 polymorphic microsatellite markers. We predicted the distribution of the species for the present, LGM, and near future (RCP 8.5, no climate mitigation) using Species Distribution Models (SDMs).
Results
SDMs revealed southward range shifts during the LGM and potential poleward expansions in the future. Genetic diversity of Phyllospadix torreyi decreases from north to south, but in Phyllospadix scouleri the trend is variable. Phyllospadix scouleri displays signals of genome admixture at the southernmost and northernmost edges of its distribution.
Main Conclusions
The genetic patterns observed in the present reveal the influence of climate-driven range shifts in the past and suggest further consequences of climate change in the future, with potential loss of unique gene pools. This study also shows that investigating climate links to present genetic information at multiple timescales can establish a historical context for analyses of the future evolutionary history of populations.
1 INTRODUCTION
Understanding climate effects on genetic diversity and structure is a major goal of population genetics and ecology (Eckert et al., 2008). In the Northern Hemisphere, the extreme climate oscillations of the Quaternary (2.6 MYA to present) impacted the geographical ranges of marine temperate species (Maggs et al., 2008). During glacial periods, populations were pushed to southward habitats of suitable climatic conditions, creating refugial areas of persistence protected from the harsh northern glacial climates. Conversely, during warmer interglacial periods, range expansions might have occurred poleward. These range shifts have shaped population genetic diversity and structure, particularly at marginal populations located near the current edges of species ranges (Hewitt, 1999), where rich distinctive gene pools are often found (Assis et al., 2014; Neiva et al., 2015). Indeed, long-term persistent populations in refugial areas exhibit distinctive genetic signals, such as private alleles that evolved locally and were not involved in subsequent range expansions (Assis et al., 2016; Diekmann & Serrao, 2012; Hewitt, 1999). However, ongoing climate change is posing threats to such refugia that may contain most of a species gene pool (Assis, Araújo, & Serrão, 2018; Assis, Serrão, et al., 2018). The potential loss of uniquely diverse populations may endanger overall genetic diversity and possibly long-term survival and adaptability (Assis, Araújo, & Serrão, 2018; Nicastro et al., 2013) as reduced genetic diversity can exacerbate the effects of inbreeding and reduce reproductive fitness, thereby raising the danger of extinction (Hoffmann et al., 2017).
Among coastal marine systems, seagrasses are foundation species that form the basis of one of the most productive and important communities. Specifically, seagrass ecosystems serve as vital habitats, harbouring a diverse range of marine life, including various fish species, invertebrates, and other organisms (Nordlund et al., 2018). Moreover, seagrasses play a crucial role in providing essential ecosystem services. These services encompass carbon sequestration, shoreline stabilization, and water filtration, contributing significantly to the overall health and resilience of coastal environments (Cullen-Unsworth & Unsworth, 2013; de los Santos et al., 2020). However, ongoing climatic change has resulted in local losses of seagrass habitat (Chefaoui et al., 2018) and associated distributional shifts (Chefaoui et al., 2021). This is especially concerning in temperate locations where some seagrasses have already been impacted near their low-latitude margins (Hyndes et al., 2017; Valle et al., 2014). These shifts may reduce seagrass population area and subsequently their resilience, genetic diversity, species richness, biomass, and productivity (Zimmerman, 2021). Given their global ecological importance, identifying regions of higher and unique seagrass genetic diversity, and projecting the impacts of climate change is of key relevance, particularly among seagrasses where there is little exploration of this important foundation genus (Melo-Merino et al., 2020).
Climate change has had a significant impact on intertidal species distributions, as coastal habitats are more exposed to its effects (Harley et al., 2006; Kunze et al., 2021). Phyllospadix is a widely distributed seagrass genus found in the temperate North Pacific region (sensu Short et al., 2007). It comprises five perennial dioecious species (Short et al., 2016), of which Phyllospadix scouleri, Phyllospadix torreyi and Phyllospadix serrulatus are distributed along the Northeast Pacific (NE Pacific; Phillips, 1979; Spalding et al., 2003). In the Northwest Pacific, Phyllospadix iwatensis and Phyllospadix japonica are distributed along the coastlines of Korea and Japan (Spalding et al., 2003). Unlike most seagrass species that grow on soft bottoms, Phyllospadix spp. occurs on wave exposed rocky substrates in the low intertidal and shallow subtidal zones (Hemminga & Duarte, 2000). Inhabiting shallow waters (down to 5 m depth), Phyllospadix species are frequently subjected to sharp environmental fluctuations, particularly intense tidal changes (Ramírez-García et al., 1998). Despite their ability to tolerate substantial tidal environmental variation, such as abrupt exposure to air during tidal fluctuations, these species might thus be close to ecophysiological tolerance limits during such tidal variations, which could render them particularly susceptible to climate-induced range shifts and their effects on genetic diversity along the temperate North Pacific bioregion, as shown for other marine forest-forming species (Grech et al., 2012; Song et al., 2021; Zhang et al., 2019).
Here, we use surfgrass, the only rocky shore seagrass genus in the world, as a model system to investigate the impacts of past and future climatic events on the distribution and genetic diversity of two foundation species. We aim to determine whether past climate-driven range shifts have influenced the present genetic diversity of Phyllospadix, if there are genetic hotspots at persistence zones with higher conservation value, and if these are at risk of future extirpations under predicted future climate scenarios. Specifically, we hypothesize that: (1) populations that have persisted during past climatic extremes currently harbour higher endemic genetic diversity; (2) populations in recently available habitats exhibit lower and non-endemic diversity; (3) populations at the lower latitude range limits are threatened by future climate change.
2 MATERIALS AND METHODS
2.1 Study area and focal species
The present study is focused on the NE Pacific coast, along the distributional ranges of P. torreyi and P. scouleri, from the Baja California Peninsula to Alaska. Samples of P. torreyi were obtained from San Juanico (26.2 N, 112.4 W, Mexico) to Fogarty Creek (44.8 N, 124.1 W, Oregon) and P. scouleri was sampled from Government Point (34.4 N, 120.5 W, California) to Vancouver Island (48.6 N, 124.5 W, Canada) (Table S1). To further check possible admixture between east and west Pacific Phyllospadix populations, we sampled three populations of P. iwatensis in Hokkaido Island (Japan; Table S2). Across sampling areas, approximately 30 individuals were sampled haphazardly at low tide along approximately 30 m, keeping a minimum distance of approximately 1 m between each sampling unit at each location along 23 sites (see Tables S1 and S2). All sampled plants were blotted dry and preserved dry in silica gel.
2.2 Species distributions models
Species Distribution Modelling (SDMs) were built for P. torreyi and P. scouleri with two machine learning algorithms, Boosted Regression Trees (BRT) and adaptive boosting (AdaBoost). These are well-suited for modelling complex interactions and non-linear relationships and have been shown to have high predictive performance while reducing overfitting through proper hyper-parametrization and forcing of monotonic responses (Elith & Leathwick, 2007; Hofner et al., 2011).
Biologically meaningful predictor variables for seagrasses (Krause-Jensen et al., 2020), namely temperature, nitrate, salinity and sea-ice, were extracted from Bio-ORACLE (Assis, Tyberghein, et al., 2018), a dataset providing global marine data layers for ecological modelling at a 0.08° resolution (approx. 9 km at the equator). These predictor variables were obtained for present-day conditions, the Last Glacial Maximum (LGM), and the extreme Representative Concentration Pathway (RCP 8.5) future scenario of increased carbon emissions with no climate mitigation policies, as detailed in a previous publication utilizing the Bio-ORACLE pipelines (Assis et al., 2023) (see Data availability statement). The choice of RCP 8.5 helps to estimate the potential impact of the most extreme predicted climate change scenario on species' distributions (Assis et al., 2022; Gouvêa et al., 2022). The predictors chosen reflected essential resources (nutrients as nitrate) as well as factors affecting physiology (salinity and temperature) and perturbation (sea ice cover). The LGM predictors considered the weighting of different Atmospheric-Ocean General Circulation Models (AOGCM; Assis, Tyberghein, et al., 2018) as well as the potential −120 m sea-level change during the LGM (Assis et al., 2016, 2018; Peltier & Solheim, 2004).
Because the focal species are intertidal, predictors were clipped to the regions defined by NE Pacific coastlines. Presence records for the two species were extracted from two biodiversity information facilities, GBIF (2021a, 2021b) and OBIS (https://www.obis.org/), and from the fine-tuned dataset of marine forest species (Assis et al., 2020). An equal number of pseudo-absences was randomly generated in locations where no presences were recorded (Assis, Serrão, et al., 2018). To reduce the potential effect of autocorrelation and sampling bias in the models, we filtered the records based on the minimum distance at which predictors were not significantly correlated (Figures S7–S9).
This distance was inferred by building a correlogram per species that identified the variability of Pearson's correlation of predictors with increasing marine distances (Krause-Jensen et al., 2020). This process reduced the initial 941 records of P. scouleri to 195, and the initial 568 records of P. torreyi to 87.
A 6-fold cross-validation approach was implemented, where independent blocks were used to find the optimal combination of hyperparameters, specifically, the number of trees (ranging from 50 to 1000, step 50), tree complexity (ranging from 1 to 6), and learning rate (0.01, 0.005, and 0.001) for BRT (Elith & Leathwick, 2007) and shrinkage (from 0.25 to 1, step 0.25), degrees of freedom (from 1 to 12, step 1) and number of interactions (from 50 to 250, step 50) for AdaBoost (Hofner et al., 2011). The performance of the model was evaluated with the area under the curve (AUC) of the receiver operating characteristic curve and True Skill Statistics (TSS), which is determined by the sum of sensitivity (true presence rate) and specificity (true absence rate) minus one (Allouche et al., 2006) and the Boyce index, which is a proper metric for presence-pseudo-absence models (Boyce et al., 2002). To reduce overfitting, negative monotonic responses were forced for maximum temperature and ice thickness, and positive responses for nitrate, salinity and minimum temperature (Hofner et al., 2011). By implementing such modelling constraints, the main hypotheses are (1) for predictor variables forced with negative responses (e.g., maximum temperature), there is an extreme maximum value from which species' probability of occurrence is zero, and (2) for predictor variables forced with positive responses (e.g., minimum temperature), there is an extreme minimum value below which species' probability of occurrence is zero. The relative importance of each predictor was assessed by measuring the increase in AUC when adding each predictor to the alternative model (Assis, Serrão, et al., 2018).
To develop parsimonious models per algorithm, with higher transferability potential, a stepwise approach was used, starting with a full model, and iteratively removing predictors from the least to the most contributory until the reduction in AUC was significant. This step was performed using a Wilcoxon Rank Sum test on the AUC values generated in the cross-validation procedure. Maps reflecting species' potential distribution for the present-day, the LGM and future conditions were created by averaging (i.e., ensemble modelling; Araújo & New, 2007) the responses of both BRT and AdaBoost parsimonious models. These were then reclassified into binomial maps showing the presence or absence of both species by using a threshold that maximized TSS (Allouche et al., 2006). To assess for past range shifts, we compared the modelled distributions for the LGM and the present and identified refugial areas where the species was predicted to occur in both time periods.
2.3 Primer design, microsatellite amplification and genotyping
Genomic DNA for microsatellite development was extracted from 500 mg of dried tissue from a pool of 97 P. torreyi individuals and 500 mg from 66 P. scouleri individual samples. These samples were used to run 454 sequencing (Biocant, Cantanhede, Portugal). Simple di- to hexanucleotide microsatellite loci motifs were tested for primer design using Primer 3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). A total set of 38 and 27 pairs of primers were designed for P. torreyi and P. scouleri, respectively, and tested for amplification. To determine the applicability and amplification success of the microsatellites, we tested them on two individual samples from five populations each of P. torreyi and P. scouleri. From the initial set, a subset of 38 loci for P. torreyi and 20 for P. scouleri, respectively, were chosen due to their amplification success. These were evaluated for their ease of scoring and polymorphism using primers labelled with fluorophores, and 11 were finally selected due to their number of repeats, similar melting temperature allowing multiplexing and cross-amplification success (primer sequences and microsatellite motifs listed in Table S3, supplementary material).
Genomic DNA was extracted using NucleoSpin96 PlantKit II (Macherey-Nagel, Germany). PCR reactions were performed in a total volume of 15 μL, containing 1× Colourless GoTaq Flexi Buffer (Promega, Madison, WI, USA), 2 mM MgCl2, 10 μM forward and reverse primers, 0.2 mM of dNTPs, 1U GoTaq G2 Flexi DNA Polymerase (Promega, USA) and 5 μL of template DNA. Cycling conditions consisted of an initial denaturation step at 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, annealing temperature (T a, Table S3) for 30 s, extension at 72°C for 30 s and a final extension at 72°C for 5 min. Amplified fragments were combined with 0.25 μL GeneScanTM 500 LIZ Size Standard (Applied Biosystems, UK) and 9.75 μL of Hi-Di formamide, followed by 5 min denaturation at 95°C and their analysis were performed on an ABI3130xl automated DNA sequencer (Applied Biosystems, Waltham, MA, USA).
Although P. torreyi and P. scouleri were genotyped for 11 microsatellite loci (Table S3), P. iwatensis populations were only genotyped for a set of eight loci (Psc07, Pto07, Pto13, Pto15, Pto25, Pto29, Pto31 and Pto35) as amplification failed for the additional three loci.
2.4 Microsatellite data analysis
Fragments were scored using STRAND (Veterinary Genetics Laboratory, University of California, Davis; http://www.vgl.ucdavis.edu/STRand) and binned using the R package ‘MsatAllele’ (Alberto, 2009). Prior to analysis, the final dataset was run in Micro-Checker 2.2.3 software (van Oosterhout et al., 2004) to test for the presence of null alleles and test Hardy–Weinberg equilibrium and run a linkage disequilibrium test. No evidence of null alleles or linkage disequilibrium was found for any of the 11 loci; therefore, they were all considered suitable markers for the analyses of population structure.
Seagrass species occupy space via rhizome growth, a type of clonal growth where genets (i.e., genetic individuals coming from distinct seeds) form ramets (i.e., units capable of independent life if separated, formed by shoots, associated roots, and a piece of rhizome). Therefore, when seagrasses are sampled, duplicate genotypes (clones) may be collected that derive from a single seed. To assess the clonality of each population, the package RClone (Bailleul et al., 2016) was used to assess the probability that identical multi-locus genotypes (MLGs) are the product of the same or separate reproductive events. The probability of finding identical MLGs resulting from distinct sexual reproductive events (Psex) was computed for each population (Arnaud-Haond et al., 2007) to check if individuals with the same MLG were truly clones. When Psex was lower than the threshold value (fixed at 0.01), two identical MLGs were considered to be the same clone. Based on the number of individuals (N) and genets (G) sampled, clonal diversity in each population was calculated using the clonal richness index (R), (R = [G − 1]/[N − 1]), ranging from 0 (one single clone) to 1 (when all samples analysed correspond to a different genet). Only unique genets per population were used for genetic diversity analyses to avoid biasing allelic frequencies, that for the research questions of this study, should represent the population of genets rather than the population of ramets.
2.5 Genetic diversity and population genetic structure
To compare population genetic diversity and differentiation, all populations must have the same loci amplified. Two datasets were therefore constructed for genetic analysis as only eight loci amplified for P. iwatensis out of the 11 that were used to amplify P. torreyi and P. scouleri. Dataset 1 includes P. torreyi and P. scouleri populations (11 loci), and dataset 2 encompasses populations for all three species (eight loci). Additionally, for higher resolution analysis of population differentiation within each species, separate genetic analyses were performed for species in dataset 1 individually (P. torreyi and P. scouleri). A total of 593 Phyllospadix samples were genotyped (304 P. torreyi and 289 P. scouleri samples) for dataset 1 (11 loci).
Summary statistics of genetic diversity were calculated for each population using GENETIX 4.05 (Laboratoire Génome, Populations, Interactions, Université de Montpellier II; http://kimura.univ-montp2.fr/genetix). This included the mean number of alleles per locus (allelic richness), the non-biased expected heterozygosity (H E), the observed heterozygosity (H O), the number of private alleles, and the multi-locus inbreeding coefficients (F IS). Sample sizes were also standardized to the smallest in any population using 104 randomizations.
Population genetic structure was inferred using STRUCTURE 2.3 (Pritchard Lab, Stanford University; http://pritchardlab.stanford.edu/structure.html) without any prior population assignments. Structure was run for both datasets and for P. torreyi and P. scouleri individually. The estimated number of genetic clusters, K, was estimated from the mean log likelihood for each K value and the ΔK statistic (Earl, 2012) to identify the optimal number of groups. This was analysed considering a range of K values from 1 to 23. Ten replicates per K value were used to verify the consistency with a 50,000 burn-in followed by 500,000 MCMC replicates per iteration. All jobs were run in parallel on multiple cores using the R package ‘ParallelStructure’ (Besnier & Glover, 2013). The best K was estimated using the program Structure Harvester which ranks K's according to Evanno method (Earl, 2012). Structure analyses were complemented with a factorial correspondence analysis (FCA) implemented in GENETIX 4.05.2 (Belkhir et al., 1996). To quantify pairwise differentiation between populations, we calculated F ST and Jost's D distances using the software GENODIVE (Meirmans & van Tienderen, 2004).
3 RESULTS
3.1 Species distribution models
The ensemble of both BRT and AdBoost algorithms resulted in high performance predictions, largely matching the known distribution of the two species (Figure 1; maps depicting the probability of occurrence available in Figures S10 and S11). Specifically, for P. scouleri AUC was 0.99, TSS was 0.93 and Boyce was 0.89, while for P. torreyi AUC was 0.99, TSS was 0.91 and Boyce was 0.73. The models considered numerous biologically meaningful predictor variables, from which temperature (maximum and minimum) were key in explaining the distribution of both species (Figures S1, S2, S10 and S11), in line with additional studies based on species distribution modelling of seagrasses (Gouvêa et al., 2024).
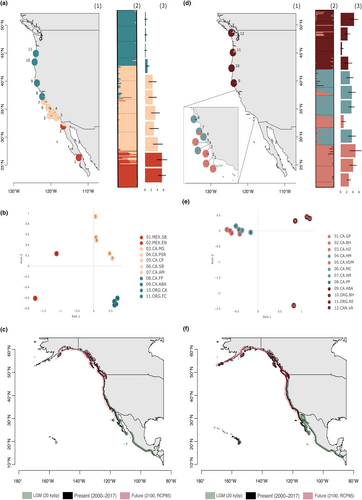
The models hindcasted significant equatorward range shifts during the Last Glacial Maximum (LGM). The extreme colder environments might have contracted poleward range margins down to Oregon (P. scouleri) and California on the West coast of the USA (P. torreyi). Concomitantly, lower latitude range margins might have remained stable or even expanded along Central American latitudes (Figure 1). In contrast, the future scenario of increased carbon emissions (RCP 8.5) projected poleward range expansions with potential habitat losses at the lower latitude range margins of Baja California Sur for both species (Figure 1).
3.2 Population genetic and genotypic diversity
Global and endemic genetic diversity generally decreased from north to south for P. torreyi but not for P. scouleri (Figure 1; Table S1). Local diversity (private alleles) ranged from high in southern populations of P. torreyi to an almost complete lack of endemism in the most northern populations (Figure 1a). The two species differed particularly in their contrasting northernmost range, where P. torreyi displayed very low within-population diversities whereas P. scouleri showed some of the highest diversity metrics in the north (Table S1).
More than half of the populations from both species had significant heterozygote excess relative to the expectation under random mating (Table S1, significant negative F IS). For dataset 2, P. iwatensis revealed lower genetic diversity relative to the two eastern species (e.g., mean A = 2.5; Â 7 = 2.06 ± 0.1 and PÂ = 0.37 ± 0.75; Table S2) and significant heterozygote excess (negative F IS) in all populations (Table S2).
When P sex values were higher than 0.01, the identical MLGs were considered derived from independent sexual reproduction events (i.e., distinct seeds), resulting in a total of 494 unique genotypes for dataset 1 (G = 255 for P. torreyi and G = 239 for P. scouleri; Table S1). The mean global genotypic richness was relatively high for both species (R = 0.88 and R = 0.92; Table S1), indicating predominant sexual reproduction. The western species P. iwatensis had slightly lower clonal richness, but still a larger proportion of the sampled shoots were derived from sexual reproduction rather than clonal propagation (R = 0.74, Table S2).
3.3 Genetic differentiation and structure
The structure results for the three species combined (dataset 2) showed differentiation between the three species, the same genetic structure for P. torreyi and P. scouleri (as in dataset 1) and P. iwatensis forming a distinctive cluster. The northernmost population of the eastern Pacific (Vancouver, P. scouleri) had individuals with a proportion of the genome assigned to the same group as the western Pacific species, P. iwatensis, in addition to the admixture already previously identified with its more southerly sister species, P. torreyi (Figure 2). The FCA retrieved similar results supporting the differentiation between the western versus eastern species along the first axis, and among the two eastern species along the second axis (Figure 2).
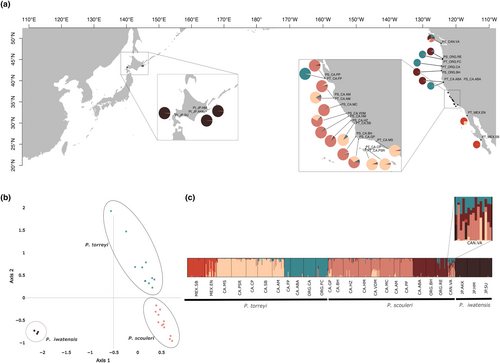
Structure analysis including all Eastern Pacific samples of both species revealed, at the first hierarchal level, a separation between two species (Figure S3). However, some populations morphologically identified as P. scouleri showed genetic admixture from P. torreyi (Figures S3 and S4), specifically at the two range extremes of P. scouleri (its southernmost and northernmost limits) and at an additional site (San Simeon) in the middle of the distribution of both species.
The subsequent hierarchical level of genetic structure (K = 5) divided south, centre, and northern populations for both species (Figure 1a,b,d,e), with some admixture as previously shown at the main split into two species K = 2. The same three groups per species were corroborated when structure analyses were run for each species separately (Figure 1a,e). P. torreyi populations were divided into (1) Mexico group (south cluster); (2) South California group (centre cluster); and (3) North California/Oregon group (north cluster) (Figure 1a,b). The same number of clusters were found for P. scouleri: (1) South California (south cluster); (2) Centre California (centre cluster); and (3) Oregon/Vancouver (north cluster), with south and centre clusters having an intermingled transition region (Figure 1d,e).
All populations within each species were significantly differentiated (p < 0.01) for both F ST and Jost'D estimates (Figures S5 and S6). The pairwise differentiation values within P. torreyi and P. scouleri were lower between southern populations, whereas the northern populations against all others had the highest differentiation (Figures S5 and S6).
4 DISCUSSION
This study shows a remarkable contrast among the population genetic diversity patterns of two sister species that differ slightly in their thermal ranges. It shows how hindcasted past range shifts, triggered by climate change along the NE Pacific coast, might have shaped the present genetic diversity and structure of Phyllospadix populations and where this effect is not evident. It also provides a baseline to assess expected further climate-driven changes and losses. Our empirical genetic data combined with hindcasting predictions from SDM show unexpected differences in the distribution of population genetic variability along the ranges of two sister species, which can be explained by differences in their thermal ranges. Contrary to expectations, the genetic patterns observed in P. scouleri do not align with the anticipated outcomes based on climate-driven range shifts. While genetic diversity of P. torreyi decreases from north to south, the trend in P. scouleri is variable. Additionally, P. scouleri exhibits signals of genome admixture at both edges of its distribution. This raises questions about the extent to which historical range shifts have influenced the current genetic diversity and structure of this species. Although contemporary effects might influence the distribution of genetic variability along a species range, the long-term accumulation of unique genetic diversity by mutation accumulation (i.e., private alleles) requires long-term persistence of populations. This is the case of P. torreyi, where the distribution of private alleles corresponds to expectations from range shifts, whereas heterozygosity and standardized allelic richness exhibit contrasting patterns that can be influenced by several other contemporary processes. The objective of this study, assessing the effects of long-term population persistence, is thus expected to be reflected in the distribution of private alleles. This makes it especially interesting that the two sister species have such a striking contrast, reflecting their thermal tolerances.
The geographical coverage was limited due to access to remote areas, introducing constraints to the inferences possible in this study. Despite this limitation, our research results reveal novel information on the distribution of intraspecies genetic biodiversity of the targeted seagrass species along their sampled distribution, enhancing our understanding of genetic diversity and structure of P. torreyi and P. scouleri.
Findings based on analyses of both independent datasets are discussed below starting with the potential effects of past range shifts (LGM) followed by the observed patterns of genetic diversity and the potential consequences of increased carbon emissions to the future distribution and diversity of Phyllospadix populations.
4.1 Genetic signatures left by past climatic changes
This study shows that the expectations of effects of past range shifts inferred by distribution models can be found in the current genetic variability across the distributional range of surfgrass species P. torreyi and shows that the same patterns were not observed in the sister species P. scouleri that has a colder affinity range distribution (as illustrated in the models of ranges). The contrasting results observed between species are a novel interesting result of this study, as the small thermal affinity differences between them correspond to large differences in private genetic diversity, with many unique alleles having accumulated in the southern region. Populations able to persist in any of these refugial areas should have accumulated a larger amount of new private genetic diversity and genetic distances across populations, in contrast with regions that suffered local extinctions followed by recent settlement (Jacobs et al., 2004; Mann & Hamilton, 1995). This is why private alleles are the important variable to locate regions of long-term persistence to reflect past stability rather than contemporary influences.
In contrast to northern populations, the southern edge populations of P. torreyi, located on the Baja California Peninsula in Mexico, were more genetically unique (private alleles, Table S1) and differentiated (FCA, Figure 1). The decline in the private alleles with increasing latitude contrasts with expected heterozygosity, particularly in the southernmost Baja California population which is less diverse (Table S1). Higher diversity in more ancient populations at southern range margins is expected in temperate species of the northern hemisphere, as long-term population persistence may have been critical in preserving and accumulating genetic differences (Hampe & Petit, 2005; Hewitt, 1999, 2004) with no notable effects from drift, local extinctions or bottlenecks. Population genetic diversity decreasing from lower to higher latitudes corroborates patterns in other intertidal vegetation species (e.g., Assis, Serrão, et al., 2018; Neiva et al., 2015) and across subtidal marine vegetation as well (e.g., Assis et al., 2016; Diekmann & Serrao, 2012; Johansson et al., 2015; Neiva et al., 2020), all likely to have been affected by the glacial cycles at higher latitudes.
Despite the expectation of lower genetic diversity at higher latitudes (Eckert et al., 2008; Excoffier et al., 2009) due to genetic bottlenecks associated with expansion after colder periods (Marko, 2004), this pattern was not observed in P. scouleri. We proposed and analysed two hypotheses that could explain the high diversity at its northernmost range: (1) admixture of genetically divergent entities such as introgression with western Pacific species of Phyllospadix, or (2) long-term persistence in this area, which might have remained ice-free up until the late Pleistocene when the glaciers retreated (Jacobs et al., 2004; Mann & Hamilton, 1995). The northernmost population analysed herein revealed higher private allelic richness but also shared genome assignment with the western Pacific group, suggesting admixture, thereby supporting the hypotheses of hybridization. However, it's important to note that the evidence for hybridization is somewhat weak, with only a small proportion of individuals showing evidence of hybridization with P. iwatensis. On the other hand, the distribution models suggest the possibility of persistence somewhere nearby, as the expected distribution of P. scouleri during the LGM closely aligns with the location of the northernmost population. Therefore, while hybridization remains a possibility, the evidence also hints at the possibility of long-term persistence nearby. The alternative hypothesis of incomplete lineage sorting (ILS) is less supported by the evidence because ILS is not expected to affect only the northernmost edge population. This pattern has been found for other NE Pacific species (Carrara et al., 2007; Jacobs et al., 2004).
4.2 Future predictions
The Intergovernmental Panel on Climate Change (IPCC) reported that the global temperature is expected to surpass 1.5°C above pre-industrial levels in the next decades, potentially exposing population and species to detrimental conditions (IPCC, 2022). Accordingly, local population extinctions, shifts, or losses of unique genetic diversity are projected (Chefaoui et al., 2018; Gouvêa et al., 2024). Specifically for our model species, recent observations show that the warming trend of the Baja California peninsula, Mexico, is in line with the RCP 8.5 scenario (Martínez-Austria & Jano-Pérez, 2021). Our projections estimate that both Phyllospadix species might have similar trailing edges in the near future as a consequence of range shifts. Due to unsuitable conditions throughout the southern part of the Baja California peninsula, the models project a northward shift of the trailing edge to the Northern more temperate region of Baja California (~30° N) by the end of the century. Baja California Sur, showing high present-day endemic (i.e., private alleles) genetic diversity, lining up with the hypothesis of providing a climatic refugium, is predicted to eventually lose populations there, resulting in a climatic impact of the loss of the global genetic diversity pool of P. torreyi. Such loss, dependent on the pace of future range changes, would be irreversible. Moreover, private alleles generally support the potential for species to undergo selection and thereby adapt to future environmental changes, as well as to avoid inbreeding depression (Hampe & Petit, 2005; Petit et al., 1998). Additionally, considering the role of Phyllospadix as a foundation species, the regional loss of populations may generate important negative cascading effects on the entire associated tidal community (Shelton, 2010). Therefore, the southern populations of P. torreyi have important priority conservation value for hosting the most unique genetic diversity found in this study.
On the other hand, the projected northward shifts in the suitable habitats of both Phyllospadix species may allow the colonization of new areas, particularly along Canada and Alaska. The projections for the leading-edge differed between both species, with P. torreyi potentially experiencing greater poleward expansions along the Gulf of Alaska (to ~57° N) and spreading to new colder and higher-latitude areas with unknown but predictable possible founder effects creating novel mosaics of genetic diversity and structure. In contrast, for P. scouleri, the poleward expansion is projected to be less extensive along this coastline, but its colder tolerance suggests that it might hypothetically find opportunities to expand further west into Russian coastlines, although such habitats are not part of the focus of the present study.
Based on projections, neither Phyllospadix species is in danger of extinction, despite the previously mentioned negative effects on their genetic diversity levels and localized loss of ecosystem services. However, because only a part of the genetic diversity is expected to shift, the spread into new regions may result in a high frequency of particular genetic variants due to allele surfing (Excoffier & Ray, 2008; Neiva et al., 2010). Species may be able to migrate and follow suitable conditions that better fit their niche if the pace of climate change allows (Ackerly, 2003; Hewitt, 2000). This process will result in a gradient of increasing genetic diversity toward the leading edge (McInerny et al., 2009). Moreover, it is important to point out that given the short time frame under the RCP 8.5 projection (i.e., until 2100), species that might have limited dispersal traits, such as the Phyllospadix genus (Kendrick et al., 2012), will require a larger amount of time for effective gene flow, increasing the likelihood that alleles become extinct before reaching new areas (Knutsen et al., 2013).
5 CONCLUSION
In this study we provide evidence of past climate change effects on genetic diversity and structure. Genetic patterns revealed in this study provide useful information to understand and predict the influence of climate-driven range shifts of the past and future and for population conservation planning and management along the NE Pacific coast. As the region of Baja California peninsula, a well-known biogeographic transition zone, has shown unique genetic diversity and was forecasted to be most affected by future climate change, this region has important conservation value. Thus, it is important to consider the influence of contemporary ecological conditions and potential obstacles, like ocean currents, that may hinder population dispersal, especially in the face of rapidly changing climatic conditions. While our study primarily focused on the genetic imprints of climate-driven range shifts, we recognize the need for future research to integrate contemporary drivers of connectivity. This approach would provide a more comprehensive understanding of how both historical and contemporary drivers contribute to surfgrass population genetic diversity and structure. Nonetheless, we establish an historical context for future studies that further wish to examine the geographical patterns and evolutionary history of these two marine foundation species, by investigating climate associations with present genetic information at multiple timescales. This first inference of how past climatic oscillations are reflected in the present genetic variation for these Phyllospadix species raises the question of whether this baseline genetic imprint of the past may shape future forecasted responses of populations to climate change while they shift northwards.
ACKNOWLEDGEMENTS
We thank Marta Valente (CCMAR sequencing laboratory) for genotyping samples. All the study protocol was performed in accordance with relevant national and international guidelines. Samples were collected under licence and logistic support of the local authorities in each country during the missions of the projects involved. This work was supported by FCT—Foundation for Science and Technology (Portugal) through UIDB/04326/2020, UIDP/04326/2020, LA/P/0101/2020, PTDC/BIA-CBI/6515/2020, the Individual Call to Scientific Employment Stimulus 2022.00861.CEECIND and the scholarships SFRH/BD/138182/2018, COVID/BD/152937/2022, and by a Pew Charitable Trusts through a Pew Fellowship in Marine Conservation awarded to EAS, the David and Lucile Packard Foundation and Bureau of Ocean Energy Management to PR.
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Open Research
DATA AVAILABILITY STATEMENT
The datasets, the code, and raster layers used in species distribution modelling are openly available in Figshare at: https://figshare.com/s/6fd408a5862d77027793.
REFERENCES
BIOSKETCH
Ana Isabel Tavares is a PhD student at CCMAR—Centre of Marine Sciences at the University of Algarve, Portugal. Her research focuses on the study of genetic biodiversity, population connectivity and evolution of seagrass species, with the aim to understand their evolutionary history in different regions with contrasting oceanographic and seascape characteristics.
Author contributions: All authors contributed to the conceptualization of the study and approved the submitted version. The study was designed by EAS, PR, GAP, MN, LL, AIT, JA. Samples were collected by LA, PR, EAS, GAP. Genetic data analyses were conducted by AIT, NC, CP. Modelling was conducted by JA. The manuscript was drafted by AIT, EAS, JA, GAP. All authors revised and approved the manuscript.




